The importance of providing open and transparent solutions to use, share, inspect, and reproduce mass spectrometry (MS) data analysis is becoming more apparent. To address these challenges, we developed a collaborative interactive web application, the GNPS Dashboard (https://gnps-lcms.ucsd.edu), to enable remote and synchronous collaborative research in a common analysis environment for MS data analysis (similar to collaborative text editing in browser-based word processors, e.g. Google Docs).
The disruptions to office and laboratory workspaces resulting from the COVID-19 pandemic, including campus closures, telework, and stay-at-home orders, increased the need for online collaborative approaches to perform scientific research. Most solutions for the analysis of mass spectrometry (MS) data are geared for exploration on a local workstation that lack synchronous collaborative data exploration, requiring specific software packages, knowledge of data storage locations, familiarity with file transfer protocols, and, often, conversion of data into compatible formats1–4. Commercial data exploration solutions usually lack universality and are costly, creating additional hurdles to data accessibility, preventing collaborators, reviewers, and readers of scientific publications from inspecting MS data to verify interpretations. However, commercial software offer deep access to proprietary vendor specific data formats and unique data analysis capabilities, e.g. creation of private databases. More broadly, as scientific data is becoming more complex, scientific reproducibility is a challenge. Therefore, there is a growing mandate by both funding bodies and journals to make mass spectrometry data (and other data) FAIR adherent (findable, accessible, interoperable, and reusable).
To empower remote collaborations or classroom teaching, the GNPS Dashboard includes leader-follower synchronization (real-time updates from one lead user) and fully collaborative synchronization (real-time updates from multiple users). Followers can disconnect the synchronization to continue the analysis from where the leader left off without needing to reload the data. The GNPS Dashboard leader-follower paradigm has already been used in classroom teaching by at least five institutions, including undergraduate institutions (Supplementary Note 1, 2). The fully collaborative synchronization enables multi-user visualization and data exploration similar to online synchronous collaborative document editing. For example, users can initiate a collaborative session with two or more people on any web-accessible device (Dashboard Collaboration Start Link, Instructions, Video Tutorial, and Video Application). A snapshot of the collaborative work is automatically created and can be shared with collaborators or included in publications. The final state of the analysis is saved together with every discrete action, enabling inspection of the evolution of data analysis.
The GNPS Dashboard includes tools that facilitate the exploration of liquid and gas chromatography-mass spectrometry data for collaborative examination, and hands-on teaching of MS concepts using private and publicly available MS data, including files stored in the MS data repositories GNPS/MassIVE5, MetaboLights6, ProteomeXchange7, and Metabolomics Workbench8 (Fig. 1, Link to Instructions and Video Tutorial). All public data from compatible repositories can be selected, using the GNPS dataset explorer (https://gnps-explorer.ucsd.edu/). Files not deposited in public MS repositories can be uploaded through a drag-and-drop option for file transfer. mzXML, mzML, CDF, and raw formats are compatible with GNPS Dashboard. Via deep linking from the GNPS platform, the GNPS Dashboard serves as a data explorer and hub for further data analysis including Molecular Networking5, GC-MS deconvolution9, in silico annotation, and MASST10 (Supplementary Note 2).
Figure 1: Overview of the GNPS Dashboard in the Data/Analysis Ecosystem.
