Abstract
SMARCA4/BRG1 encodes for one of two mutually exclusive ATPases present in mammalian SWI/SNF chromatin remodeling complexes and is frequently mutated in human lung adenocarcinoma. However, the functional consequences of SMARCA4 mutation on tumor initiation, progression, and chromatin regulation in lung cancer remain poorly understood. Here, we demonstrate that loss of Smarca4 sensitizes CCSP+ cells within the lung in a cell-type dependent fashion to malignant transformation and tumor progression, resulting in highly advanced dedifferentiated tumors and increased metastatic incidence. Consistent with these phenotypes, Smarca4-deficient primary tumors lack lung lineage transcription factor activities and resemble a metastatic cell state. Mechanistically, we show that Smarca4 loss impairs the function of all three classes of SWI/SNF complexes, resulting in decreased chromatin accessibility at lung lineage motifs and ultimately accelerating tumor progression. Thus, we propose that the SWI/SNF complex – via Smarca4 – acts as a gatekeeper for lineage-specific cellular transformation and metastasis during lung cancer evolution.
Keywords: lung adenocarcinoma, SWI/SNF, Smarca4/Brg1, mouse models, chromatin remodeling
Introduction
Genes encoding for components of the mammalian ATP-dependent chromatin remodeling complex, SWI/SNF (also known as BAF), are among the most commonly mutated targets in cancer (1). However, their exact contributions to tumorigenesis are not well understood in many cancer types. Such lack of understanding reflects the complexity of SWI/SNF function due to its cell type-specific roles and the heterogeneity of SWI/SNF complexes within a cell at a given time (2). Previous studies have shown highly context-specific roles for SWI/SNF in tumor progression (3,4). These studies strongly emphasize the need for a deeper mechanistic understanding of the impact of precise SWI/SNF mutations on the complex’s function and tumor cell biology in order to devise effective therapeutic strategies tailored to specific SWI/SNF mutations and tumor types.
SMARCA4 (BRG1) encodes for one of two mutually exclusive ATPases of SWI/SNF complexes (5) and is among the most frequently mutated genes (6) in non-small cell lung cancer (NSCLC), occurring at a frequency of 10% (7,8). Non-small cell lung cancers harboring SMARCA4 mutations are predominantly of the lung adenocarcinoma (LUAD) subtype (8). Among SMARCA4 mutations, truncating and missense mutations are the most prevalent and these can be monoallelic or biallelic (7–9). Of these, inactivating SMARCA4 alterations that result in the complete absence of SMARCA4 protein expression, such as truncating mutations, are associated with the poorest outcomes in patient survival (8,9).
Previous studies in mouse models have examined the functional consequences of SMARCA4 inactivation on lung cancer progression. In a carcinogen-induced model of lung cancer, loss of one allele of Smarca4 at tumor initiation promoted tumorigenesis, while loss of both alleles had no detectable effect (10). Interestingly, two independent studies using a genetically engineered mouse model (GEMM) of Kras-driven lung adenocarcinoma produced conflicting findings with loss of Smarca4 resulting in a negative or positive effect on tumorigenesis (11,12). These results raise questions about the tumor suppressive functions of SMARCA4 in the lung, and whether SMARCA4 inactivating mutations and the loss of its protein expression observed in human patients confer a functional advantage in tumor initiation or progression. Furthermore, despite SMARCA4’s well-described role as a core catalytic component of the SWI/SNF chromatin remodeling complex, studies investigating its functions in chromatin regulation in lung cancers have been limited to NSCLC cell lines (13–15). As such, the direct consequences of Smarca4 inactivation on SWI/SNF function on chromatin regulation during lung cancer evolution is unknown.
Here, we address the impact of SMARCA4/Smarca4 inactivation on tumor initiation and progression, chromatin accessibility, and SWI/SNF function in lung adenocarcinoma using a combination of GEMMs, patient-derived xenograft (PDX) models, and epigenomic profiling. We demonstrate a tumor suppressive function for Smarca4 that is dependent on the cell type in which the mutation occurs. We further identify transcription factor (TF) programs altered in the context of SMARCA4 loss. In particular, our studies reveal that Smarca4-deficient tumors effectively lose lung lineage transcription factor activities and harbor features of dedifferentiation, reminiscent of a metastatic cell state. We further determine the underlying mechanism behind these aberrant cell states as a consequence of altered SWI/SNF function upon Smarca4 inactivation. Collectively, this work provides key insights into SMARCA4 function in tumor initiation and progression, chromatin state, and SWI/SNF function in lung cancer. More broadly, our data have implications for understanding the patterns of cancer-associated mutations in subtypes of human cancer.
