An inducible CDK6–INK4 complex blocks drug binding and impairs efficacy for current-generation CDK4/6 inhibitors, but harbors sensitivity to potent CDK4/6 degrader compounds that represent a promising strategy for next-generation cell-cycle inhibitors.
Abstract
Cyclin-dependent kinases 4 and 6 (CDK4/6) represent a major therapeutic vulnerability for breast cancer. The kinases are clinically targeted via ATP competitive inhibitors (CDK4/6i); however, drug resistance commonly emerges over time. To understand CDK4/6i resistance, we surveyed over 1,300 breast cancers and identified several genetic alterations (e.g., FAT1, PTEN, or ARID1A loss) converging on upregulation of CDK6. Mechanistically, we demonstrate CDK6 causes resistance by inducing and binding CDK inhibitor INK4 proteins (e.g., p18INK4C). In vitro binding and kinase assays together with physical modeling reveal that the p18INK4C–cyclin D–CDK6 complex occludes CDK4/6i binding while only weakly suppressing ATP binding. Suppression of INK4 expression or its binding to CDK6 restores CDK4/6i sensitivity. To overcome this constraint, we developed bifunctional degraders conjugating palbociclib with E3 ligands. Two resulting lead compounds potently degraded CDK4/6, leading to substantial antitumor effects in vivo, demonstrating the promising therapeutic potential for retargeting CDK4/6 despite CDK4/6i resistance.
Significance:
CDK4/6 kinase activation represents a common mechanism by which oncogenic signaling induces proliferation and is potentially targetable by ATP competitive inhibitors. We identify a CDK6–INK4 complex that is resilient to current-generation inhibitors and develop a new strategy for more effective inhibition of CDK4/6 kinases.
This article is highlighted in the In This Issue feature, p. 275
Introduction
Antagonism of oncogenic kinases through small-molecule ATP-competitive inhibitors has been a widely successful strategy for anticancer therapy. Unfortunately, drug resistance is frequently encountered and is often commonly due to genetic alterations that lead to kinase reactivation and drug insensitivity (1, 2). Among the most common mechanisms of kinase reactivation are somatic point mutations in the target kinase domain that remodel the drug binding pocket, thereby increasing its relative affinity to ATP over drugs. Examples of such mutations include T790M in EGFR-driven lung cancer (3) and T315I in BCR–ABL-driven leukemia (4, 5), among many others. ATP-competitive CDK4/6 kinase inhibitors (CDK4/6i) have led to major improvements in the survival of patients with breast cancer and have shown early signs of efficacy in several other cancer types (6). Resistance to these drugs has been studied by many groups, but surprisingly, not a single instance of point mutation in CDK4 or CDK6 kinase has been described from thousands of tumors sequenced (5–8). In previous studies, we and others have found that upregulation of CDK6 promotes resistance to CDK4/6i (8–10). In this study, we investigate the basis for CDK6-mediated resistance and find that upregulation of this kinase leads to a distinctive mechanism to increase the relative affinity of the kinase toward ATP over drug. Overexpression of CDK6 causes the upregulation of and binding to INK4 proteins, which then serve as strong competitive inhibitors of the drug but only weak competitive inhibitors of ATP. Thus, overexpression of CDK6 leads to a kinase-weak but drug-insensitive complex that persistently drives G1 progression and cancer growth. To overcome this challenge, we developed a novel set of bifunctional degraders that can effectively bind CDK6, in part through allosteric contacts. In particular, a degrader that is able to bind and degrade both CDK4 and CDK6 is identified and demonstrates activity in cancers resistant to first-generation CDK4/6i.
Results
INK4 Proteins Interact with CDK6 in CDK4/6i-Resistant Cells
Previous studies have identified upregulation of wild-type (WT) CDK6 expression as a recurrent mechanism by which tumors restore cell proliferation during CDK4/6i therapy (8–10). To determine if overexpression of CDK6 leads to reactivation of G1 checkpoint kinase activity, we immunoprecipitated CDK4 and CDK6 from isogenic drug-sensitive (MCF7 parental cells with low CDK6) and drug-resistant (MCF7 FAT1-loss cells with high CDK6) cells (8) and assayed their kinase activity using RB substrate (Fig. 1A and B; Supplementary Fig. S1A). As expected, drug-sensitive cells displayed higher basal levels of expression and activity of CDK4 compared with CDK6. By contrast, drug-resistant CDK6-high cells had similar levels of CDK4 and CDK6 kinase activity. Pretreatment of cells with abemaciclib potently inhibited the kinase activity of CDK4 in both sensitive (84% reduced compared with untreated) and resistant (82%) cells but could only partially reduce CDK6 activity (48%) in resistant cells despite the near-equal IC50 derived from using recombinant CDK4 (2 nmol/L) and CDK6 (5 nmol/L) kinases, as previously reported (11). As the composition of CDK4/6 complexes with specific members (e.g., D-cyclin, p16, p21, p27) can modify kinase activity and drug response (12–17), we investigated CDK4- and CDK6-interacting proteins by immunoprecipitation (IP) of CDK4 and CDK6 from drug-sensitive and drug-resistant cells followed by mass spectrometry (MS). Across three replicates, we identified seven proteins that were found in association with CDK6, but not CDK4, in CDK4/6i-resistant cells (Fig. 1C). Among proteins known to interact with CDK4/6 and regulate cell cycle, the INK4 proteins p15INK4B and p18INK4C (parental cells lack endogenous p16INK4A) appeared as the top two that associated with CDK6 but not CDK4 (Fig. 1D and E). We verified these findings by IP and immunoblotting and again found that both p15INK4B and p18INK4C associate with CDK6 in the resistant CDK6-high MCF7 FAT1CR cells (Fig. 1F; Supplementary Fig. S1B). We verified this strong association in other INK4+/CDK6-expressing breast cancer cell lines that were resistant to CDK4/6i (Supplementary Fig. S1C and S1D). Of note, we observed that the INK4-associated CDK6 was an active kinase by demonstrating its phosphorylation of RB (Supplementary Fig. S1E). Consistent with previous data, expression of the INK4 proteins was also upregulated in resistant cells compared with sensitive cells (8). Finally, we conducted an unbiased screen of over 1,000 cell lines (PRISM) and found INK4 overexpression (along with RB1 loss) to be among the top genomic alterations associated with CDK4/6i resistance (Fig. 1G). These data reveal that INK4 proteins strongly associate with CDK6 in CDK6-high cells that are resistant to CDK4/6i (18).
Figure 1.
INK4–CDK6 complex promotes resistance to CDK4/6i in cells. A, Schematic for analysis of CDK4 and CDK6 interactions and activity via coimmunoprecipitation (co-IP) followed by ADP-Glo kinase assays and mass spectrometry. B, ADP-Glo kinase assay showing immunoprecipitated CDK4 and CDK6 (IP-CDK4 and IP-CDK6) kinase activity from MCF7 parental and CDK6-high cells [cells with FAT1 CRISPR knockout (CR) that have high CDK6 expression, previously shown to have resistance to CDK4/6i; ref. 8], with or without 100 nmol/L abemaciclib treatment. Data are shown as mean + SD of three biologically independent samples. P values were determined by unpaired two-sided Student t test. RLU, relative luminescence units. C, Venn diagram showing the number of unique proteins identified by mass spectrometry coimmunoprecipitated from IP-CDK4 and IP-CDK6 in FAT1-loss cells. Percentages were calculated by number of proteins identified in each subgroup divided by total proteins identified by IP of either CDK4 or CDK6. Data are shown as means of three replicates. D, Pathway analysis by Gene Ontology of proteins interacting with CDK6 but not CDK4 in the FAT1-loss cells. The proteins were grouped by their putative biological functions. E, Unique peptide counts of cyclin-dependent kinases and their endogenous inhibitor proteins identified in the co-IP/mass spectrometry associated with CDK4 or CDK6 in the FAT1-loss cells. N = 2. F, Co-IP and immunoblotting reveal association of p15INK4B and p18INK4C with CDK6, but not CDK4, in CDK6-high cells. G, Cell line screening results showing that models with high CDKN2A or low RB1 mRNA expression are correlated with poor response to palbociclib. H, Interface residues in CDK6 in close proximity with INK4 isoforms based on previous INK4-bound CDK6 structures in the Protein Data Bank (ref. 65; no available structure for p15INK4B). CDK6-HA was immunoprecipitated using HA beads in parental MCF7 cells and MCF7 cells expressing HA-WT-CDK6-, HA-V16D-, and R31C-mutant CDK6 (disrupted INK4–CDK6 interaction) or HA-K43M/D163N-mutant CDK6 (kinase dead), and interaction with INK4 proteins was determined by immunoblotting. I, Disruption of INK4s and CDK6 binding or impairment of CDK6 kinase activity restores the sensitivity of CDK6-overexpressing cells to CDK4/6i. Cells were treated with DMSO or 100 nmol/L abemaciclib for 24 hours prior to collection. J, Percentage of cell viability of cells overexpressing WT CDK6 or R31C- or D163N-mutant CDK6 treated with increasing concentrations of abemaciclib compared with parental cells. IC50 values were recorded on day 5 following treatment. Data are shown as mean ± SD; n = 6. K, Knockdown of p15INK4B and p18INK4C in FAT1-loss cells promotes suppression of RB phosphorylation in response to abemaciclib to a similar extent as in parental cells. Cells were collected 24 hours after 100 nmol/L abemaciclib treatment. Representative blots are shown, which were repeated independently three times. L, The growth rate of p15INK4B and p18INK4C knockout in FAT1-loss cells was inhibited by 100 nmol/L abemaciclib. The cell viability was recorded at day 14 and day 21. ****, P < 0.0001. Data are shown as mean ± SD; n = 6. See also Supplementary Fig. S1.
Interaction of INK4s and CDK6 Promotes Resistance to CDK4/6i
Based on previous crystallographic structures of CDK6–INK4 (19, 20), we selected candidate residues in CDK6 that are in proximity of the INK4 binding site and performed site-directed mutagenesis of apparent CDK6–INK4 interface residues. By coimmunoprecipitation, we confirmed that V16D and R31C alterations disrupted the interaction of CDK6 with p15INK4B and p18INK4C but with intact kinase activity (Fig. 1H; Supplementary Fig. S1F). By contrast, classical kinase-dead mutations (K43M and D163N), far from the interface, did not disrupt the interaction. Consistent with a functional role for the INK4 interaction in drug resistance, mutations in the CDK6–INK4 interface decreased phosphorylation of RB and downstream signaling in response to abemaciclib and palbociclib to the same extent as kinase-dead mutations (Fig. 1I). Cell viability assays also showed restored cellular sensitivity to abemaciclib in cells expressing mutant forms of CDK6 impaired at binding INK4 (R31C or V16D; Fig. 1J). These finding were recapitulated in multiple estrogen receptor (ER)–positive breast cancer cell lines, including MCF7, T47D, ZR-75-1, CAMA1, and EFM19 (Supplementary Fig. S1G–S1N). Moreover, overexpression of WT but not mutant forms of CDK6 (R31C or D163N) reduced the accumulation of cells in G1 after treatment with abemaciclib or palbociclib (Supplementary Fig. S1O–S1Q). To confirm the role of INK4 proteins in mediating drug resistance, we genetically knocked out CDKN2B (p15) and CDKN2C (p18) in CDK6-high cells and found that p18INK4C loss could partially restore the responsiveness of RB phosphorylation to abemaciclib treatment, whereas loss of both p15INK4B and p18INK4C could do so almost entirely (Fig. 1K). Concordantly, long-term growth assays demonstrated that the loss of INK4 proteins rendered CDK6-high cells sensitive to CDK4/6i. (Fig. 1L; Supplementary Fig. S1R and S1S). Conversely, overexpression of p16INK4A in T47D cells lowered the potency of both abemaciclib and palbociclib (Supplementary Fig. S1T and S1U). Taken together, these data implicate a drug-insensitive INK4–CDK6 complex in driving persistent RB phosphorylation in CDK4/6i-resistant tumors.
