Abstract
Newcastle disease virus (NDV) is an economically important pathogen of poultry that may cause clinical disease that ranges from a mild respiratory syndrome to a virulent form with high mortality, depending on an isolate's pathotype. Infections with virulent NDV strains are required to be reported by member nations to the Office of International Epizootes (OIE). The primary determinant for virulence among NDV isolates is the presence or absence of dibasic amino acids in the fusion (F) protein cleavage activation site. Along with biological virulence determinations as the definitive tests, OIE accepts reporting of the F protein cleavage site sequence of NDV isolates as a virulence criterion. Nucleotide sequence data for many NDV isolates recently isolated from infected chickens and other avian species worldwide have been deposited in GenBank. Consequently, viral genomic information surrounding the F protein cleavage site coding sequence was used to develop a heteroduplex mobility assay (HMA) to aid in further identification of molecular markers as predictors of NDV virulence. Using common vaccine strains as a reference, we were able to distinguish virulent viruses among NDV isolates that correlated with phylogenetic analysis of the nucleotide sequence. This technique was also used to examine NDV isolates not previously characterized. We were able to distinguish vaccine-like viruses from other isolates potentially virulent for chickens. This technique will help improve international harmonization of veterinary biologics as set forth by the OIE and the Veterinary International Cooperation on Harmonization of Technical Requirements of Veterinary Medicinal Products. Ultimately, the HMA could be used for initial screening among a large number of isolates and rapid identification of potentially virulent NDV that continue to threaten commercial poultry worldwide.
Newcastle disease virus (NDV) is a member of the Paramyxoviridae family and has been designated avian paramyxovirus-1. Outbreaks of Newcastle disease were first reported among poultry in Java, Indonesia, and England during 1926. It is currently a worldwide problem and all orders of birds have been reported to be capable of infection with NDV (1). Infectious virus may be transmitted by ingestion or inhalation, which is the basis of mass application vaccination procedures for poultry (25). Isolates of NDV may be categorized into three main pathotypes depending on severity of disease following chicken inoculation (1, 2). Lentogenic isolates are of low virulence and cause mild respiratory or enteric infections. Viruses of intermediate virulence that cause primarily respiratory disease are termed mesogenic, while virulent viruses that cause high mortality are termed velogenic. Velogenic NDV can be classified as neurotropic or viscerotropic based on clinical manifestations (1). Virulent NDV isolates are List A pathogens, and it is compulsory that reports of its isolation be made to the Office of International Epizootes (OIE) (29).
The principle molecular determinant for NDV pathogenicity is reported to be the fusion (F) protein cleavage site amino acid sequence (11, 26, 28) and the ability of various cellular proteases to cleave the F protein of different pathotypes (12, 30). Dibasic amino acids surrounding glutamine (Q) at position 114 are present in the F protein cleavage site of mesogenic or velogenic strains, while lentogenic NDV isolates lack this motif (11, 26). The presence of dibasic amino acids in the F protein sequence allows for systemic spread of velogenic NDV, whereas replication of lentogenic NDV is limited to mucosal surfaces of the host (30). This is a major factor in differentiating velogenic and mesogenic NDV from lentogenic NDV isolates in cell culture. All NDV isolates will replicate in chicken embryo kidney cells (18), presumably due to the presence of a required protease (30). However, lentogens must have proteases added to cell cultures for replication in avian fibroblasts or mammalian cell types, whereas mesogenic and velogenic NDV do not have this requirement (18, 28).
Traditional biological pathotyping of NDV field isolates is determined by embryo and chicken inoculation (2). The mean death time in eggs and intracerebral pathogenicity index (ICPI) differentiate low-virulent lentogens from mesogens of intermediate virulence and highly virulent velogens. The intravenous pathogenicity index differentiates mesogens from velogens, and intracloacal inoculation is used to differentiate viscerotropic velogens from neurotropic velogens (1). Antigenic differences occur among strains (34), and monoclonal antibodies (3, 31) have been used to identify at least 13 antigenic NDV groups (1). Most isolates within a group are of a similar pathotype, but the results do not provide a reliable alternative to conventional live-animal pathotyping. Assays including hemagglutination inhibition (HI), virus neutralization, neuraminidase inhibition, hemolysis inhibition, and enzyme-linked immunosorbent assay have also been used to identify NDV (2, 16, 44).
Reverse transcription (RT) coupled to PCR (RT-PCR) has been used by several investigators to amplify F gene sequences of many NDV isolates obtained worldwide (7, 17, 23, 35, 36, 45). Amplification products were analyzed by gel electrophoresis before and after digestion with restriction enzymes, giving somewhat inconsistent results (17). Collins et al. (7) amplified the F gene cleavage activation site and deduced the F protein cleavage site amino acid sequence from nucleotide sequences of the RT-PCR product. However, several primer sets have been needed to amplify sequences from a variety of strains (7). Slot blot hybridization assays with an oligonucleotide probe to a conserved region of the F gene have also been used to potentially identify RNA from several strains of NDV (15).