Results
Smarca4 mutation has divergent effects on lung tumor suppression
To determine the impact of Smarca4 loss on tumor initiation and progression, we first sought to model SMARCA4 inactivation in a defined and relevant genetic system. KRAS is the most frequent oncogene co-mutated with SMARCA4 (35%) in contrast to EGFR, which tends to be mutated in SMARCA4 wild-type lung cancers (7,8). Mutations in the tumor suppressor TP53 also occur at a high frequency (56%) among SMARCA4-mutant tumors (8). Given the spectrum of these co-occurring mutations, we crossed a floxed allele of Smarca4 (16) into a well-characterized mouse model (17) of lung adenocarcinoma (KrasLSL-G12D/+; Trp53fl/fl, KP; Fig. 1A). In this model, concomitant activation of oncogenic Kras, deletion of the tumor suppressor Trp53, and deletion of exons encoding for the ATPase domain of Smarca4 occur in the lungs of mice upon intratracheal delivery of adenoviral Cre recombinase. For these experiments, we used adenoviruses in which Cre expression is driven by the Sftpc (surfactant-associated protein C, SPC) promoter (18), the activity of which is observed in alveolar type II (AT2) cells – one of the presumed cells-of-origin of lung adenocarcinomas (19).
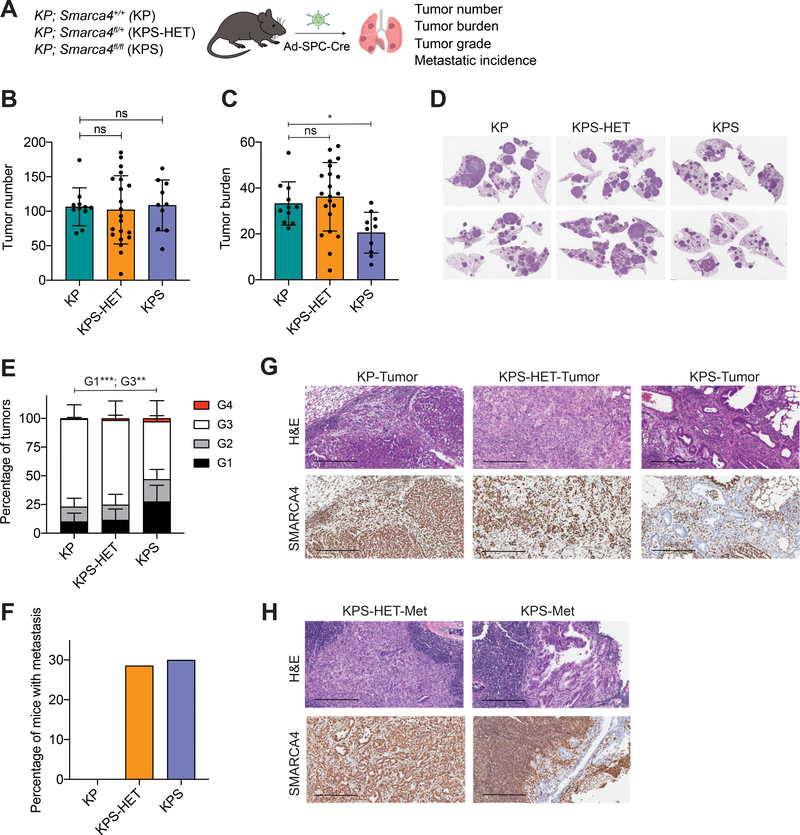
Figure 1. Smarca4 inactivation has divergent effects on lung tumor suppression.
(A) Schematic diagram of the experimental strategy of tumor initiation in SPC-expressing cells in the lungs of KP (n=11), KPS-HET (n=21), and KPS (n=10) animals using adenoviral SPC-Cre (Ad-SPC-Cre) and subsequent analysis 17 weeks post-infection. (B) Quantification of tumor number (number of tumors per mouse) in KP (n=11), KPS-HET (n=21), and KPS (n=10) animals 17 weeks post-infection. Data are mean ± s.d. One-way ANOVA: not significant (ns). (C) Quantification of tumor burden (% tumor area/lung area per mouse) in KP (n=11), KPS-HET (n=21), and KPS (n=10) animals 17 weeks post-infection. Data are mean ± s.d. One-way ANOVA: F(2, 39) = 5.475, p = 0.0080; Dunnett’s multiple comparisons test: *adjusted p-value = 0.0443. (D) Representative hematoxylin and eosin (H&E) staining of tumor-bearing lungs of KP, KPS-HET, and KPS animals 17 weeks post-infection. (E) Distribution of histological tumor grades (% tumor grade area/total tumor area where G1 = Grade 1, G2 = Grade 2, G3 = Grade 3 and G4 = Grade 4) in KP (n=11), KPS-HET (n=21), and KPS (n=10) animals 17 weeks post-infection. Data are mean ± s.d. One-way ANOVA (G1): F(2, 39) = 9.702, p = 0.0004; Dunnett’s multiple comparisons test (G1): ***adjusted p-value = 0.0009; one-way ANOVA (G3): F(2, 39) = 8.772, p = 0.0007; Dunnett’s multiple comparisons test (G3): **adjusted p-value = 0.0013. (F) Percentage of KP (n=11), KPS-HET (n=21), and KPS (n=10) mice with metastasis 17 weeks post-infection. Chi-square: ns, p = 0.1305. (G) Representative H&E and SMARCA4 staining of advanced tumors in KP, KPS-HET, and KPS animals. Scale bars: 300 μm. (H) Representative H&E and SMARCA4 staining of metastases in KPS-HET and KPS animals. Scale bars: 300 μm.