The INK4–CDK6 Complex Is Insensitive to CDK4/6i
To further establish the role of the INK4 interaction in mediating the CDK4/6i insensitivity of CDK6, we used recombinant CDK6–cyclin D3 and p18INK4C and performed an in vitro kinase assay (Fig. 2A and B; Supplementary Fig. S2A–S2C). As expected, abemaciclib potently inhibited CDK6–cyclin D3 kinase activity with an IC50 of 8 nmol/L (Supplementary Fig. S2B), approximating published reports (21), and addition of recombinant p18INK4C protein inhibited CDK6 kinase activity (Supplementary Fig. S2C). Although addition of p18 did lower CDK6–cyclin D activity, it also prevented the near-complete suppression by abemaciclib observed in the absence of p18 (Fig. 2A and B; Supplementary Fig. S2D). Immunoblotting for RB phosphorylation confirmed that preincubation with p18 impairs abemaciclib inhibition of CDK6 activity (Fig. 2C).
Figure 2.
INK4–CDK6 complexes are insensitive to CDK4/6i. A,In vitro kinase assay using recombinant CDK6–cyclin D3 and RB substrate demonstrates that preincubation of the complex with p18 (purple) prevents complete inhibition of kinase activity by abemaciclib. Data are shown as mean ± SD of two biological replicates. RLU, relative luminescence units. B, Effect of preincubation of p18 on CDK6–cyclin D3 in vitro kinase activity. Data are shown as mean ± SD of two biological replicates. C, Assay of CDK6–cyclin D3 kinase activity and response to p18 by immunoblotting demonstrating that p18 impairs the ability of abemaciclib to inhibit CDK6 phosphorylation of RB. D, Computational modeling of the effect of p18 binding to the CDK6 binding pocket expressed as volume change for abemaciclib (top) or AMP-PNP (bottom). Structures of CDK6–cyclin complex before and after p18 binding are represented by crystallographic structures with PDB IDs 2EUF and 1G3N (shown in ribbons). The binding pockets were approximated by spheres (shown in green and indicated by red arrows; shown in purple and indicated by green arrows). The volume of each binding pocket was quantified using the total volume of the corresponding set of spheres, and percentage of changes was calculated. E, The table summarizes the changes of binding pocket volume for two CDK4/6i (palbociclib and abemaciclib) and AMP-PNP upon binding of INK4s (p16, p18, p19). F, MST assay of CDK6 binding to abemaciclib showing the change in Kd as a result of p18 binding (red). Data are shown as mean ± SD of two independent measurements. See also Supplementary Fig. S2.
To elucidate structural mechanisms underlying the effect of INK4 proteins on CDK6 drug inhibition, we inspected existing CDK6 structures alone [Protein Data Bank (PDB) ID: 2EUF; ref. 22] or in complex with INK4s (listed in the table of Fig. 1H). Structural superimposition indicates that the N-lobe of CDK6 is twisted toward cyclin D upon binding of INK4 (Fig. 2D). We found that INK4 (p16, p18, or p19) binding to CDK6 caused distortion of the N-lobe of CDK6, thereby more significantly decreasing the effective binding pocket volume for CDK4/6i than for AMP-PNP, a nonhydrolyzable analogue of ATP (Fig. 2D). This effect is most prominent in p18-bound CDK6, in which p18 binding causes a drastic reduction of the CDK4/6i binding volume (−87.65% for abemaciclib and −85.03% for palbociclib) but minimally affects the binding volume of AMP-PNP (+0.54%; Fig. 2D). The p16-bound CDK6 (−87.44%/−7.64%) and p19-bound CDK6 (−62.36%/−32.05%) led to similar reductions in abemaciclib and AMP-PNP binding volumes (Fig. 2E). The reduction of binding pocket volume upon association of INK4 compellingly explains the impaired CDK4/6i inhibition and residual kinase activity in the presence of INK4 observed in our biochemical assays. To directly test whether p18INK4C alters the binding affinity of CDK4/6i to CDK6, we performed microscale thermophoresis (MST) assays and found the Kd of abemaciclib to CDK6 was increased fourfold in the presence of p18INK4C (Fig. 2F). Taken together, these data reveal that the addition of p18INK4C to the cyclin D–CDK6 complex suppresses CDK4/6i binding, likely mediating drug resistance.
Multiple Genetic Alterations Lead to CDK6-Mediated Resistance in Patients
To define the prevalence of the CDK6-high, CDK4/6i-resistant state in clinically relevant samples, we analyzed CDK6 and INK4 protein expression by immunohistochemistry, using a panel of patient-derived ER+ breast cancer xenografts (Fig. 3A). We found that among 14 distinct models, eight models displayed intense CDK6 staining. Of these, seven of eight were found to be resistant to CDK4/6i (Fig. 3B). Based on our prior work establishing Hippo pathway suppression as a mechanism of CDK6 upregulation (8), we further analyzed FAT1 and nuclear YAP protein levels in these samples and indeed found that a subset of high-CDK6 tumors featured low FAT1, high nuclear YAP1, and high p15/p18 (Fig. 3C; Supplementary Fig. S3A). However, high CDK6 was found in some FAT1 WT tumors as well, suggesting that additional genetic alterations might promote high CDK6 expression. Indeed, both PTEN and ARID1A have been implicated as potential regulators of this pathway; therefore, we tested whether loss of these might also increase CDK6 abundance (23, 24). Knockdown of either PTEN or ARID1A in CDK4/6i-sensitive cell lines led to upregulation of CDK6 expression and resistance to abemaciclib (Fig. 3D–G; Supplementary Fig. S3B). Given the canonical role of PTEN in suppressing AKT activation, we tested the effects of the AKT inhibitor MK-2206 and found its administration suppressed the expression of CDK6 in PTEN knockdown cells (Fig. 3H). With respect to ARID1A and its known role in sequestering YAP (24), we found that knockdown of YAP reduced CDK6 expression in the ARID1A knockdown cells (Fig. 3I). Moreover, we found an induction of INK4 protein expression concomitant with CDK6 upregulation in both PTEN and ARID1A knockdown cells (Fig. 3H and I), suggesting that coordinate upregulation of INK4 and CDK6 may be responsible for mediating drug resistance in this context. Given these results, we analyzed the genomic landscape of 1,366 metastatic tumors from 1,115 patients with hormone receptor (HR)+/HER2− metastatic breast cancer (MSK-IMPACT) and found that 190 out of 1,115 (17%) patients showed at least one of the genetic alterations that might be associated with CDK6 upregulation and resistance to CDK4/6i (Fig. 3J). These findings demonstrate that mutations that can promote a CDK6–INK4 complex represent a significant cohort of patients in whom current ATP-competitive CDK4/6i may prove ineffective.
Figure 3.
Multiple genetic alterations promote CDK6-mediated resistance in patients. A, IHC of FAT1, CDK6, YAP, p15INK4B, and p18INK4C in representative patient-derived xenograft (PDX) models that are sensitive or resistant to CDK4/6i. B, Number of cases that show high or low CDK6 in PDX models that are sensitive or resistant to CDK4/6i. Immunoreactive score (IRS) >2 is recorded as high CDK6 expression. C, IRS of CDK6, nuclear YAP, FAT1, p15, and p18 staining in sensitive and resistant PDX models. D and E, Immunoblotting demonstrating that knockdown of PTEN or ARID1A in MCF7 cells promotes upregulation of CDK6 and resistance to 100 nmol/L abemaciclib (abema) treatment. Cells were treated for 24 hours prior to collection. ns, not significant. F, Cell viability (percentage of control cells) plots showing that both PTEN knockdown cells have decreased sensitivity to abemaciclib compared with parental cells. IC50 values were recorded on day 5. Data are shown as mean ± SD; n = 6. G, Cell viability (percentage of control cells) plots showing that ARID1A knockdown cells have decreased sensitivity to abemaciclib compared with parental cells. Knockdown of YAP1 in shARID1A cells restores its sensitivity to abemaciclib. IC50 values were recorded on day 7. Data are shown as mean ± SD; n = 6. H, Immunoblotting showing inhibition of AKT (2 μmol/L MK-2206) suppresses induction of CDK6 expression in PTEN knockdown cells. I, Immunoblotting showing that knockdown of YAP1 in shARID1A cells decreases CDK6 expression. All blots were repeated at least three times, and representative blots are shown. J, The pattern, frequency, and type of genomic alterations in CDK6-associated genes in 1,366 metastatic tumors from 1,115 patients with HR+/HER2− metastatic breast cancer. A total of 190 cases show at least one of the genetic alterations associated with CDK6 upregulation. See also Supplementary Fig. S3.