The OIE now accepts the F protein cleavage site sequence as an alternative virulence criterion along with ICPI determinations for NDV pathotyping. International harmonization by veterinary biologics manufacturers is a major concern to facilitate international trade. Goals set forth will include mutual recognition of testing, development of standards for government control laboratories, mutual recognition of inspection for biologic products release, coordinated reviews of product applications, and harmonization of licensing requirements. These will be addressed via the Veterinary International Cooperation on Harmonization of Technical Requirements of Veterinary Medicinal Products (VICH) (10).
The heteroduplex mobility assay (HMA) (22) has proven to be a useful technique to categorize measles (20), polio (5), influenza (46), and human immunodeficiency (8, 27) viruses. This technique can be used to distinguish between measles viruses from different phylogenetic groups to as low as a difference of 2.9% sequence identity (20). In our laboratory we have utilized degenerate oligonucleotide primers to reliably amplify sequences that encode the F protein cleavage activation site by RT-PCR using NDV genomic RNA as a template (35, 36). Therefore, we utilized an HMA, following use of NDV genomic RNA as a template for RT-PCR, to detect differences in the F protein cleavage site coding sequences among various isolates. This will help improve rapid NDV diagnostics and epidemiology that directly addresses needs set forth by the OIE and VICH to facilitate international trade harmonization.
MATERIALS AND METHODS
Viruses.
Reference NDV strains tested were described in detail previously or are referenced therein (35, 36) and are listed in Table 1. Strains B1, La Sota, VGGA, and Queensland/V4 were analyzed as lentogenic vaccine strains used in the poultry industry worldwide. Two mesogenic isolates of intermediate virulence that were used for the analysis included the vaccine strain Roakin and the Kimber virus. Several velogenic viruses also included were the previously characterized isolates Herts33, Italy/Milano, TexasGB, Largo, turkey/ND, and an NDV isolate obtained from cormorants and designated cormorant/MN (35, 36). Viruses previously not characterized by nucleotide sequencing that were included for analysis were isolates from the 1998 outbreak of Newcastle disease in Australia (42), a virulent virus from the United Kingdom, Essex70, two viruses (VF 74-9 and VF 74-27) isolated prior to an outbreak in Northern Ireland during the 1970s (13), and a lentogenic isolate from chickens in the southeastern United States (GA2918). These NDV isolates are also listed in Table 1.
TABLE 1.
NDV isolates examined by HMA in this study
| Isolate nomenclaturea | Nameb | Pathotypec |
|---|---|---|
| Chicken/United States/B1/48 | B1 | Lentogenic |
| Chicken/United States/LaSota/46 | LaSota | Lentogenic |
| Chicken/Northern Ireland/Ulster/64 | Ulster | Lentogenic |
| Chicken/Australia/QV4/66 | QV4 | Lentogenic |
| Chicken/Northern Ireland/VF74-27/74 | VF74-27 | Lentogenic |
| Turkey/United States/VGGA/87 | VGGA | Lentogenic |
| Chicken/Australia/9809-14-1110/98 | 14-1110 | Lentogenic |
| Chicken/United States (Ga.)/2918/99 | GA2918 | Lentogenic |
| Chicken/United States/Roakin/48 | Roakin | Mesogenic |
| Chicken/United States/Kimber/47 | Kimber | Mesogenic |
| Chicken/United Kingdom/Herts/33 | Herts33 | Viscerotropic |
| Chicken/Italy/Milano/45 | Italy/Milano | Viscerotropic |
| Chicken/United States (Tex.)/GB/48 | TexasGB | Neurotropic |
| Chicken/United Kingdom/Essex/70 | Essex70 | Viscerotropic |
| Mixed species/United States/Largo/71 | Largo | Viscerotropic |
| Chicken/Northern Ireland/VF74-9/74 | VF74-9 | Viscerotropic |
| Cockatiel/United States (Fla.)/FL/80 | FL80 | Viscerotropic |
| Cormorant/United States (Minn.)/40068/92 | cormorant/MN | Neurotropic |
| Turkey/United States (N.Dak.)/43084/92 | turkey/ND | Neurotropic |
| Chicken/Australia/9809-19-1107/98 | 19-1107 | Neurotropic |
Isolates are presented as species type/country(state) of origin/accession number or common name/year of isolation.
Given name in present report.
Viscerotropic is viscerotropic velogenic; neurotropic is neurotropic velogenic.
Propagation of isolates, RNA extraction, and RT-PCR amplification.
All NDV isolates were propagated in embryonated chicken eggs (2) and maintained at the Southeast Poultry Research Laboratory as master stocks. These stocks are passaged once at low titer for experimental purposes. Viral genomic RNAs were purified by acid-phenol extraction directly from allantoic fluid (6, 35), and purified RNA was stored at −70°C in ethanol. Viral RNA from infectious allantoic fluid (0.5 μg) was reverse transcribed with random primers (19) and cDNA was amplified by PCR using 3 U of Amplitaq polymerase (Perkin-Elmer) (21, 32) with 100 pmol of the sense (5′-CCTTGGTGAITCTATCCGIAGG-3′) and antisense (5′-CTGCCACTGCTAGTTGIGATAATCC-3′) primers as previously reported (35, 36). These primers represent F gene sequences surrounding those encoding the F protein cleavage site.
HMA.