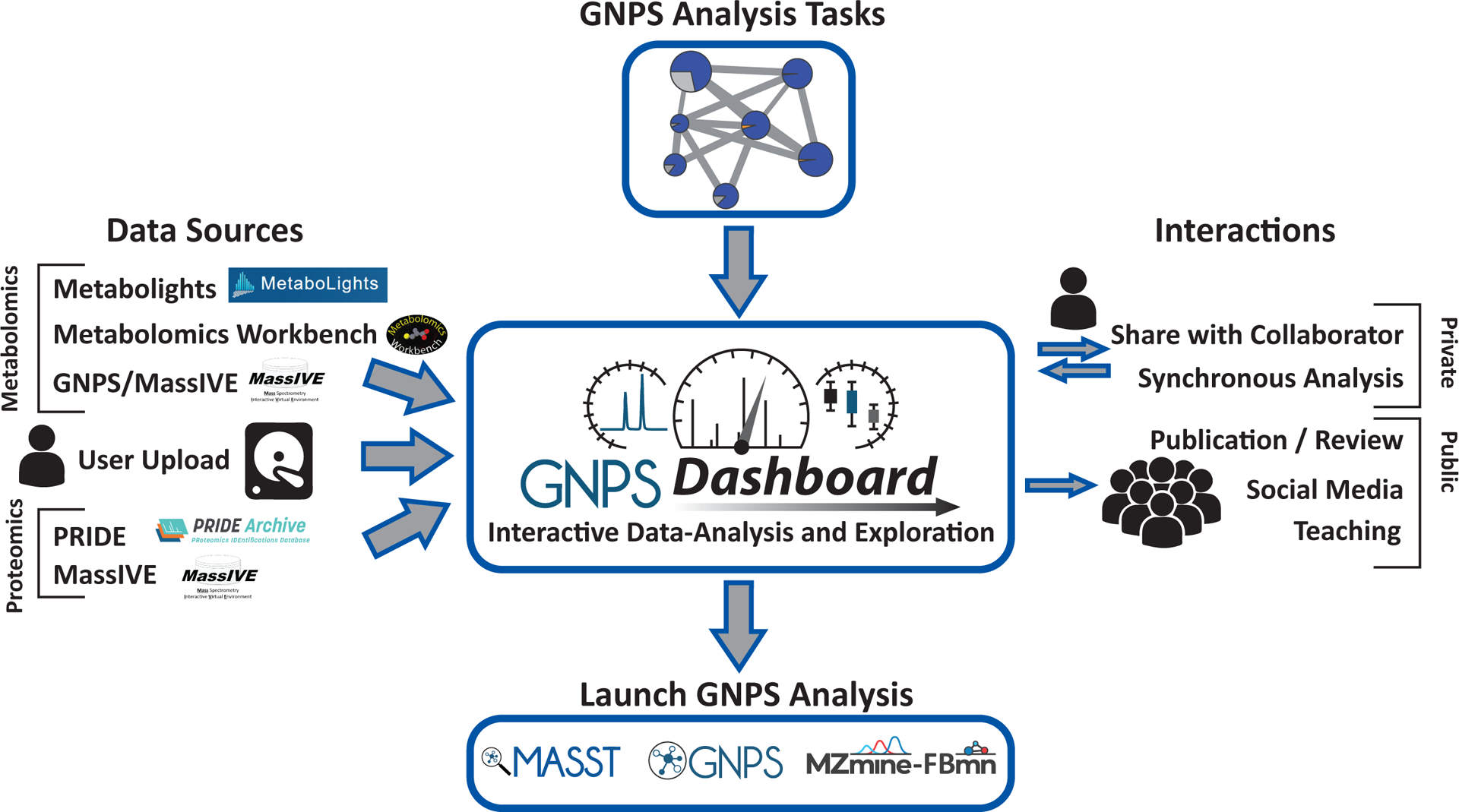
The GNPS Dashboard has been integrated into the web-accessible mass spectrometry ecosystem. The top and left panels show how the GNPS Dashboard can ingress datasets from public metabolomics resources: MetaboLights, Metabolomics Workbench, and GNPS/MassIVE, proteomics resources: PRIDE and MassIVE, private user uploads, and private GNPS analysis tasks. The bottom panel shows that directly out of the GNPS Dashboard, users can launch downstream analysis on their data. Finally, the right panel shows how the GNPS Dashboard and its visualizations can interact with the wider community with reproducible URL or QR code link outs from any analysis and teaching/synchronous collaboration modes for real-time interactivity.
The GNPS Dashboard’s visualization enable inspection of Total Ion Chromatograms, retention time versus m/z heat map, extracted ion chromatograms, tandem mass spectra for inspection/visualization of individual compounds, and quantitative comparison of the peak abundances of two groups as box-plots all with publication-quality figures. The dashboard aids peer review of scientific manuscripts and inspect public quantitative mass spectrometry data to validate published results. Beyond visualization and analysis of MS data, the dashboard supports the development of other bioinformatics tools that do not have their own web-enabled user interfaces. Documentation and tutorials are available in Supplementary Note 1 and Supplementary Fig. 1.