Degraders Targeting CDK6 Complexes Inhibit Resistant Cells
As the INK4–CDK6 complex confers resistance to the current generation of CDK4/6i, we explored the potential of other compounds to target this pathway. Recently, bifunctional degraders [proteolysis targeting chimera (PROTAC)] have emerged as a promising approach to target “undruggable” proteins and overcome resistance to small-molecule inhibitors (25, 26). We previously identified the selective CDK6 degrader BSJ-03-123 (27) and here examined its effect in CDK6-high, CDK4/6i-resistant cells. BSJ-03-123 led to dose-dependent degradation of CDK6 but had no effect on CDK4. As a result, BSJ-03-123 could inhibit the phosphorylation of RB and expression of downstream cell-cycle signaling components (e.g., cyclin A2 and E2F1) in CDK4/6i-resistant cells (Fig. 4A). However, when the same cells were grown in long-term culture with effective CDK6 inhibition or knockdown (9), we did not observe significant growth suppression due to preserved CDK4 activity (Supplementary Fig. S4A). We thus generated a panel of CDK4/6-selective degraders by linking palbociclib to Cereblon (CRBN) binding (28) or von Hippel-Landau (VHL) binding ligands (Supplementary Fig. S4B). All the chemical synthesis information is provided in Supplementary Data. Among these, BSJ-05-017 and BSJ-03-096 showed the highest potency in degrading CDK4 and CDK6, acting at doses as low as 10 nmol/L (Fig. 4B; Supplementary Fig. S4C and S4D). The two compounds demonstrated effective inhibition of the phosphorylation of RB and the expression of E2F1–cyclin A2 in both CDK4/6i-sensitive and CDK4/6i-resistant cells (Fig. 4B and C; Supplementary Fig. S4E and S4F). The degradation of CDK4 and CDK6 was abolished due to loss of binding to VHL with a reversal of the two chiral centers in the VHL ligand (Supplementary Fig. S4G; ref. 29). To assess the therapeutic potential of BSJ-05-017 in CDK6-driven cells, we performed cell proliferation assays and found BSJ-05-017 to be equipotent in suppressing proliferation (Fig. 4D; Supplementary Fig. S4H) and inducing cell-cycle arrest and senescence (Fig. 4E; Supplementary Fig. S4I) of CDK4/6i-sensitive and CDK4/6i-resistant breast cancer cells. To understand how these degraders could induce degradation and do so despite the presence of INK4, we manually constructed atomic-level molecular models of CDK4/6 complexed with degraders: E3 ligase adapter pairs (BSJ-03-123: CRBN, BSJ-05-017: VHL) in the presence of p18 or p27 and cyclin D based on existing crystallographic data and previously reported PROTAC degrader binding models (12, 22). The E3 ligase adapters shifted ∼0.5 nm from their initial conformation to adopt a new stable conformation in all four models (Supplementary Fig. S4J). To dissect the binding patterns of each of the ternary complex models, hydrogen bonds between the CDKs and the E3 ligase adapters or the degraders were identified for each simulation trajectory (Supplementary Fig. S4K and S4L). Comparing the CRBN/CDK4 versus CRBN/CDK6 conformations, it appears that CDK4 and CDK6 are engaging distinct regions of the CRBN surface with minimal overlap. The CDK6 binding region on the CRBN surface is closer to the binding pocket of BSJ-03-123, and one of the key residues identified here, H353, was previously reported to be important for CRBN to recruit and interact with various substrates (30–32). By contrast, CDK4 engages a set of CRBN residues that are distal to the degrader binding pocket. This difference potentially explains the selective degradation of CDK6 over CDK4 induced by BSJ-03-123. To investigate how both BSJ-03-123 and BSJ-05-017 target CDK6, we examined the modeled binding modes of the degrader warhead to CDK6 in complex with E3 ligase adapters in detail. There is minimal interaction between BSJ-03-123 and the kinase binding pocket (no interaction with CDK6 and only one h-bond with the hinge region V101 in CDK4; top row of Supplementary Fig. S4L). Instead, the stabilization of the ternary complex and the effective degradation of CDK6 appear to result from protein–protein interactions between CRBN and CDK6. In the case of the BSJ-05-017 (bottom row of Supplementary Fig. S4L), the degrader appears to partially interact with the kinase binding pocket [F164 in CDK6 and D163 in CDK4, both residues in the Asp–Phe–Gly (DFG) motif] despite the distorted binding pocket and DFG-out conformation of CDK6 in the presence of INK4. In this case, both the degrader–CDK interactions and VHL–CDK interactions contribute to the stabilization of the complex, explaining the robust CDK degradation by BSJ-05-017 observed experimentally. The capacity of these molecules to effectively degrade CDK6 is also consistent with prior reports that even PROTAC compounds with weak ligand binding affinity for the target protein can still achieve formation of a stable complex through additional interactions, leading to potent protein degradation (33).
Figure 4.
Compounds targeting the CDK6–INK4 complex inhibit CDK4/6i-resistant tumors. A, Immunoblotting of MCF7 parental cells and cells with high CDK6 expression [CDK6-overexpressing (OE) cells and CDK6-high cells with FAT1 loss] treated for 24 hours with increasing concentrations of bifunctional degrader compound, BSJ-03-123, demonstrating dose-dependent targeting of CDK6 but not CDK4. B, Assessment of a panel of degrader compounds that target CDK4 and/or CDK6. Immunoblotting after 24-hour drug treatment (500 nmol/L) in FAT1-loss cells shows varying selectivity for CDK4 versus CDK6. Representative blots from three independent experiments are shown. Among them, BSJ-05-017 and BSJ-03-096 show the most significant degradation of both CDK4 and CDK6. C, Immunoblot depicting dose–response effects of BSJ-05-017 in both CDK4/6i-sensitive (left) and CDK4/6i-resistant (right) cells in comparison with palbociclib (500 nmol/L) after 24-hour treatment. D, Percentage of growth plot showing that BSJ-05-017 inhibits sensitive MCF7 parental and resistant CDK6-high cells with equal potency, whereas palbociclib shows only partial inhibition of resistant cells. IC50 values were recorded at day 7. Data are shown as mean ± SD; n = 6. E, Assay for drug-induced senescence (Senescence Green) demonstrating number of senescence marker–positive cells induced by 8 days of treatment with DMSO, BSJ-05-017 (500 nmol/L), abemaciclib (100 nmol/L), and palbociclib (500 nmol/L). BSJ-05-017 induced a significantly higher number of cells into senescence compared with abemaciclib or palbociclib in CDK6-high cells. F, Immunoblotting showing the degradation of CDK4/6 and decreased phospho-RB1 and E2F1 levels in CDK6-high (FAT1 loss) tumor-bearing mice administered 25 mg/kg BSJ-05-017 intraperitoneally. Tumors were collected 6 hours after 3 consecutive days of vehicle or BSJ-05-017 treatment (n = 2). G, Growth curve plots of cell-derived xenografts of MCF7 parental, CDK6-overexpressing, and PTEN-loss cells. Mice were treated with vehicle, ribociclib (25 mg/kg, orally), BSJ-05-017 (50 mg/kg, i.p.), or BSJ-03-096 (50 mg/kg, orally) daily for 25 to 35 days. Tumor volumes were recorded every 3 to 4 days. Data are shown as mean ± SD; n = 4. See also Supplementary Fig. S4.
To ascertain the potential for in vivo use of BSJ-05-017 and BSJ-03-096, we assessed its pharmacokinetic (PK) properties following a single dose in mice intraperitoneally or per os (PO). Both BSJ-05-017 and BSJ-03-096 displayed high drug exposure in plasma, achieving a Cmax of 2.6 μmol/L and 0.9 μmol/L, respectively, as well as good metabolic stability, as near equivalent to ribociclib at 20 mg/kg in a previous report (34). At 24 hours postdosing, the compound's concentration remained near 100 nmol/L, still above the IC50 for in vitro CDK4/6 degradation (Supplementary Fig. S4M). Given these promising results, we next evaluated the in vivo effects of both compounds in CDK6-low and CDK6-high cell-derived xenografts. Compared with the vehicle control, BSJ-05-017 induced near-complete degradation of CDK4 and CDK6, leading to significant suppression of RB1 phosphorylation and E2F1 expression (Fig. 4F). In the long term, MCF7 parental cell–derived xenografts with low CDK6 expression were sensitive to ribociclib, BSJ-05-017, and BSJ-03-096, whereas CDK6-overexpressing and shPTEN xenografts were durably inhibited by BSJ-03-096 (−68.9% and −54.9%) and BSJ-05-017 (−64.9% and −47.4%) while showing tumor outgrowth after initial response to ribociclib (Fig. 4G). These results reveal that in multiple models of CDK4/6i resistance, more potent and complete inhibition of the CDK4/6 kinases has substantial antitumor effects.
Discussion
In this report, we uncover how CDK6 evades kinase inhibition by current-generation ATP-competitive CDK4/6i through the cloaking of its ATP binding pocket by INK4 proteins. The results reveal a surprising mechanism of drug resistance to a broadly effective therapy by means of induction of an endogenous tumor suppressor and weak inhibitor of the very oncogenic kinase that the drug is targeting. Through a structural understanding of how this complex generates drug-insensitive signaling, we identify a strategy toward next-generation therapies that can overcome this CDK6–INK4 complex and exert more uniform CDK4/6 inhibition.
The finding of induction of CDK6 as a recurrent mechanism of resistance to CDK4/6i (8–10, 35) is surprising inasmuch as the drugs all potently bind and antagonize both recombinant CDK4 and CDK6. To resolve this conundrum, we investigated the protein complexes of CDK4 and CDK6 present in sensitive and resistant cells. Kinase pulldowns from CDK4/6i-sensitive models manifest high levels of intrinsic CDK4 activity that is CDK4/6i sensitive but express very little CDK6 (Fig. 1). By contrast, CDK4/6i-resistant models manifest high expression of CDK6, and this activity is CDK4/6i resistant. These results highlighted a critical difference between CDK4 and CDK6 present in resistant cells, which we resolved through study of the protein–protein interaction profiles of these kinases. We identified an abundant CDK6–INK4 complex in multiple CDK4/6i-resistant ER+ breast cancer cell lines, which was notable given the known binding site for INK4 at the entrance to the CDK4/6 ATP binding pocket (36–38). We established the significance of this interaction in mediating CDK4/6i resistance by blocking the interaction through mutation of CDK6 or knockdown of INK4, both of which eliminated drug resistance. Moreover, we reconstituted the complex using recombinant CDK6 and p18INK4C, demonstrating that addition of INK4 could almost completely prevent CDK4/6i from suppressing CDK6 activity. These data point to a CDK6–INK4 complex that harbors low but drug-insensitive kinase activity. Such a result is analogous to other forms of drug resistance, in which moving from complete to only partial kinase blockade is enough to allow tumor escape (3, 39).
To develop therapies capable of inhibiting this INK4–CDK6 complex, we generated structural models that could inform how INK4 was affecting drug and ATP binding. Taking advantage of a panel of INK4- or drug-bound CDK4 and CDK6 crystal structures, we were able to develop refined molecular simulations of how INK4 might alter ATP or CDK4/6i binding. The results readily explained the biochemical findings, revealing that INK4 proteins cause distortion of the N-lobe of CDK6 in a manner that disrupts the CDK4/6i binding pocket volume up to 80 times more than it does to the ATP pocket volume. Biophysical measurements by MST confirmed that p18INK4C indeed decreased CDK6 binding affinity to CDK4/6i. These findings suggest that new strategies toward inhibiting CDK6–INK4 might turn attention to locations other than the currently targeted drug binding pocket.
One approach to potentially enhance inhibition of refractory targets is through PROTAC molecules that induce target degradation (27, 34). This emerges due to (i) potentially different requirements for the affinity necessary for a drug to inhibit enzymatic activity compared with drug inducing E3 ligase recognition, (ii) vulnerabilities of certain proteins based on their half-lives to degradation compared with catalytic inhibition, and (iii) potential allosteric interactions between the larger PROTAC molecules with other portions of the target driving binding and efficacy. To test this approach for CDK6–INK4, we generated a small library of selective and potent CDK4/6 PROTACs and identified compounds that could specifically target CDK4, CDK6, or both. Using these compounds in our sensitive and resistant models, we demonstrated that degradation of CDK6 is feasible. Importantly, effective inhibition of both CDK4 and CDK6 through degradation led to potent tumor growth inhibition in CDK4/6i-resistant tumor models. Whether achieving such levels of degradation will be safe and tolerable in the clinic is unknown, as other bifunctional degraders (e.g., targeting androgen receptor) are only beginning to be assessed in early-phase trials; however, the data in murine models suggest that intraperitoneal and PO dosing can lead to effective circulating drug concentrations and target inhibition in xenografted tumors. Importantly, the findings convey that those alternative strategies that seek to overcome the INK4–CDK6 complex may represent a substantial therapeutic advance and even lead to efficacy in tumor types in which expression of CDK6 is intrinsically high. Notable in this regard is that many of the “acquired” genetic alterations that lead to CDK6 upregulation in ER+ breast cancer (Fig. 3) are present as early events in cancers such as lung, endometrial, or prostate cancer.