The HMA method (22) used was modified from a protocol developed for human immunodeficiency virus studies (8). Amplification products of 254 bp obtained from each isolate listed in Table 1 were mixed with an equal amount of either the RT-PCR product of NDV B1 or NDV Ulster in a 10-μl volume. The mix was denatured at 95°C for 5 min and immediately chilled on wet ice. Gel loading buffer was then added, and samples were separated by electrophoresis at 250 V in 1× TBE buffer (100 mM Tris, 100 mM boric acid, 2 mM EDTA, pH 8) for 5 h. Electrophoresis was completed using a mutation detection enhancement gel matrix (FMC Bioproducts, Rockland, Maine) at 1× concentration according to the manufacturer's protocol. Urea was added to the gel matrix at 15% to increase resolution. Gels were stained in ethidium bromide and photographed over a UV transilluminator.
Nucleotide sequencing and phylogenetic analysis.
Following RT-PCR, double-stranded sequencing (33) with fluorescently labeled dideoxynucleotides (Applied Biosystems Inc.) was completed with an automated sequencer (38). Following alignment of nucleotide sequences, phylogenetic analysis was completed (39) as described elsewhere (35, 36).
Nucleotide sequence accession numbers.
Nucleotide sequences of NDV isolates not previously characterized have been deposited in GenBank with accession numbers AF355274 through AF355279.
RESULTS
Viruses examined by HMA.
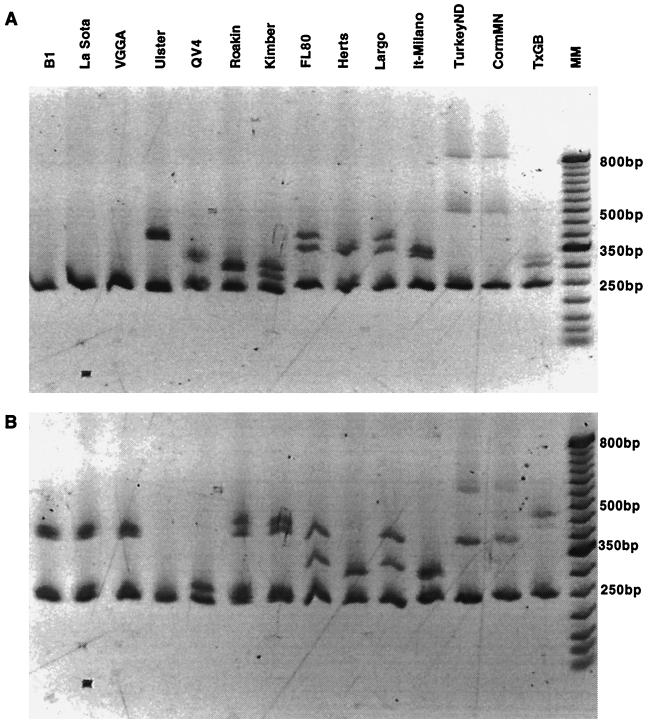
Isolates of NDV chosen for analysis represented well-characterized viruses that have been previously described and also several recently obtained isolates (Table 1). These viruses included all pathotypes with various chronological and geographical origins. Isolates not previously characterized and utilized for HMA included viruses obtained from an outbreak in Northern Ireland during the mid-1970s (VF74-9 and VF 74-27) along with an NDV isolated during an outbreak in Australia during 1998 (19-1107 and 14-1110). A lentogenic field isolate from the United States (GA2918) was also included for analysis. The HMA patterns were obtained by using two different lentogenic vaccine viruses, B1 and Ulster, as the control or reference viruses against which other NDV isolates were compared (Fig. 1A and B).
FIG. 1.
HMA for reference NDV isolates. (A) Utilization of the B1 vaccine type virus as the reference. (B) Utilization of the Ulster vaccine type virus as the reference. Virus isolates are presented in Table 1.
HMA of reference NDV isolates.
The most common vaccine virus utilized worldwide is the B1 type live virus. When the amplification product from B1 was mixed with the amplification product from the VGGA and LaSota type vaccine products, homoduplexes were obtained (Fig. 1A). This is not unexpected since the nucleotide sequences for these amplification products share 98% identity with the nucleotide differences occurring at external positions of the amplification product sequence. Two other lentogens, Ulster and QV4, also used as potential vaccine viruses, produced heteroduplex patterns dissimilar from one another when the B1 amplification product was used as the reference (Fig. 1A). This is reflected by the 89% nucleotide sequence identity between the B1 amplification product and the sequences for Ulster and QV4 NDV isolates. Also, the nucleotide mismatches among these isolates occurred throughout the amplification products, with several substitutions found in the interior of the F protein cleavage site coding region.
Three potentially virulent viruses isolated from the United States that are chronologically related to B1 were included for analysis by HMA. The mesogenic Roakin and Kimber viruses along with the neurovirulent TexasGB strain produced heteroduplexes following annealing of their respective RT-PCR products with the B1 amplification product (Fig. 1A). Nucleotide sequences of the amplification products from these NDV isolates shared 94% identity with the B1 vaccine strain. Although this is a relatively high sequence identity, nucleotide mismatches were present throughout the amplification product. The majority of nucleotide differences occurred at interior positions encoding basic amino acids present in the F protein cleavage site of more-virulent NDV isolates. Highly virulent viruses isolated outside the United States and chronologically akin to the B1 vaccine virus included the Herts33 and Italy/Milano strains. These viruses exhibited similar HMA patterns (Fig. 1A) and had identities in the nucleotide sequences encoding the F protein cleavage site of 85 and 86%, respectively, relative to the B1 amplification product.