The GNPS Dashboard encodes the visualization in a shareable URL, which empowers users to share the exact same visualization with colleagues, thus reducing miscommunication and improving data transparency, for example, during (remote) meetings. Every visualization and analysis result can be shared via a URL that will re-launch the original data visualization on their device along with the history of the analysis (up to 1000 steps per session). Users can share these links with collaborators and embed them in publications, presentations, or social media posts (e.g., Example Tweet). The final visualization can also be shared as a Quick Response (QR) code. Anyone with a link or QR code can build upon the analysis and re-share their additions. Links and QR codes will remain valid for data that has been archived in a public repository.
Together, the capabilities of the GNPS Dashboard should help improve MS data accessibility/interoperability, lower barriers of entry for mass spectrometry data analysis in the research environment and the classroom, encourage data transparency and sharing, and strengthen the reproducibility of MS data analysis.
Supplementary Material
Acknowledgments
This work was, in part, supported by the National Institutes of Health (NIH) with grant numbers U19AG063744, U2CDK119886, OT2 OD030544, GM107550, R03CA211211, R24GM127667, 1R01LM013115 and P41GM103484, the National Science Foundation (NSF) with grant IOS-1656475 and ABI 1759980, and the Gordon and Betty Moore Foundation (GBMF7622). DP was supported by the Deutsche Forschungsgemeinschaft through the CMFI Cluster of Excellence (EXC 2124). VVP was supported by the NIH R35GM128690. NB and BP were supported by NIH P41 GM103484. MTM was supported by NSF grant CHE-1845230. DB was supported by NSF grant DUE 16-25354.The I.M laboratory was supported by grants from the European Research Council (No. 640384) and from the Israel Science Foundation (ISF No. 1947/19). TRN, BB, and KL were supported by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. AWP and DACJ were supported by NIH R00 GM 118762. This research was supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences of the NIH (ES103363-01).
Furthermore, we would like to thank Tristan de Rond, Laura-Isobel McCall, Wout Bittremieux, Kelly Weldon, and Emily Gentry for testing the software and suggesting updates. We would like to thank Vagisha Sharma, for working on the standalone MSView app during her masters degree research 12 years ago, which provided some visualization inspiration for the GNPS Dashboard. We thank Claire O’Donovan and the team at MetaboLights for developing a well-documented API. Lastly, we thank all members of the research community who make their data publicly accessible, which contributes to open, transparent, and reproducible science.
Footnotes
Code availability
While the GNPS Dashboard is accessible as a free public web service, it is possible to locally install the GNPS Dashboard to function with local data sources, making collaborative analysis and sharing possible, privately, within an institution when necessary. The source code are available through the GNPS web environment and GitHub, enabling quick installation on local servers.
The GNPS Dashboard source code can be found on GitHub: https://github.com/mwang87/GNPS_LCMSDashboard under a modified UCSD BSD License.
The GNPS Dataset Explorer source code can be found on GitHub: https://github.com/mwang87/GNPS_DatasetExplorer under an MIT License.
Further implementation details can be found in Supplementary Methods.
Competing Interests
PCD is a scientific advisor of Sirenas, Galileo, Cybele, and scientific advisor and co-founder of Ometa Labs LLC and Enveda with approval by the UC San Diego. MW is a founder of Ometa Labs LLC. TRN is an advisor of Brightseed Bio.
References
- 1.Pluskal T, Castillo S, Villar-Briones A & Orešič M BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsugawa H et al. Nat. Methods 12, 523–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Röst HL et al. Nat. Methods 13, 741–748 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Huang Y-C, Tremouilhac P, Nguyen A, Jung N & Bräse S. l J. Cheminformatics 13, 8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M et al. Nat. Biotechnol 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haug K et al. Nucleic Acids Res. 48, D440–D444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vizcaíno JA et al. Nat. Biotechnol 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sud M et al. Nucleic Acids Res. 44, D463–D470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aksenov AA et al. Nat. Biotechnol 39, 169–173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M et al. Nat. Biotechnol 38, 23–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