The efficacy of the compounds is particularly notable in providing the very first proof that “retargeting” CDK4/6 kinases can show efficacy even after resistance to first-generation CDK4/6i emerges. This may be of relevance for several mechanisms of resistance to first-generation CDK4/6i, as we find a variety of genetic alterations can lead to induction of CDK6, including FAT1 loss (8), PTEN loss (40–42), and ARID1A loss (43). Beyond these alterations, recent literature has pointed to resistance to CDK4/6i through genetic alterations in CCNE1 (44), RB1 (45), and FGFR1 (46). The extent to which these and other alterations (other than RB1) maintain a CDK4/6-dependent checkpoint will need to be investigated using such new-generation CDK4/6i as presented here. Taken together, our results imply that use of current-generation CDK4/6i should be informed by the presence or absence of CDK6–INK4 complexes and reveal the broader therapeutic potential for strategies that promote complete downregulation of both CDK4 and CDK6.
Methods
Cell Lines
MCF7, T47D, CAMA1, ZR-75-1, EFM19, and BT474 cell lines were obtained from the American Type Culture Collection. HEK293T was a gift from Ping Chi's lab. MCF7 cells were maintained in DMEM/F12 medium. T47D, ZR-75-1, EFM19, and BT474 cells were maintained in RPMI medium. CAMA1 cells were maintained in DMEM. All media were supplemented with 10% FBS, 2 mmol/L L-glutamine, 20 U/mL penicillin, and 20 μg/mL streptomycin. All cell lines were authenticated by short tandem repeat genotyping and tested negative for Mycoplasma contamination. All cell lines were cultured and collected within 10 passages.
Drugs and Reagents
Abemaciclib (LY2835219) and palbociclib (PD-0332991) were obtained from Selleck Chemicals and TargetMol. We obtained ribociclib (LEE011) from Novartis. These drugs were dissolved in dimethyl sulfoxide. Phospho-RB1 (Ser780; #8180), phospho-RB1 (Ser807/811; #8516), RB1 (#9309), cyclin D1 (#2978), CDK6 (#3136), CDK4 (#12790), CDK2 (#2546), E2F1 (#3742), cyclin A2 (#4656), cyclin E2 (#4132), YAP (#14074), TAZ (#4883), p18 (#2896), and β-actin (#4970) antibodies were purchased from Cell Signaling Technology. FAT1 (#ab190242) and p15INK4B (ab53034) antibodies were purchased from Abcam. Recombinant human CDK6–cyclin D3 (C35-10H) and CDK4–cyclin D3 (C31-18G) were purchased from SignalChem. RB1 protein (#ab56270) was purchased from Abcam. ADP-Glo Kinase Assay Kit (V6930) was purchased from Promega.
Coimmunoprecipitation
Coimmunoprecipitation–MS
Cell pellets were lysed in coimmunoprecipitation lysis buffer (Pierce; #87787) supplemented with 1× protease and phosphatase inhibitors (#78444; Pierce). After a 10-minute incubation on ice, lysates were centrifuged at maximum speed for 10 minutes at 4°C, and the supernatants were obtained for the measurement of protein concentration. Then, 1 mg of lysates was immunoprecipitated by incubating 1 μg CDK4 (#sc-23896; Santa Cruz Biotechnology) or CDK6 (#sc-177-G; Santa Cruz Biotechnology) antibody at 4°C overnight. Next, 20 μL of chromatin immunoprecipitation (ChIP)–grade magnetic beads (Thermo Fisher Scientific) was added into each IP tube and incubated for 2 hours. IP samples were washed three times with IP lysis buffer, resuspended in 2× LDS sample buffer (Invitrogen), and boiled for 5 minutes at 100°C before loading onto SDS-PAGE gels. The gel was stained with SimplyBlue safestain (Thermo Fisher Scientific) and used for MS. MS was conducted through the Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) platform. MS raw files were converted into MGF by Proteome Discover (Thermo Scientific) and processed using Mascot 2.4 (Matrix Science) by searching against the UniProt human database supplemented with common contaminant proteins. Mascot data were assembled by Scaffold and X!-Tandem software, and search criteria for identification were four minimum peptides and 1% FDR at the peptide and protein levels. Scaffold_4.8.3 was used to visualize and analyze the MS data. A protein threshold above 99% and peptide threshold above 95% were used to isolate proteins of interest. Gene Ontology analysis was performed using the Gene Ontology website (http://geneontology.org/).
IP–In Vitro Kinase Assay
For the IP-kinase assay, cells were lysed on ice for 10 minutes in kinase lysis buffer [20 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1 mmol/L Na2EDTA, 1 mmol/L EGTA, 1% Triton, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, 1 μg/mL leupeptin, from Cell Signaling Technology, #9803] supplemented with 1× protease and phosphatase inhibitors. Lysates were collected as described above. Then, 300 μg of cell lysates was incubated with 1 μg CDK4 (#sc-23896; Santa Cruz Biotechnology) or CDK6 (#sc-177-G; Santa Cruz Biotechnology) antibody at 4°C overnight. Next, 20 μL of ChIP-grade magnetic beads (Thermo Fisher Scientific) was added into each IP tube and incubated for 2 hours. IP samples were washed two times with kinase lysis buffer and two times with kinase reaction buffer [40 mmol/L Tris-HCl (pH 7.5), 20 mmol/L MgCl2, 0.1 mg/mL BSA, from SignalChem, #K03-09, with 50 μmol/L DTT added freshly]. Then, 100 μL of kinase reaction buffer with 0.5 μg of recombinant human RB1 protein and 100 μmol/L ATP was added into each tube. The kinase reaction system was incubated at 30°C for 30 minutes on a thermomixer. Next, 20 μL of reaction mixture (without beads) was mixed with 20 μL ADP-Glo reagent and incubated for 1 hour at room temperature. Then, 40 μL of kinase detection reagent was added and incubated for 40 minutes at room temperature. Samples were read on the Glomax luminometer (Promega), and kinase activities were calculated. The remaining reaction mixture (without beads) was denatured by LDS and DTT, and Western blotting was performed to detect phosphorylation of RB protein. The remaining proteins on beads were eluted by 2× LDS buffer, and Western blotting was used to confirm the kinase pulldown.
In Vitro Kinase Assay Using Recombinant Proteins
An in vitro kinase assay was performed in a final volume of 5 μL kinase buffer (#K03-09; SignalChem) supplemented with 50 μmol/L DTT, 100 μmol/L ATP, and 5 ng/μL RB1 recombinant protein. CDK4–cyclin D3 (#C31-18G; SignalChem) and CDK6–cyclin D3 (#C35-10G; SignalChem) were used as kinases. CDK4/6i and/or INK4 proteins were preincubated with the kinases for 10 minutes at room temperature before adding ATP and RB1 substrate. After a 1-hour incubation at room temperature, 5 μL ADP-Glo reagent was added to the kinase reaction mixture and incubated for 1 hour, followed by incubating with 10 μL kinase detection reagent for 40 minutes. The luminescence was detected on SpectraMax iD5 microplate reader.
Cloning and Plasmids
LentiCRISPRv2 or lenti-sgRNA backbone was used for generating knockout cell lines. LentiCRISPRv2 puro, lentiCRISPRv2 hygro, and lenti-sgRNA neo were gifts from Brett Stringer (plasmids #98290, #98291, and #104992; Addgene). Single-guide RNAs were designed through MIT CRISPR Designer (crispr.mit.edu), and the sequences are as follows: FAT1-CRISPR, CACGGTGACGTTGTACTCGG; CDKN2B (p15)-CRISPR, ACGGAGTCAACCGTTTCGGG and CTCCACTAGTCCCCGCGCCG; CDKN2C (p18)-CRISPR, GAATGACAGCGAAACCAGTT and TTAACATCGAGGATAATGAA; and PTEN-CRISPR, TCATCTGGATTATAGACCAG. Instructions for using the lentiCRISPRv2 plasmids are as described by the Zhang laboratory (https://media.addgene.org/cms/filer_public/53/09/53091cde-b1ee-47ee-97cf-9b3b05d290f2/lenticrisprv2-and-lentiguide-oligo-cloning-protocol.pdf). Oligos were annealed and ligated with BsmBI-digested lentiviral vector. Then, the ligation system was transformed into Stbl3 bacteria, and plasmids were extracted for sequencing.
pLKO-PTEN-shRNA-1320 and pLKO-PTEN-shRNA-3001 were gifts from Todd Waldman (plasmids #25638 and #25639; Addgene). We obtained them from Dr. Neal Rosen's lab. Other short hairpin RNA (shRNA) sequences used are as follows: Renilla-sh, TGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGA; ARID1A-sh, TGCTGTTGACAGTGAGCGCAAGCGAGACACAGCTATTTAATAGTGAAGCCACAGATGTATTAAATAGCTGTGTCTCGCTTTTGCCTACTGCCTCGGA; and YAP-sh, TGCTGTTGACAGTGAGCGCTAGGTTGATCACTCATAATAATAGTGAAGCCACAGATGTATTATTATGAGTGATCAACCTATTGCCTACTGCCTCGGA. Renilla, ARID1A, and YAP1 shRNAs were put into mir-E, an optimized microRNA backbone, as previously described (47). Briefly, hairpin ultramers were amplified and put into lentiviral SGEP or SGEN vectors, which were gifts from the Charles Sawyers lab. Proper insertions were verified by Sanger sequencing. ARID1A siRNA was purchased from Invitrogen (#4392420). pDONR223-CDK6 was cloned into MSCV-N-Flag-HA-IRES-PURO (a gift from William Hahn and David Root; #23688; Addgene) and pLenti PGK Neo DEST (w531-1; a gift from Eric Campeau and Paul Kaufman; plasmid #19067; Addgene) using the Gateway LR Clonase II Enzyme Mix (Invitrogen; ref. 9). Single-site mutagenesis was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (#200522; Agilent Technologies). Proper mutations were verified by Sanger sequencing.
Lentiviral and Retroviral Infection and Generation of Stable Cell Lines
HEK293T cells were transfected with 4.5 μg of lentiviral vector, 4.5 μg psPAX2/pCL-Ampho, and 1 μg pVSVG with 40 μL X-tremeGENE HP (Roche) according to the manufacturer's protocol. Conditioned medium containing recombinant lentivirus was collected 48 hours after transfection and filtered through nonpyrogenic filters with a pore size of 0.45 μmol/L (Merck Millipore). Samples of these supernatants were applied immediately to target cells together with Polybrene (Sigma-Aldrich) at a final concentration of 8 μg/mL, and supernatants were incubated with cells for 12 hours. After infection, cells were placed in fresh growth medium and cultured as usual. Selection with 2 μg/mL puromycin (Thermo Fisher Scientific), 1 mg/mL G418 (InvivoGen), or 200 μg/mL hygromycin (InvivoGen) was initiated 48 hours after infection.
Cell Viability Assay
Cell viability was measured by Resazurin (R&D Systems) as described previously (48). Briefly, 1,500 cells were seeded in a 96-well plate and allowed to recover overnight. Cells were treated with drugs at day 0. Resazurin was added to the cells 4 hours prior to the measurements on day 3, day 5, and day 7. Fluorescent intensity was measured using a microplate reader (SpectraMax M5; Molecular Devices). IC50 was calculated by GraphPad Prism 7.0 (GraphPad Software) using a sigmoidal regression model.