Two more recently obtained viruses from psittacine-type birds that were included for analysis were the Largo and FL80 isolates. These viscerotropic velogenic viruses shared only 85% identity with the nucleotide sequence encoding the F protein cleavage site of the B1 virus. The nucleotide sequence of the RT-PCR product sequences of Largo and FL80 shared 96% identity and produced very similar HMA patterns when the B1 amplification product was used as the reference (Fig. 1A). The turkey/ND virus had a similar HMA pattern to the cormorant/MN isolate (Fig. 1A) when using the B1 virus RT-PCR product as a reference to hybridize with the amplification products synthesized from genomic RNA of these isolates. These two viruses shared 100% nucleotide sequence identity in the F protein cleavage site sequence and only 86% identity with the B1 sequence.
A second reference virus, Ulster, was also used to examine HMA patterns produced among previously characterized lentogenic NDV isolates (Fig. 1B). As stated, nucleotide sequences encoding the F protein cleavage site of Ulster shared 90% identity with the B1, LaSota, and VGGA viruses. These NDV vaccine isolates had similar HMA patterns when using the Ulster amplification product as a reference relative to the homoduplex produced by Ulster with itself. Another lentogenic NDV not common to the United States, QV4, shared 93% sequence identity with Ulster nucleotide sequences encoding the F protein cleavage site. This comparison also produced a distinctive HMA pattern (Fig. 1B), due to the presence of mismatches occurring throughout the amplification products obtained for Ulster and QV4.
The same potentially virulent NDV isolates used to examine HMA patterns using B1 as a reference were repeated using the Ulster virus RT-PCR product as a reference (Fig. 1B). The Roakin, Kimber, and TexasGB viruses shared 89% sequence identity with the Ulster virus nucleotide sequences encoding the F protein cleavage site sequence. Nucleotide sequence differences occurred throughout the RT-PCR product. These mismatches included those nucleotides coding for basic amino acids at the cleavage site at interior locations of the amplification product. The Roakin and Kimber viruses had very similar HMA patterns, while the TexasGB virus had a slightly different pattern. The Herts33 and Italy/Milano viruses had similar HMA patterns and shared 90 and 89% sequence identity, respectively, with the Ulster nucleotide sequences encoding the F protein cleavage site sequence. The Largo and FL80 viruses had similar HMA patterns (Fig. 1B) and shared 96% sequence identity with each other but only 88% sequence identity with the Ulster RT-PCR product. The turkey/ND and cormorant/MN isolates that have 100% sequence identity had similar HMA patterns (Fig. 1B) when using Ulster as a reference. Ulster shared 86% identity in the nucleotide sequences encoding the F protein cleavage site. As with the other viruses exhibiting differences in their HMA patterns, the nucleotide differences were present throughout the amplification product.
HMA examination of NDV isolates not previously characterized.
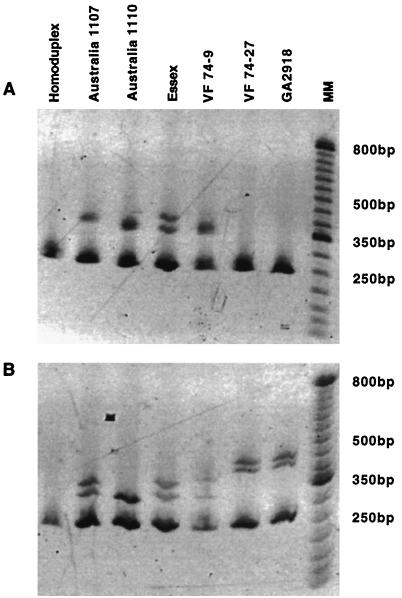
To ascertain the feasibility of the HMA to differentiate newly acquired NDV isolates, amplification products of B1 (Fig. 2A) and Ulster (Fig. 2B) were utilized as a reference to hybridize with RT-PCR products from a select group of viruses not previously characterized. These NDV isolates included viruses obtained during an outbreak of highly virulent Newcastle disease during 1974 in Northern Ireland (VF74-9), along with another virulent NDV from the United Kingdom, Essex70, isolated four years earlier. Also included for comparison were two viruses isolated during 1998 in Australia from the outbreak of neurovirulent Newcastle disease among domestic chickens (19-1107 and 14-1110) and a lentogenic field isolate (GA2918) from the southeastern United States (Table 1).
FIG. 2.
HMA for NDV isolates not previously characterized. (A) Utilization of the B1 vaccine type virus as the reference. (B) Utilization of the Ulster vaccine type virus as the reference. Virus isolates are presented in Table 1.
When B1 was used as the reference virus, two homoduplex patterns were obtained with isolates VF74-27 from Northern Ireland and GA2918 (Fig. 2A). Consequently, both of these NDV isolates have F protein nucleotide coding sequences with a high percentage of identity to the B1 vaccine type virus and are likely lentogenic. The GA2918 isolate was obtained during 1999 in the United States in Georgia and had a very low ICPI value (0.00) equivalent to its classification as a lentogen. These two NDV isolates had the same HMA pattern when analyzed using the Ulster virus as a reference (Fig. 2B) and were very similar to the vaccine-like viruses B1, LaSota, and VGGA (Fig. 1B). Sequence analysis of the amplification products revealed that the RT-PCR products from isolates VF74-27 and GA2918 shared 98% identity with B1 and 89% with Ulster, substantiating the close relationship to the B1-type NDV.
The viscerotropic VF74-9 NDV isolated during a virulent outbreak in Northern Ireland during 1974 had an HMA pattern unlike that of its lentogenic counterpart isolated at the same time and with the same geographical origin. Patterns were different between these two viruses whether using the B1 amplification product (Fig. 2A) or the Ulster virus (Fig. 2B) as a reference. Also, the Essex70 virus (Fig. 2A and B) obtained in the United Kingdom prior to the Northern Ireland outbreak had an HMA pattern somewhat dissimilar to that of the VF74-9 isolate. Consequently, these were possibly two separate viral populations of virulent NDV. However, nucleotide sequence analysis of the amplification products from the F protein cleavage sites revealed that the Essex70 virus shared 99% identity with that of the VF74-9 viral sequences. Two nucleotide sequence differences occurred at interior positions within the amplification product.
Viruses obtained from an outbreak of neurovirulent Newcastle disease in Australia during 1998 were analyzed using both B1 (Fig. 2A) and Ulster (Fig. 2B) as reference isolates. In both cases, homoduplexes were not obtained. A suspected precursor virus, 14-1110, with an ICPI value less than 0.7, had a distinctive HMA pattern compared to the virulent 19-1107 isolate that had an ICPI value of 1.7, indicating a highly virulent NDV. The pattern obtained for the lentogenic 14-1110 virus was very similar to the pattern obtained for QV4 when using either B1 (Fig. 1A) or Ulster (Fig. 1B) as the reference. The 14-1110 viral nucleotide sequence of the amplification product for the F protein cleavage site shared 96% identity with QV4, while there was 95% sequence identity between 19-1107 and QV4. There are only four nucleotide sequence differences between 14-1110 and 19-1107 and two of these occur at codons for a G-to-K substitution and the L-for-F substitution in the fusion protein.
Phylogenetics and predicted amino acid sequence of the F protein cleavage site.
Nucleotide sequences comprising the amplification product encoding the F protein cleavage site and surrounding region of the genome were aligned, followed by phylogenetic analysis (Fig. 3A). The NDV isolates examined separated into two principle groups. The first clade of NDV isolates was composed of very diverse viruses with origins worldwide that included both neurotropic and viscerotropic velogenic NDV. The majority of highly virulent NDV isolates in clade I may have had viruses related to Herts33 or Italy/Milano as potential progenitors. Virulent viruses such as Essex70 and isolate VF74-9 from the Northern Ireland Newcastle disease outbreak were most closely related to viruses isolated from psittacine birds in the United States during 1971. Viruses associated with an outbreak of neurovirulent Newcastle disease among cormorants and a remote free-range turkey flock in the United States were also more related to virulent viruses isolated during the 1970s. The neurovirulent 19-1107 NDV isolated from an outbreak in Australia during 1998 is directly related to a potential lentogenic precursor isolate, 14-1110. Clade II was composed of viruses closely related to the B1 and LaSota vaccine viruses. Neurovirulent viruses isolated in the United States prior to 1970, such as TexasGB, were among these viruses but were separated phylogenetically from the lentogenic vaccine and field isolates.
FIG. 3.
Phylogenetic relationships and predicted amino acid sequences of the F protein cleavage site among NDV isolates examined by HMA. (A) Nucleotide sequences of the amplification products used for HMAs were aligned. Subsequently, an unrooted tree was generated by parsimony analysis. The number of nucleotide differences is provided for each branch point. Bootstrap confidence limits following 1,000 replications are presented in parentheses for the major informative branches, while the predominant NDV clades are designated I and II. (B) Alignment of the predicted amino acid sequences obtained from the amplification products for each isolate.
Alignment of the predicted amino acid sequences surrounding the F protein cleavage site illustrate the heterogeneity found among the genomes of various NDV isolates (Fig. 3B). The lentogenic B1 vaccine-type viruses had the characteristic 109SGGGRQGR/LIG119 sequence, while the Ulster/QV4 vaccine-type viruses had a K for R substitution at position 113 of the cleavage site. Many of the virulent viruses had the sequence 109SGGRRQKR/FIG119 at the fusion protein cleavage site, although three velogenic viruses, Herts33, Italy/Milano, and 19-1107, had an R-for-K substitution at residue 115. All the virulent viruses isolated since 1970 had a V-for-I substitution at position 118 following the cleavage site, and two viruses from the outbreak among cormorants in North America, cormorant/MN and turkey/ND, had an R-for-G substitution at position 110. However, the virulent virus from Australia isolated during the 1998 Newcastle disease outbreak had the I at residue 118. There are a greater number of synonymous changes throughout the nucleotide sequences of the amplification products which also contribute to heteroduplex patterns observed among isolates that share many of the same amino acid sequences.