Western Blotting
Cell lysates were collected in RIPA buffer (Thermo Fisher Scientific) supplemented with protease and phosphatase inhibitors (Pierce). Protein concentration was quantified by using the BCA kit (Fisher Scientific). Then, 60 to 100 μg of protein lysates were loaded onto 4% to 12% SDS-PAGE gels (Invitrogen) for electrophoresis and transferred onto nitrocellular membranes. Blots were blocked with Intercept (TBS) Blocking Buffer (#927-60001; LI-COR Bioscience) and incubated with primary antibody at 4°C overnight. Secondary antibodies conjugated with fluorescence (#926-68071 and #926-32210; LI-COR Bioscience) were incubated for 1 hour at room temperature, and blots were scanned by Odyssey Clx Imaging System from LI-COR Bioscience.
IHC
IHC was performed on formalin-fixed, paraffin-embedded tumor specimens from patient-derived xenografts provided by Dr. Violeta Serra from VHIO in Barcelona, Spain. A standard multimer/diaminobenzidine (DAB) detection protocol was performed on a Ventana BenchMark ULTRA Automated stainer, as previously described (8), with appropriate negative and positive controls. Then, 2 μg/mL FAT1 (#ab190242; Abcam), 1 μg/mL YAP (#14074; Cell Signaling Technology), 1 μg/mL CDK6 (#HPA002637; Sigma-Aldrich), 1 μg/mL p15 (#MAB6798; R&D Systems) and 1 μg/mL p18 (#2896; Cell Signaling Technology) antibodies were used. Images were taken under a Leica DMi8 microscope and evaluated by a pathologist at Memorial Sloan Kettering Cancer Center (MSKCC). Quantification of the staining was based on the percentage of positive staining and staining intensity at the indicated location. The immunoreactive scores were recorded as previously described (49, 50).
Computational Structural Analysis
INK4–CDK6 Interface Analysis
We superposed three crystallographic structures in the PDB database of CDK6–INK4 (PDB IDs: 1BI7, 1BI8, 1G3N; refs. 19, 20) using UCSF-Chimera v1.14 (51), and CDK6 residues in proximity of INK4 (≤2.7 Å) were selected (listed in Fig. 2A) as candidates for mutagenesis experiments.
Quantification of the Change in CDK6 Binding Pocket Volume upon INK4 Binding
Besides the existing INK4-bound CDK6 structures as listed in the table in Fig. 2A, we also selected PDB ID 2EUF (22) as the structure that cognately binds to palbociclib without INK4 bound. Structural superposition was performed using UCSF-Chimera v1.14. For each structure, we docked AMP-PNP and palbociclib into the CDK6 binding pocket using the flexible docking protocol in the software DOCK6.9 (52). We also used DOCK6.9 to generate spheres (i.e., probes along the protein surface) and selected those that were occupied by the predicted binding pose of the ligand to approximate the volume of the binding pocket. We elected to use the docked pose for AMP-PNP and the superposed pose for palbociclib due to incomplete pose generation in the distorted binding pocket. Input and raw docking results can be found at the following GitHub repository: https://github.com/choderalab/CDK_PROTAC.
Manual Construction of the Structural Models of the Ternary Complex
We used various existing structures (WT, human proteins) from the PDB to construct the complex. The catalytic domains of CDK6 and CDK4 were from PDB IDs 1G3N and 3G33, respectively. CRBN was from PDB ID 5FQD, and VHL was from PDB ID 5T35. Cyclin D1 was from PDB ID 6P8E. For each PROTAC degrader, first, the two warheads were docked to the binding pocket of the E3 ligase adapter and CDK4 (superposed for CDK6 due to the distorted binding pocket). Each palbociclib posed in the CDK binding pockets was then relaxed with a short (20 ns) molecular dynamics simulation (at 310.15 K, 1.0 atm, 4 fs time steps with heavy hydrogen masses, to further open the pocket to increase compatibility with the degrader linker) using OpenMM package v7.4.2 (53). Then the docked poses for the two warheads were superimposed to common rotatable bonds in an extended pose of the degrader linker using UCSF-Chimera v1.14. Once clashes in the protein targets were eliminated by manual rotation and reorientation of side chains, the two warheads and the linker were manually bonded.
Molecular Dynamics Simulations and Postanalysis
The manually constructed model structures were solvated in TIP3P water (54) and neutralized with an amount of NaCl equivalent to the ionic strength of 20 mmol/L MgCl2 to match the experimental condition. The small-molecule ligands were parameterized using the GAFF force field (55) and the AM1-BCC charging method (56) implemented in the software package Antechamber (57). Molecular dynamics simulations were run using the Amber14SB force field (58) through the OpenMM package v7.4.2 (53). Short equilibration (5 ns) was performed before the production run (ended up with ∼300 ns) using the Langevin integrator at 400.15 K and 1.0 atm with a time step of 4 fs (using heavy hydrogens with a mass of 4 atomic mass units). The arbitrarily high temperature (127°C) was used for the simulations to ensure that the complexes were not trapped in initial conformations and were able to reach reasonable equilibration. Trajectories from the simulations were postanalyzed (imaged on one of the protein components and converted to the pdb format) using MDTraj v1.9.4 (59) and visualized using the software package PyMOL v2.2.0. Hydrogen bonds in each trajectory were identified using the Baker–Hubbard criterion in MDTraj, and the union of the three sets of the most frequently observed hydrogen bonds was identified. Coordinates for the equilibrated structures of the four ternary complex models can be found at the following GitHub repository: https://github.com/choderalab/CDK_PROTAC.
MST Assay
MST assay was done by Reaction Biology Corp. Briefly, protein CDK6 was labeled using the Protein Labeling Kit RED-NHS (NanoTemper Technologies). The labeling reaction was performed according to the manufacturer's instructions in the supplied labeling buffer applying a concentration of 15 μmol/L protein (molar dye/protein ratio ≈3:1) at room temperature for 30 minutes. Unreacted dye was removed with the supplied dye removal column equilibrated with storage buffer [50 mmol/L Hepes, pH 7.5; 500 mmol/L NaCl; 10% glycerol; 0.25 mmol/L Tris (2-carboxy-ethyl)-phosphin-HCl; and 0.01% Tween 20]. The degree of labeling was determined using UV-visible spectrophotometry at 650 and 280 nm. A degree of labeling of 0.6 was achieved. The labeled protein CDK6 was adjusted to 12 nmol/L with assay buffer (20 mmol/L K phosphate, pH 8.0; 50 mmol/L NaCl; and 0.05% Pluronic). Then, 250 nmol/L p18 was preincubated with CDK6 for 15 minutes prior to the addition of ligand. The ligand abemaciclib was dissolved in assay buffer, and a series of sixteen 1:1 dilutions was prepared using the same buffer, producing ligand concentrations ranging from 122 pmol/L to 4 μmol/L. Each ligand dilution was mixed with one volume of labeled protein, resulting in a final labeled CDK6 concentration of 6 nmol/L and final ligand concentrations ranging from 61 pmol/L to 2 nmol/L. After a 20-minute incubation, the samples were loaded into standard Monolith NT.115 Capillaries (NanoTemper Technologies). MST was measured using a Monolith NT.115 instrument (NanoTemper Technologies) at an ambient temperature of 25°C. Instrument parameters were adjusted to 10% LED power and medium MST power. Data of three independently pipetted measurements were analyzed (MO.Affinity Analysis software version 2.1.3; NanoTemper Technologies) using the signal from an MST-on time of 5 seconds. The data were expressed as baseline-corrected normalized fluorescence, ΔFnorm [‰]. To obtain ΔFnorm, the baseline Fnorm value is subtracted from all data points of the same curve. (The baseline Fnorm value is equivalent to the mean Fnorm value of the unbound target, usually in capillaries 14–16, and is given by the MO.Affinity Analysis software as the “unbound” value when a fit is performed.)
Synthesis of PROTAC Degraders and Palbociclib–Biotin
The synthesis of the degrader library is described in detail in the Supplementary Materials. For the synthesis of biotin-labeled palbociclib, a solution of tert-butyl (2-(2-(2-(2-(4-(6-((6-acetyl-8-cyclopentyl-5-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl)amino)pyridin-3-yl)piperazin-1-yl)ethoxy)ethoxy)ethoxy)ethyl)carbamate (3, 30.5 mg, 0.0422 mmol) in dichloromethane (DCM; 1 mL) was added to trifluoroacetic acid (1 mL), and the resulting solution was stirred for 0.5 hours at room temperature. The mixture was concentrated to give the primary amine intermediate, which was directly dissolved into 1.0 mL dimethylformamide (DMF), followed by addition of biotin (10.3 mg, 0.0422 mmol), Hexafluorophosphate Azabenzotriazole Tetramethyl Uronium (HATU; 24 mg, 0.063 mmol), and N,N-diisopropylethylamine (DIPEA; 0.058 mL, 0.269 mmol). The resulting mixture was stirred at room temperature for 0.5 hours, then purified by reverse-phase HPLC (5%–95% MeOH in H2O) to give BSJ-03-163 as a yellow solid (26.1 mg, 81%). LC/MS indicated m/z 849 [M+1]. All the chemical synthesis information is provided in Supplementary Data.
Flow Cytometry: Senescence Green and Cell Cycle
Senescence Analysis
Cells were treated with DMSO, abemaciclib (100 nmol/L), palbociclib (500 nmol/L), and BSJ-05-017 (500 nmol/L) for 8 days. Cells were harvested using trypsin/EDTA and then stained with the CellEvent Senescence Green Flow Cytometry Assay Kit (Invitrogen) according to the manufacturer's instructions. Briefly, cells were fixed with 2% paraformaldehyde for 15 minutes at room temperature and stained with the CellEvent Senescence Green Probe for 2 hours at 37°C without CO2. After incubation, cells were washed with PBS three times and resuspended in FACS buffer (PBS with 2% FBS) for analysis on BD Biosciences LSR Fortessa using a 488-nm laser and 530-nm/30 filter (BD Biosciences). Data analysis was performed with FCS Express 7 (De Novo Software).
Cell-cycle Analysis
Cells were treated with DMSO, abemaciclib (100 nmol/L), palbociclib (500 nmol/L), and BSJ-05-017 (500 nmol/L) for 24 hours. Cells were detached from the cell culture dish with trypsin/EDTA, and then washed with PBS and fixed in 70% ice-cold EtOH overnight. Prior to staining, EtOH was removed, and cells were washed twice with FACS buffer. Cells were then resuspended in staining buffer containing 1,000 μL FACS buffer with 2 μg/mL propidium iodide (Invitrogen) and 100 μg/mL RNase A (Invitrogen). Cell-cycle profiles were measured with BD Biosciences LSR Fortessa and analyzed with FCS Express 7.
Proteomics
Molt4 cells were treated with 250 nmol/L of either of the compounds BSJ-05-017 or BSJ-03-096 (singlicate) or DMSO control (biological triplicate) for 5 hours. Cells were harvested by centrifugation and prepared for MS as described previously (28). Data were collected as reported (28). LC/MS data were analyzed using Proteome Discoverer 2.4 (Thermo Fisher Scientific) as previously described (28). Reporter ion intensities were normalized and scaled using in-house scripts in the R framework (https://www.R-project.org/). Statistical analysis was carried out using the limma package within the R framework (60).