DISCUSSION
The B1 isolate of NDV is the most widely used vaccine strain for Newcastle disease worldwide (1, 25) and it was therefore chosen as the primary standard for comparing NDV isolates using the HMA. The Ulster strain has been periodically used as a vaccine strain outside the United States (1, 25), while QV4 has been incorporated into poultry feed in developing countries as an oral vaccine due to its greater thermostability (4, 14). All the lentogenic NDV field isolates analyzed during this study were most similar to the B1 strain and not to the Ulster-type vaccine. This reflects what has been determined previously (24) and also represents the wide use of this virus in major commercial poultry operations. The lentogenic VF74-27 virus was isolated during an outbreak of highly virulent Newcastle disease in Northern Ireland during 1974 and indicates that B1 vaccine-type viruses were being used during that epidemic. However, no lentogenic isolates shared sequence identity with the Ulster-type viruses; this may be due to the fact that live vaccines of this type are not extensively utilized in North America or Europe. Lentogenic NDV examined that originated outside North America were the Australian QV4 (37) and 14-1110 (42) strains. These two viruses had similar HMA patterns, and the 14-1110 isolate is believed to be the progenitor of virulent viruses that caused the major Newcastle disease outbreak in Australia during 1998, represented by isolate 19-1107 (42). This was confirmed by the phylogenetic relationship of 19-1107 with the QV4 and 14-1110 viruses.
Virulent NDV isolates had the most heterogeneous HMA patterns, which demonstrates the high sequence variation not only with the lentogenic vaccine-like NDV strains but also among highly virulent viruses themselves. This high sequence variation among virulent NDV isolates has been reported by several investigators (7, 23, 36, 45). Phylogenetically, these viruses represent isolates obtained from a wide variety of avian species with different geographic origins at various time points. A virulent virus isolated from chickens during an outbreak of highly virulent Newcastle disease in Northern Ireland during 1974 was most closely related to a virus isolated during 1970 in the United Kingdom. Both these viruses were, in turn, closely related to a psittacine isolate from 1970 and certainly may all be related to pandemic viruses circulating worldwide at that time (1). Specifically, a highly virulent viscerotropic virus from a psittacine bird was epidemiologically linked to the major outbreak in the United States during the early 1970s (41). These genetically heterogenous NDV isolates in clade I are not related to neurovirulent viruses, such as TexasGB, of clade II that were present in the United States prior to 1970.
The nucleotide sequence variation among virulent NDV isolates indicates that multiple lineages of NDV are circulating (23, 36); this is not that surprising, considering the nature of viruses that posses RNA genomes (9). It is important to note that although the HMA patterns were not specifically similar among even the closely related viruses during our study, this may be due to several factors. It is known that not only the percentage sequence identity may affect the HMA patterns obtained but also the relative positions of nucleotide mismatches may play a role. Lack of base pairing at interior positions of the two strands of DNA are more likely to affect the pattern obtained than those bases not forming exact pairs at exterior positions of the sequence (40). Consequently, amplification products with high sequence identity may either form homoduplexes if those nonpaired bases are at the exterior positions or form heteroduplexes if at interior positions. Although the RT-PCR products obtained for the Essex70 and VF74-9 viruses share 99% sequence identity, the different HMA patterns obtained were probably due to nucleotide substitutions occurring at interior positions. This is most certainly the case for the Australian NDV isolates and points to the utility of the technique, since these viruses were closely related but produced different HMA patterns. The nucleotide differences between these two sets of NDV isolates are primarily in the F protein cleavage site sequence located at an interior location of the amplification product. Consequently, discernible patterns were produced with the HMA by using primer sequences utilized by our laboratory.
Large fragments encompassing the entire measles virus nucleoprotein gene (1,683 bp) did not provide the required resolution when using the HMA relative to a 589-bp fragment (20). Also, hepatitis C virus genotyping utilizing HMA has been accomplished using a 178-bp fragment following RT-PCR (43). Therefore, we used a 254-bp product that was produced following amplification of coding sequences surrounding the F protein cleavage site of the NDV genome, which was developed for phylogenetic classification (35, 36). This satisfies the OIE requirements for NDV pathotyping and identification of the cleavage site when reporting isolates during outbreaks (29). The HMA was superior to restriction endonuclease analysis of amplification products because it is not dependant on a restricted number of site-specific sequences. Consequently, HMA utilized for NDV was found to be a reliable and rapid screening technique to select isolates for subsequent nucleotide sequence analysis to determine molecular phylogenetic relationships and for reporting of the F protein cleavage site.
ACKNOWLEDGMENTS
We acknowledge the excellent technical assistance of Joyce Bennett and Phillip Curry.
These investigations were supported by the USDA Scientific Cooperation Research Program, Research and Scientific Exchanges Division, Foreign Agricultural Service (grant no. X01-4510-62-751007-49) to B.S.S. for support of A.B. at the Southeast Poultry Research Laboratory and by the USDA Agricultural Research Service (CRIS project no. 6612-3200-021-00D-092).