In Vivo Studies
PK Study
We performed a PK study in the Drug Metabolism and Pharmacokinetics (DMPK) Core facility at Scripps Florida (https://www.scripps.edu/science-and-medicine/cores-and-services/dmpk-core/index.html). C57BL/6 male mice were dosed with BSJ-05-017 solution formulation (i.p., 5/95 DMSO/10% captisol, dose 25 mg/kg) and BSJ-03-096 solution formulation (PO, 5/95 DMSO/10%, dose 10 mg/kg). Blood samples were collected at 0.08, 0.25, 0.5, 1, 2, 4, 6, 8, and 24 hours. The blood samples were collected from sets of three mice at each time point in microcentrifuge tubes containing K2EDTA as an anticoagulant. Plasma samples were separated by centrifugation and stored below −70°C until bioanalysis. All samples were processed for analysis by precipitation using acetonitrile and analyzed with a partially validated LC/MS/MS method [lower limit of quantification (LLOQ), –1.22 ng/mL for i.v. and PO; LLOQ, –5.02 ng/mL for i.p.]. PK parameters were calculated using the noncompartmental analysis tool of WinNonlin Enterprise software (version 6.3). All procedures were approved by the Scripps Florida Institutional Animal Care and Use Committee (IACUC), and the Scripps Vivarium is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Pharmacodynamic Study
NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (NSG) mice were obtained from The Jackson Laboratory (stock no. 005557). Each mouse was injected with FAT1-loss cells subcutaneously 1 week after the implantation of estradiol pellets (25 mg). After the tumors reached 200 mm3, mice were treated for 3 consecutive days with BSJ-05-017 at 25 mg/kg. Tumors were collected at 6 hours. Lysates were prepared by homogenization in SDS-lysis buffer (∼1 mL/mg tissue; 50 mmol/L Tris-HCl, pH 7.4; 2% SDS) and boiled for 10 minutes, followed by brief sonication as described previously (61). Lysates were cleared by centrifugation at 14,000 × g for 10 minutes, and the supernatant was collected for Western blotting.
Efficacy Study
NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (NSG) mice were obtained from The Jackson Laboratory (stock no. 005557). Each mouse was injected with MCF7 parental, CDK6-ovexpressing, or PTEN loss cells subcutaneously 1 week after the implantation of estradiol pellets (25 mg). After the tumors reached 150 to 200 mm3, mice were treated at a 5 days on/2 days off schedule for 25 to 35 days with ribociclib at 25 mg/kg (orally), BSJ-05-017 at 50 mg/kg (i.v.), and BSJ-03-096 at 50 mg/kg (orally). Tumor volumes were recorded every 3 to 4 days. Mice were sacrificed if tumors reached 1,000 mm3 or at the end of the experiment. Tumors were collected and processed as described above.
The pharmacodynamic and efficacy studies were approved by the IACUC review board at MSKCC.
Human Participants
A total of 1,366 metastatic tumors from 1,115 patients with HR+/HER2− metastatic breast cancer who underwent prospective clinical genomic profiling between April 2014 and March 2020 were analyzed. This study was approved by the MSKCC Institutional Review Board, and all patients provided written informed consent for tumor sequencing and review of patient medical records for detailed demographic, pathologic, and treatment information (NCT01775072). Detailed clinicopathologic data were obtained for each sample.
Prospective Sequencing and Analysis
For all 1,366 tumors, matched tumor and normal DNA samples were extracted from either representative formalin-fixed, paraffin-embedded tumor biopsy samples or mononuclear cells from peripheral blood, respectively. All specimens underwent massively parallel next-generation sequencing in a laboratory certified by Clinical Laboratory Improvement Amendments using MSK-IMPACT, an FDA-authorized hybridization capture-based next-generation sequencing assay, which analyzes all protein-coding exons and selected intronic and regulatory regions of 341 to 468 cancer-associated genes, all as previously described (62–64). Somatic mutations, DNA copy-number alterations, and structural rearrangements were identified as previously described (63), and all mutations were manually reviewed.
PRISM Cell Line Screening
Cell Lines
The current PRISM cell set consists of 931 cell lines representing more than 45 lineages, including both adherent and suspension/hematopoietic cell lines. These cell lines largely overlap with and reflect the diversity of the Cancer Cell Line Encyclopedia (CCLE) cell lines (see https://portals.broadinstitute.org/ccle). Cell lines were grown in RPMI 10% FBS without phenol red for adherent lines and RPMI 20% FBS without phenol red for suspension lines. Parental cell lines were stably infected with a unique 24-nucleotide DNA barcode via lentiviral transduction and blasticidin selection. After selection, barcoded cell lines were expanded and QCed (Mycoplasma contamination test, an SNP test for confirming cell line identity, and barcode ID confirmation). Passing barcoded lines were then pooled (20–25 cell lines per pool) based on doubling time and frozen in assay-ready vials.
PRISM Screening
Test compounds were added to 384-well plates and run at 8-point dose with threefold dilutions in triplicate. These assay-ready plates were then seeded with the thawed cell line pools. Adherent cell pools were plated at 1,250 cells per well, whereas suspension and mixed adherent/suspension pools were plated at 2,000 cells per well. Treated cells were incubated for 5 days and then lysed. Lysate plates were collapsed together prior to barcode amplification and detection.
Barcode Amplification and Detection
Each cell line's unique barcode was located at the end of the blasticidin resistance gene and expressed as mRNA. These mRNAs were then captured by using magnetic particles that recognized polyA sequences. mRNA was then reverse-transcribed into cDNA, and then the sequence containing the unique PRISM barcode was amplified using PCR. Finally, Luminex beads that recognized the specific barcode sequences in the cell set were hybridized to the PCR products and then detected using a Luminex scanner, which reported the signal as a median fluorescent intensity.
Biomarker Identification
After data processing, we explored the univariate associations between the PRISM sensitivity profiles and the genomic features or genetic dependencies. In particular, we computed the Pearson correlations and associated P values. Correlations and P values for log-viability values at each dose, AUC scores, and logIC50 values were tabulated. For each data set, the q values were computed from P values using the Benjamini–Hochberg algorithm. Associations with q values above 0.1 were filtered out, and q values below 1e-20 were plotted at 1e-20 for plot readability. Univariate models were run on available feature sets, including CCLE genomic characterization data such as gene expression, cell lineage, mutation, copy number, metabolomics, and proteomics, as well as loss-of-function genetic perturbation (both RNAi and CRISPR) data from the Dependency Map. In addition to these data sets, viability data from the PRISM drug repurposing project were used as a feature set for univariate analysis. For discrete data, such as mutation and lineage, a t test was done to determine differential sensitivities. For continuous data, such as gene expression, correlations between sensitivity and the characteristic of interest were calculated to determine any association.
Data Availability Statement
The computational structural information can be found in the following GitHub repository: https://github.com/choderalab/CDK_PROTAC. The genomic sequencing data have been deposited in the following repository: https://cbioportal.mskcc.org/study/summary?id=breast_ink4_msk_2021.
Authors' Disclosures
Q. Li reports a patent for CDK4/6 degraders pending. B. Jiang reports a patent for CDK4/6 degraders pending. Z. Li reports other support from SEMA4 outside the submitted work. P. Razavi reports grants from Grail, Illumina, Invitae/Archer Dx; grants and personal fees from Novartis and Epic Sciences; and personal fees from Foundation Medicine, Inivata, Tempus Labs, Natera, and AstraZeneca outside the submitted work. K.A. Donovan reports grants from NIH during the conduct of the study, as well as personal fees from Kronos Bio outside the submitted work. J.S. Reis-Filho reports personal fees from Paige, Repare Therapeutics, Goldman Sachs, Grupo Oncoclinicas, Roche Tissue Diagnostics, Genentech, Roche, In Vicro, and Eli Lilly outside the submitted work. E.S. Fischer reports personal fees and other support from Jengu Therapeutics, Neomorph, Inc., and Civetta Therapeutics; other support from C4 Therapeutics; grants from Novartis and Ajax; and personal fees from EcoR1 Capital and Sanofi outside the submitted work. N. Rosen reports personal fees and other support from Beigene, MAPCure, and Zai Lab; other support from Kura and Effector; personal fees from Tarveda, Ribon, and Concarlo; grants, personal fees, and other support from AstraZeneca, Boehringer, and Pfizer/Array; and grants from Revolution Medicines during the conduct of the study. V. Serra reports grants from Susan G. Komen Foundation (CCR15330331) and Instituto de Salud Carlos III (CPII19/00033) during the conduct of the study, as well as grants from Novartis, Genentech, and from AstraZeneca outside the submitted work. A. Koff reports grants from NCI during the conduct of the study, as well as other support from Atropos Therapeutics and Eli Lilly and Company outside the submitted work. J.D. Chodera reports grants from National Institutes of Health during the conduct of the study, as well as grants from National Science Foundation, Parker Institute for Cancer Immunotherapy, Relay Therapeutics, Silicon Therapeutics, EMD Serono (Merck KGaA), AstraZeneca, Vir Biotechnology, Bayer, XtaPi, Molecular Sciences Software Institute, Starr Cancer Consortium, Open Force Field Consortium, and Cycle for Survival, other support from Charité – Universitätsmedizin, University of Edinburgh, Freie Universität Berlin, and Gesellschaft Deutscher Chemiker, personal fees and other support from Boehringer Ingelheim and University of South Florida, and personal fees from Foresite Labs, OpenEye Scientific, Interline Therapeutics, Blueprint Medicines, Celgene, and Astex outside the submitted work. N.S. Gray reports grants from Deerfield during the conduct of the study; personal fees from C4 Therapeutics, Allorion, Larkspur, Inception, Syros, EoCys, Jengu, and Voronoi outside the submitted work; a patent for CDK4/6 degraders pending; and is on the scientific advisory board and is an equity holder in C4 therapeutics, which works on small-molecule protein degraders, which is related to the work in this publication. S. Chandarlapaty reports grants from NIH/NCI, Breast Cancer Research Foundation (BCRF), Cancer Couch Foundation, and The Shen Family Fund during the conduct of the study; personal fees from Sanofi, Novartis, Lilly, Paige.ai, Context Therapeutics, Bristol Myers Squibb, and Sermonix and grants from Daiichi Sankyo and Paige.ai outside the submitted work; and a patent for CDK4/6 degraders pending. No disclosures were reported by the other authors.