REFERENCES
- 1.Alexander D J. Newcastle disease and other avian Paramyxoviridae infections. In: Calnek B W, Barnes H J, Beard C W, Reid W M, Yoder H W Jr, editors. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 541–570. [Google Scholar]
- 2.Alexander D J. Newcastle disease. In: Purchase H G, Arp L H, Domermuth C H, Pearson J E, editors. A laboratory manual for the isolation and identification of avian pathogens. 3rd ed. Kennett Square, Pa: American Association of Avian Pathologists, Inc.; 1989. pp. 114–120. [Google Scholar]
- 3.Alexander D J, Manville R J, Kemp P A, Parsons G, Collins M S, Brockman S, Russell P H, Lister S A. Use of monoclonal antibodies in the characterisation of avian paramyxovirus 1 (Newcastle disease virus) isolates submitted to an international reference laboratory. Avian Pathol. 1987;16:553–565. doi: 10.1080/03079458708436406. [DOI] [PubMed] [Google Scholar]
- 4.Awan M A, Otte M J, James A D. The epidemiology of Newcastle disease in rural poultry: a review. Avian Pathol. 1994;23:405–423. doi: 10.1080/03079459408419012. [DOI] [PubMed] [Google Scholar]
- 5.Chezzi C, Schoub B D. Differentiation between vaccine-related and wild-type polioviruses using a heteroduplex mobility assay. J Virol Methods. 1996;62:93–102. doi: 10.1016/s0166-0934(96)00552-6. [DOI] [PubMed] [Google Scholar]
- 6.Chomcynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate–phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Collins M S, Bashiruddin J B, Alexander D J. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch Virol. 1993;128:363–370. doi: 10.1007/BF01309446. [DOI] [PubMed] [Google Scholar]
- 8.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 9.Domingo E, Escarmis C, Seilla N, Baranowski E. Population dynamics in the evolution of RNA viruses. Adv Exp Med Biol. 1998;440:721–727. doi: 10.1007/978-1-4615-5331-1_93. [DOI] [PubMed] [Google Scholar]
- 10.Espeseth D A, Chapek M L. Report of the committee on biologics & bio/technology. Proc. 102nd Annu. Mtg. U.S. Animal Health Assoc. Richmond, Va: USAHA; 1998. pp. 112–119. [Google Scholar]
- 11.Glickman R L, Syddall R J, Iorio R M, Sheehan J P, Bratt M A. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J Virol. 1988;62:354–356. doi: 10.1128/jvi.62.1.354-356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotoh B, Ohnishi Y, Inocencio N M, Esaki E, Nakayama K, Barr P J, Thomas G, Nagai Y. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J Virol. 1992;66:6391–6397. doi: 10.1128/jvi.66.11.6391-6397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchinson H L. The control and eradication of Newcastle disease in Northern Ireland. Vet Rec. 1975;96:213–217. doi: 10.1136/vr.96.10.213. [DOI] [PubMed] [Google Scholar]
- 14.Ideris A, Ibrahim A L, Spradbrow P B. Vaccination of chickens against Newcastle disease with a food pellet vaccine. Avian Pathol. 1990;19:371–384. doi: 10.1080/03079459008418687. [DOI] [PubMed] [Google Scholar]
- 15.Jarecki-Black J C, King D J. An oligonucleotide probe that distinguishes isolates of low virulence from the more pathogenic strains of Newcastle disease virus. Avian Dis. 1993;37:724–730. [PubMed] [Google Scholar]
- 16.Jestin V, Cherbonnel M, L'Hospitalier R, Bennejean G. An ELISA blocking test using a peroxidase-labeled anti-HN monoclonal antibody for the specific titration of antibodies to avian paramyxovirus type 1. Arch Virol. 1989;105:199–208. doi: 10.1007/BF01311357. [DOI] [PubMed] [Google Scholar]
- 17.Jestin V, Jestin A. Detection of Newcastle disease virus RNA in infected allantoic fluids by in vitro enzymatic amplification (PCR) Arch Virol. 1991;118:151–161. doi: 10.1007/BF01314026. [DOI] [PubMed] [Google Scholar]
- 18.King D J. Newcastle disease virus passage in MDBK cells as an aid in detection of a virulent subpopulation. Avian Dis. 1993;37:961–969. [PubMed] [Google Scholar]
- 19.Kotewicz M L, Sampson C M, D'Alessio J M, Gerard G F. Isolation of cloned Maloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity. Nucleic Acids Res. 1988;16:265–277. doi: 10.1093/nar/16.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreis S, Whistler T. Rapid identification of measles virus strains by the heteroduplex mobility assay. Virus Res. 1997;47:197–203. doi: 10.1016/s0168-1702(96)01413-x. [DOI] [PubMed] [Google Scholar]
- 21.Lewis J G, Chang G-J, Lanciotti R S, Trent D W. Direct sequencing of large flavivirus PCR products for analysis of genome variation and molecular epidemiological investigations. J Virol Methods. 1992;38:11–24. doi: 10.1016/0166-0934(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 22.Lichten M J, Fox M S. Detection of non-homology containing heteroduplex molecules. Nucleic Acids Res. 1983;11:3959–3971. doi: 10.1093/nar/11.12.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomniczi B, Wehmann E, Herczeg J, Ballagi-Pordany A, Kaleta E F, Werner O, Meulemans G, Jorgensen P H, Mante A P, Gielkens A L, Capua I, Damoser J. Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII) Arch Virol. 1998;143:49–64. doi: 10.1007/s007050050267. [DOI] [PubMed] [Google Scholar]