Authors' Contributions
Q. Li: Conceptualization, resources, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. B. Jiang: Conceptualization, resources, data curation, formal analysis, investigation. J. Guo: Conceptualization, resources, data curation, software, investigation. H. Shao: Data curation, validation. I.S. Del Priore: Data curation. Q. Chang: Validation, investigation. R. Kudo: Data curation, investigation. Z. Li: Data curation, investigation. P. Razavi: Data curation, software. B. Liu: Data curation, methodology. A.S. Boghossian: Data curation, methodology. M.G. Rees: Data curation. M.M. Ronan: Data curation. J.A. Roth: Data curation, supervision, investigation, writing–review and editing. K.A. Donovan: Conceptualization, supervision, investigation, writing–review and editing. M. Palafox: Data curation, coauthor. J.S. Reis-Filho: Conceptualization, investigation, writing–review and editing. E. de Stanchina: Data curation, software, validation, investigation, writing–review and editing. E.S. Fischer: Data curation, software, supervision, validation, writing–review and editing. N. Rosen: Resources, supervision, validation, writing–review and editing. V. Serra: Resources, supervision, validation, methodology, writing–review and editing. A. Koff: Software, supervision, methodology, writing–review and editing. J.D. Chodera: Conceptualization, resources, software, supervision, writing–review and editing. N.S. Gray: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Chandarlapaty: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
Supplementary Material
Acknowledgments
The Chandarlapaty lab has received generous funding support for this research from the Cancer Couch Foundation, the Shen Family Fund, the Smith Fund for Cancer Research, the Breast Cancer Research Foundation, an NIH Cancer Center Support Grant (P30 CA008748), and NIH R01234361. Q. Li has received support from Translational Research Oncology Training Fellowship (MSKCC) made possible by the generous contribution of First Eagle Investment Management. V. Serra reports grants from the Susan G. Komen Foundation (CCR15330331) and Instituto de Salud Carlos III (CPII19/00033) during the conduct of the study and grants from Novartis, Genentech, and AstraZeneca outside the submitted work. The Chodera laboratory receives or has received funding from multiple sources, including the NIH and an NIH Cancer Center Support Grant (P30 CA008748), the National Science Foundation, the Parker Institute for Cancer Immunotherapy, Relay Therapeutics, Entasis Therapeutics, Silicon Therapeutics, EMD Serono (Merck KGaA), AstraZeneca, Vir Biotechnology, Bayer, XtalPi, Foresite Laboratories, the Molecular Sciences Software Institute, the Starr Cancer Consortium, the Open Force Field Consortium, Cycle for Survival, a Louis V. Gerstner Young Investigator Award, and the Sloan Kettering Institute. J. Guo acknowledges support from NIH grant R01 GM121505. J.D. Chodera acknowledges support from NIH grant P30 CA008748, NIH grant R01 GM121505, and NIH grant R01 GM132386. A complete funding history for the Chodera lab can be found at http://choderalab.org/funding, including complete funding information and grant numbers. The authors thank Dr. Marie Will and Madeline Dorso for helpful comments on the manuscript and Dr. Zhan Yao for helpful advice on the kinase assays.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
References
- 1. Lovly CM, Shaw AT. Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clin Cancer Res 2014;20:2249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat Rev Drug Discovery 2004;3:1001–10. [DOI] [PubMed] [Google Scholar]
- 3. Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KKet al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mian AA, Schüll M, Zhao Z, Oancea C, Hundertmark A, Beissert Tet al. The gatekeeper mutation T315I confers resistance against small molecules by increasing or restoring the ABL-kinase activity accompanied by aberrant transphosphorylation of endogenous BCR, even in loss-of-function mutants of BCR/ABL. Leukemia 2009;23:1614–21. [DOI] [PubMed] [Google Scholar]
- 5. Wander SA, Cohen O, Gong X, Johnson GN, Buendia-Buendia JE, Lloyd MRet al. The genomic landscape of intrinsic and acquired resistance to cyclin-dependent kinase 4/6 inhibitors in patients with hormone receptor-positive metastatic breast cancer. Cancer Discov 2020;10:1174–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamilton E, Infante JR. Targeting CDK4/6 in patients with cancer. Cancer Treat Rev 2016;45:129–38. [DOI] [PubMed] [Google Scholar]
- 7. Condorelli R, Spring L, O'Shaughnessy J, Lacroix L, Bailleux C, Scott Vet al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol 2018;29:640–5. [DOI] [PubMed] [Google Scholar]
- 8. Li Z, Razavi P, Li Q, Toy W, Liu B, Ping Cet al. Loss of the FAT1 tumor suppressor promotes resistance to CDK4/6 inhibitors via the hippo pathway. Cancer Cell 2018;34:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti Met al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 2017;36:2255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cornell L, Wander SA, Visal T, Wagle N, Shapiro GI. MicroRNA-mediated suppression of the TGF-β pathway confers transmissible and reversible CDK4/6 inhibitor resistance. Cell Rep 2019;26:2667–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol 2016;13:417–30. [DOI] [PubMed] [Google Scholar]
- 12. Guiley KZ, Stevenson JW, Lou K, Barkovich KJ, Kumarasamy V, Wijeratne TUet al. p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science 2019;366:eaaw2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev 1993;7:812–21. [DOI] [PubMed] [Google Scholar]
- 14. Blain SW, Montalvo E, Massagué J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem 1997;272:25863–72. [DOI] [PubMed] [Google Scholar]
- 15. LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HSet al. New functional activities for the p21 family of CDK inhibitors. Genes Dev 1997;11:847–62. [DOI] [PubMed] [Google Scholar]
- 16. Parry D, Mahony D, Wills K, Lees E. Cyclin D-CDK subunit arrangement is dependent on the availability of competing INK4 and p21 class inhibitors. Mol Cell Biol 1999;19:1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grimmler M, Wang Y, Mund T, Cilenšek Z, Keidel E-M, Waddell MBet al. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 2007;128:269–80. [DOI] [PubMed] [Google Scholar]
- 18. Swarbrick A, Lee CS, Sutherland RL, Musgrove EA. Cooperation of p27(Kip1) and p18(INK4c) in progestin-mediated cell cycle arrest in T-47D breast cancer cells. Mol Cell Biol 2000;20:2581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes Dev 2000;14:3115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 1998;395:237–43. [DOI] [PubMed] [Google Scholar]
- 21. Chen P, Lee NV, Hu W, Xu M, Ferre RA, Lam Het al. Spectrum and degree of CDK drug interactions predicts clinical performance. Mol Cancer Ther 2016;15:2273–81. [DOI] [PubMed] [Google Scholar]
- 22. Lu H, Schulze-Gahmen U. Toward understanding the structural basis of cyclin-dependent kinase 6 specific inhibition. J Med Chem 2006;49:3826–31. [DOI] [PubMed] [Google Scholar]
- 23. Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan Net al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 2012;14:1322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang L, Azzolin L, Di Biagio D, Zanconato F, Battilana G, Lucon Xiccato Ret al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature 2018;563:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pettersson M, Crews CM. Proteolysis targeting chimeras (PROTACs)—past, present and future. Drug Discov Today Technol 2019;31:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Konstantinidou M, Li J, Zhang B, Wang Z, Shaabani S, Ter Brake Fet al. PROTACs—a game-changing technology. Expert Opin Drug Discov 2019;14:1255–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brand M, Jiang B, Bauer S, Donovan KA, Liang Y, Wang ESet al. Homolog-selective degradation as a strategy to probe the function of CDK6 in AML. Cell Chem Biol 2019;26:300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang B, Wang ES, Donovan KA, Liang Y, Fischer ES, Zhang Tet al. Development of dual and selective degraders of cyclin-dependent kinases 4 and 6. Angew Chem Int Ed Engl 2019;58:6321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khan S, Zhang X, Lv D, Zhang Q, He Y, Zhang Pet al. A selective BCL-X(L) PROTAC degrader achieves safe and potent antitumor activity. Nat Med 2019;25:1938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petzold G, Fischer ES, Thomä NH. Structural basis of lenalidomide-induced CK1α degradation by the CRL4CRBN ubiquitin ligase. Nature 2016;532:127–30. [DOI] [PubMed] [Google Scholar]
- 31. Matyskiela ME, Lu G, Ito T, Pagarigan B, Lu C-C, Miller Ket al. A novel cereblon modulator recruits GSPT1 to the CRL4CRBN ubiquitin ligase. Nature 2016;535:252–7. [DOI] [PubMed] [Google Scholar]
- 32. Guo WH, Qi X, Yu X, Liu Y, Chung CI, Bai Fet al. Enhancing intracellular accumulation and target engagement of PROTACs with reversible covalent chemistry. Nat Commun 2020;11:4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa Set al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol 2018;25:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martinez-Chavez A, van Hoppe S, Rosing H, Lebre MC, Tibben M, Beijnen JHet al. P-glycoprotein limits ribociclib brain exposure and CYP3A4 restricts its oral bioavailability. Mol Pharm 2019;16:3842–52. [DOI] [PubMed] [Google Scholar]
- 35. Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RGet al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 2013;24:167–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adachi M, Roussel MF, Havenith K, Sherr CJ. Features of macrophage differentiation induced by p19INK4d, a specific inhibitor of cyclin D-dependent kinases. Blood 1997;90:126–37. [PubMed] [Google Scholar]
- 37. Hirai H, Roussel MF, Kato JY, Ashmun RA, Sherr CJ. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol 1995;15:2672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reynisdottir I, Massague J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev 1997;11:492–503. [DOI] [PubMed] [Google Scholar]
- 39. Yao Z, Gao Y, Su W, Yaeger R, Tao J, Na Net al. RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat Med 2019;25:284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Costa C, Wang Y, Ly A, Hosono Y, Murchie E, Walmsley CSet al. PTEN loss mediates clinical cross-resistance to CDK4/6 and PI3Kalpha inhibitors in breast cancer. Cancer Discov 2020;10:72–85. [DOI] [PubMed] [Google Scholar]
- 41. Gary JM, Simmons JK, Xu J, Zhang S, Peat TJ, Watson Net al. Hypomorphic mTOR downregulates CDK6 and delays thymic Pre-T LBL tumorigenesis. Mol Cancer Ther 2020;19:2221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu J, Duan Z, Guo W, Zeng L, Wu Y, Chen Yet al. Targeting the BRD4/FOXO3a/CDK6 axis sensitizes AKT inhibition in luminal breast cancer. Nat Commun 2018;9:5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kato S, Schwaederle M, Daniels GA, Piccioni D, Kesari S, Bazhenova Let al. Cyclin-dependent kinase pathway aberrations in diverse malignancies: clinical and molecular characteristics. Cell Cycle 2015;14:1252–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor-Harding B, Aspuria PJ, Agadjanian H, Cheon DJ, Mizuno T, Greenberg Det al. Cyclin E1 and RTK/RAS signaling drive CDK inhibitor resistance via activation of E2F and ETS. Oncotarget 2015;6:696–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet Eet al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004;3:1427–38. [PubMed] [Google Scholar]
- 46. Formisano L, Lu Y, Servetto A, Hanker AB, Jansen VM, Bauer JAet al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun 2019;10:1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth Met al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 2013;5:1704–13. [DOI] [PubMed] [Google Scholar]
- 48. Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AMet al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov 2017;7:277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Q, Wang H, Zogopoulos G, Shao Q, Dong K, Lv Fet al. Reg proteins promote acinar-to-ductal metaplasia and act as novel diagnostic and prognostic markers in pancreatic ductal adenocarcinoma. Oncotarget 2016;7:77838–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaemmerer D, Peter L, Lupp A, Schulz S, Sänger J, Baum RPet al. Comparing of IRS and Her2 as immunohistochemical scoring schemes in gastroenteropancreatic neuroendocrine tumors. Int J Clin Exp Pathol 2012;5:187–94. [PMC free article] [PubMed] [Google Scholar]
- 51. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng ECet al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
- 52. Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PTet al. DOCK 6: impact of new features and current docking performance. J Comput Chem 2015;36:1132–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eastman P, Swails J, Chodera JD, McGibbon RT, Zhao Y, Beauchamp KAet al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol 2017;13:e1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys 1983;79:926–35. [Google Scholar]
- 55. Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem 2004;25:1157–74. [DOI] [PubMed] [Google Scholar]
- 56. Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 2002;23:1623–41. [DOI] [PubMed] [Google Scholar]
- 57. Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 2006;25:247–60. [DOI] [PubMed] [Google Scholar]
- 58. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 2015;11:3696–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernández CXet al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys J 2015;109:1528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi Wet al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra Vet al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2011;19:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HRet al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed Aet al. Memorial Sloan Kettering–integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan Net al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 2018;34:427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig Het al. The protein data bank. Nucleic Acids Res 2000;28:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The computational structural information can be found in the following GitHub repository: https://github.com/choderalab/CDK_PROTAC. The genomic sequencing data have been deposited in the following repository: https://cbioportal.mskcc.org/study/summary?id=breast_ink4_msk_2021.



![Figure 1. INK4–CDK6 complex promotes resistance to CDK4/6i in cells. A, Schematic for analysis of CDK4 and CDK6 interactions and activity via coimmunoprecipitation (co-IP) followed by ADP-Glo kinase assays and mass spectrometry. B, ADP-Glo kinase assay showing immunoprecipitated CDK4 and CDK6 (IP-CDK4 and IP-CDK6) kinase activity from MCF7 parental and CDK6-high cells [cells with FAT1 CRISPR knockout (CR) that have high CDK6 expression, previously shown to have resistance to CDK4/6i; ref. 8], with or without 100 nmol/L abemaciclib treatment. Data are shown as mean + SD of three biologically independent samples. P values were determined by unpaired two-sided Student t test. RLU, relative luminescence units. C, Venn diagram showing the number of unique proteins identified by mass spectrometry coimmunoprecipitated from IP-CDK4 and IP-CDK6 in FAT1-loss cells. Percentages were calculated by number of proteins identified in each subgroup divided by total proteins identified by IP of either CDK4 or CDK6. Data are shown as means of three replicates. D, Pathway analysis by Gene Ontology of proteins interacting with CDK6 but not CDK4 in the FAT1-loss cells. The proteins were grouped by their putative biological functions. E, Unique peptide counts of cyclin-dependent kinases and their endogenous inhibitor proteins identified in the co-IP/mass spectrometry associated with CDK4 or CDK6 in the FAT1-loss cells. N = 2. F, Co-IP and immunoblotting reveal association of p15INK4B and p18INK4C with CDK6, but not CDK4, in CDK6-high cells. G, Cell line screening results showing that models with high CDKN2A or low RB1 mRNA expression are correlated with poor response to palbociclib. H, Interface residues in CDK6 in close proximity with INK4 isoforms based on previous INK4-bound CDK6 structures in the Protein Data Bank (ref. 65; no available structure for p15INK4B). CDK6-HA was immunoprecipitated using HA beads in parental MCF7 cells and MCF7 cells expressing HA-WT-CDK6-, HA-V16D-, and R31C-mutant CDK6 (disrupted INK4–CDK6 interaction) or HA-K43M/D163N-mutant CDK6 (kinase dead), and interaction with INK4 proteins was determined by immunoblotting. I, Disruption of INK4s and CDK6 binding or impairment of CDK6 kinase activity restores the sensitivity of CDK6-overexpressing cells to CDK4/6i. Cells were treated with DMSO or 100 nmol/L abemaciclib for 24 hours prior to collection. J, Percentage of cell viability of cells overexpressing WT CDK6 or R31C- or D163N-mutant CDK6 treated with increasing concentrations of abemaciclib compared with parental cells. IC50 values were recorded on day 5 following treatment. Data are shown as mean ± SD; n = 6. K, Knockdown of p15INK4B and p18INK4C in FAT1-loss cells promotes suppression of RB phosphorylation in response to abemaciclib to a similar extent as in parental cells. Cells were collected 24 hours after 100 nmol/L abemaciclib treatment. Representative blots are shown, which were repeated independently three times. L, The growth rate of p15INK4B and p18INK4C knockout in FAT1-loss cells was inhibited by 100 nmol/L abemaciclib. The cell viability was recorded at day 14 and day 21. ****, P < 0.0001. Data are shown as mean ± SD; n = 6. See also Supplementary Fig. S1.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/7d30/9762350/28719896bc2a/356fig1.jpg)
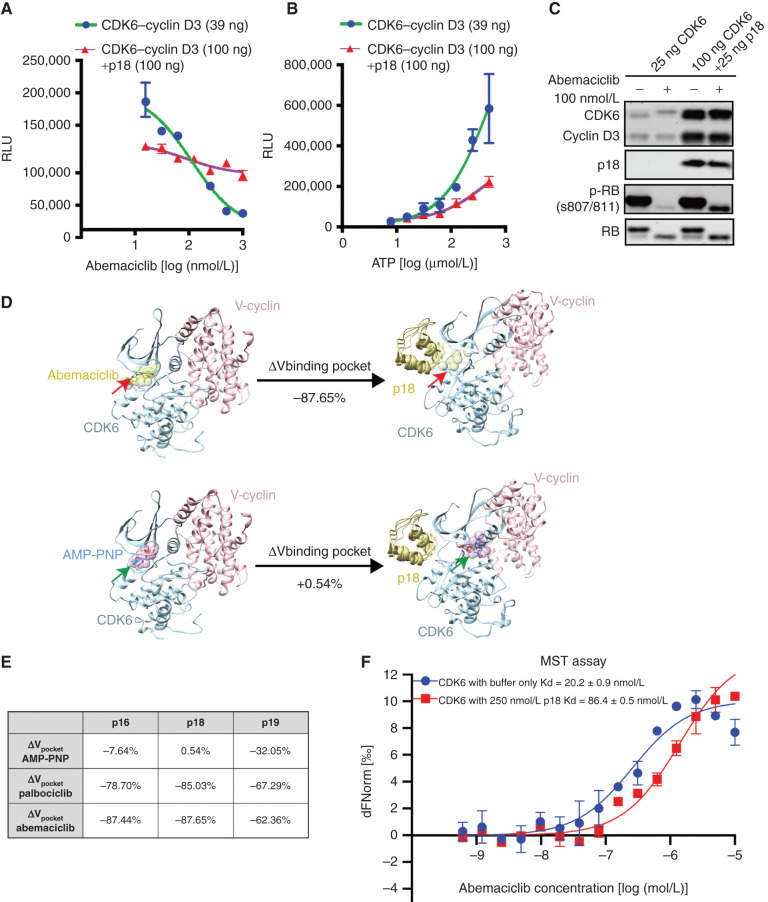
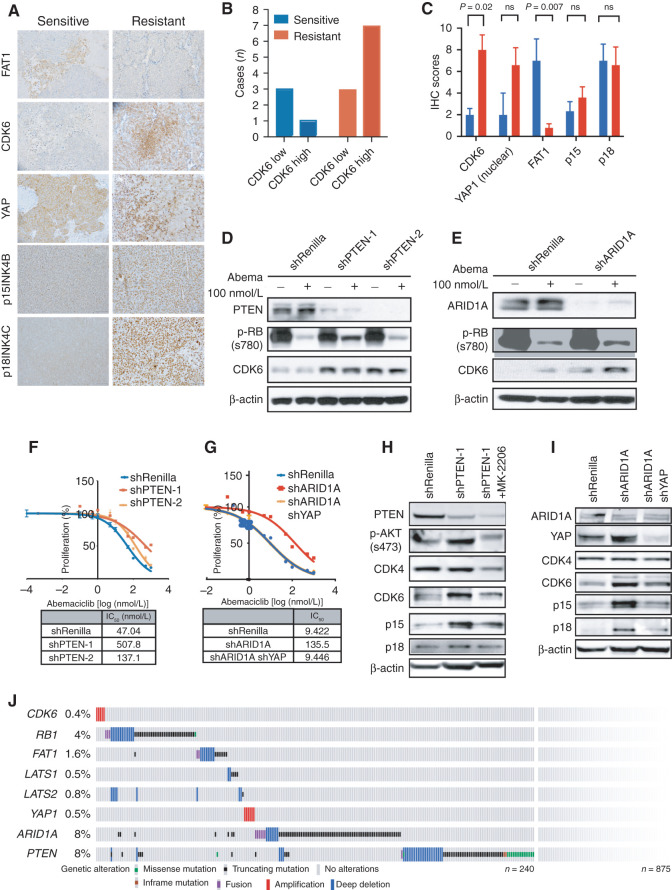
![Figure 4. Compounds targeting the CDK6–INK4 complex inhibit CDK4/6i-resistant tumors. A, Immunoblotting of MCF7 parental cells and cells with high CDK6 expression [CDK6-overexpressing (OE) cells and CDK6-high cells with FAT1 loss] treated for 24 hours with increasing concentrations of bifunctional degrader compound, BSJ-03-123, demonstrating dose-dependent targeting of CDK6 but not CDK4. B, Assessment of a panel of degrader compounds that target CDK4 and/or CDK6. Immunoblotting after 24-hour drug treatment (500 nmol/L) in FAT1-loss cells shows varying selectivity for CDK4 versus CDK6. Representative blots from three independent experiments are shown. Among them, BSJ-05-017 and BSJ-03-096 show the most significant degradation of both CDK4 and CDK6. C, Immunoblot depicting dose–response effects of BSJ-05-017 in both CDK4/6i-sensitive (left) and CDK4/6i-resistant (right) cells in comparison with palbociclib (500 nmol/L) after 24-hour treatment. D, Percentage of growth plot showing that BSJ-05-017 inhibits sensitive MCF7 parental and resistant CDK6-high cells with equal potency, whereas palbociclib shows only partial inhibition of resistant cells. IC50 values were recorded at day 7. Data are shown as mean ± SD; n = 6. E, Assay for drug-induced senescence (Senescence Green) demonstrating number of senescence marker–positive cells induced by 8 days of treatment with DMSO, BSJ-05-017 (500 nmol/L), abemaciclib (100 nmol/L), and palbociclib (500 nmol/L). BSJ-05-017 induced a significantly higher number of cells into senescence compared with abemaciclib or palbociclib in CDK6-high cells. F, Immunoblotting showing the degradation of CDK4/6 and decreased phospho-RB1 and E2F1 levels in CDK6-high (FAT1 loss) tumor-bearing mice administered 25 mg/kg BSJ-05-017 intraperitoneally. Tumors were collected 6 hours after 3 consecutive days of vehicle or BSJ-05-017 treatment (n = 2). G, Growth curve plots of cell-derived xenografts of MCF7 parental, CDK6-overexpressing, and PTEN-loss cells. Mice were treated with vehicle, ribociclib (25 mg/kg, orally), BSJ-05-017 (50 mg/kg, i.p.), or BSJ-03-096 (50 mg/kg, orally) daily for 25 to 35 days. Tumor volumes were recorded every 3 to 4 days. Data are shown as mean ± SD; n = 4. See also Supplementary Fig. S4.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/7d30/9762350/9d9ea24db08c/356fig4.jpg)