- 24.Marin M C, Villegas P, Bennett J, Seal B. Virus characterization and sequence of the fusion protein gene cleavage site of recent Newcastle disease virus field isolates. Avian Dis. 1996;40:382–390. [PubMed] [Google Scholar]
- 25.Meulmanns G. Control by vaccination. In: Alexander D J, editor. Newcastle disease. Boston, Mass: Kluwer Academic Publishers; 1988. pp. 318–332. [Google Scholar]
- 26.Millar N S, Chambers P, Emmerson P T. Nucleotide sequence of the fusion and hemagglutinin-neuraminidase glycoprotein genes of Newcastle disease virus, strain Ulster: molecular basis for variations in pathogenicity between strains. J Gen Virol. 1988;69:613–620. doi: 10.1099/0022-1317-69-3-613. [DOI] [PubMed] [Google Scholar]
- 27.Murphy G, Belda F J, Pau C P, Clewley J P, Parry J V. Discrimination of subtype B and non-subtype B strains of human immunodeficiency virus type 1 by serotyping: correlation with genotyping. J Clin Microbiol. 1999;37:1356–1360. doi: 10.1128/jcm.37.5.1356-1360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai Y, Klenk H D, Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72:494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- 29.Office International des Epizootes. Newcastle disease. International Health Code. 9th ed. 2000. http://www.oie.int/ ( http://www.oie.int/). ). [Google Scholar]
- 30.Ogasawara T, Gotoh B, Suzuki H, Asaka J, Shimokata K, Rott R, Nagai Y. Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo. EMBO J. 1992;11:467–472. doi: 10.1002/j.1460-2075.1992.tb05076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell P H, Alexander D J. Antigenic variation of Newcastle disease virus strains detected by monoclonal antibodies. Arch Virol. 1983;75:243–253. doi: 10.1007/BF01314890. [DOI] [PubMed] [Google Scholar]
- 32.Saiki R K, Scharf F, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnhem N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 33.Sanger F, Nickles S, Carlson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schloer G. Antigenic relationships among Newcastle disease virus mutants obtained from laboratory strains and from recent California isolates. Infect Immun. 1974;10:724–732. doi: 10.1128/iai.10.4.724-732.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seal B S, King D J, Bennett J D. Characterization of Newcastle disease virus isolates by reverse transcription PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J Clin Microbiol. 1995;33:2624–2630. doi: 10.1128/jcm.33.10.2624-2630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seal B S, King D J, Locke D P, Senne D A, Jackwood M W. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J Clin Microbiol. 1998;36:1141–1145. doi: 10.1128/jcm.36.4.1141-1145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons G C. The isolation of Newcastle disease virus in Queensland. Austral Vet J. 1967;43:29–30. doi: 10.1111/j.1751-0813.1967.tb04764.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith L M, Sanders J Z, Kaiser R J, Hughs P, Dodd C, Connell C R, Heines C, Kent S B H, Hood L E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:673–681. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 39.Swafford D. PAUP*: phylogenetic analysis using parsimony, version 4. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 40.Upchurch D A, Shaankarappa R, Mullins J I. Position and degree of mismatches and the mobility of DNA heteroduplexes. Nucleic Acids Res. 2000;28:E69. doi: 10.1093/nar/28.12.e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Utterback W W, Schwartz J H. Epizootiology of velogenic viscerotropic Newcastle disease in southern California, 1971–1973. J Am Vet Med Assoc. 1973;163:1080–1088. [PubMed] [Google Scholar]
- 42.Westbury H. Newcastle disease virus: an evolving pathogen? Avian Pathol. 2001;30:5–11. doi: 10.1080/03079450020023131. [DOI] [PubMed] [Google Scholar]
- 43.White P A, Zhai X, Carter I, Zhao Y, Rawlinson W D. Simplified hepatitis C virus genotyping by heteroduplex mobility analysis. J Clin Microbiol. 2000;38:477–482. doi: 10.1128/jcm.38.2.477-482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson R A, Perrotta C, Frey B, Eckroade R J. An enzyme-linked immunosorbent assay that measures protective antibody levels to Newcastle disease virus in chickens. Avian Dis. 1984;29:1079–1085. [PubMed] [Google Scholar]
- 45.Yang C-Y, Chang P-C, Hwang J-M, Shieh H K. Nucleotide sequence and phylogenetic analysis of Newcastle disease virus isolates from recent outbreaks in Taiwan. Avian Dis. 1997;41:365–373. [PubMed] [Google Scholar]
- 46.Zou S, Stansfield C, Bridge J. Identification of new influenza B virus variants by multiplex reverse transcription-PCR and the heteroduplex mobility assay. J Clin Microbiol. 1998;36:1544–1548. doi: 10.1128/jcm.36.6.1544-1548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]