The KP mouse model recapitulates the full cascade of lung adenocarcinoma development (17). Tumor-initiating cells infected with adenoviral Cre undergo hyperplasia, progression to adenomas, and finally to adenocarcinomas, which have the ability to metastasize to local and distal sites. We assessed the impact of Smarca4 loss on tumorigenesis 17 weeks post-tumor initiation using various metrics: tumor number, tumor burden, tumor grade, and metastatic incidence.
We observed no differences in the number of tumors among the three genotypes (Fig. 1B). A selection against full Smarca4 loss was evident, as shown by a decrease in overall tumor burden in KrasLSL-G12D/+; Trp53fl/fl; Smarca4fl/fl (KPS) mice compared to those with wild-type Smarca4 (KP) or reduced Smarca4 gene dosage (KrasLSL-G12D/+; Trp53fl/fl; Smarca4fl/+, KPS-HET; Fig. 1C and 1D). Histological examination of SMARCA4 protein expression in the lungs of KPS mice revealed that a considerable fraction of tumors (14–35%) across all animals retained expression (Supplementary Fig. S1A and S1B). We used laser-capture microdissection to isolate SMARCA4-positive tumors from KPS mice, performed genotyping PCR, and detected both the floxed and recombined alleles in all cases (Supplementary Fig. S1C). Tumors with retained SMARCA4 expression as a result were classified as recombination escapers and were heterozygous for Smarca4 loss. Despite a substantial fraction of tumors in KPS animals retaining SMARCA4 expression, we did detect tumors clearly absent of SMARCA4 staining. These tended to be smaller in size and were associated with decreased proliferation (measured by Ki67 staining) compared to their SMARCA4-positive counterparts (Supplementary Fig. S1D and S1E). To estimate the distribution of tumor grades in these animals in an unbiased fashion, we applied a deep-learning algorithm based on well-established criteria (17,20) that histologically classifies KP tumors by tumor grade to the lesions in the lungs of these animals. We found an increase in the fraction of early lesions (Grades 1–2) and a decrease in more advanced Grade 3 tumors in KPS mice compared to KP and KPS-HET animals (Fig. 1E). Altogether, these data indicate that full Smarca4 inactivation restrains tumor progression in the vast majority of tumors initiated from SPC+ cells.
Closer inspection of tumor-bearing lungs of KPS mice, however, revealed that these animals had the highest fraction of the most advanced type of lesions (Grade 4), despite their markedly decreased overall tumor burden (Fig. 1E). Furthermore, we observed increased frequency of metastases to the thymus and lymph node among KPS and KPS-HET mice that was higher than KP animals (Fig. 1F). Strikingly, Grade 4 lesions and metastases from KPS animals universally lacked SMARCA4 protein expression (Fig. 1G and 1H) – indicating a strong selection for full SMARCA4 loss in these highly advanced tumors and metastases.
Collectively, these results point towards a paradoxical role for Smarca4 in tumor suppression. While Smarca4 loss inhibits tumor progression in a large fraction of tumors initiated by SPC-Cre, a subset of SMARCA4-deficient transformed cells can give rise to highly advanced and metastatic tumors.
Epigenetic states of Smarca4-deficient primary tumors arising from SPC+ cells resemble metastatic cell states
Given the role of SMARCA4 in chromatin remodeling, we hypothesized that Smarca4 inactivation directly alters distinct transcription factor programs – perhaps in a cell type-specific fashion – to affect the final tumorigenic outcome. As a catalytic subunit of SWI/SNF, SMARCA4 has a key role in the ability of the complex to regulate nucleosome positioning and chromatin accessibility. Such accessibility is crucial for transcription factor binding to regulatory elements in order to specify gene expression programs that dictate cell state. To address this hypothesis and investigate the heterogeneity among Smarca4-deficient tumors in vivo, we performed the single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) on isolated cancer cells from KP, KPS-HET, and KPS animals (Fig. 2A). We performed these experiments on moribund animals to better capture the cell states spanning tumor progression – including those of high grade tumors and metastases cells – from all three SMARCA4 genotypes. In total, we generated chromatin accessibility profiles from 25,229 cells. These include 21,780 cells from the tumor-bearing lungs of 3 animals per genotype and 3,449 cells from metastatic sites (thymi and lymph nodes) of 2 animals per genotype (Supplementary Fig. S2A–S2C).
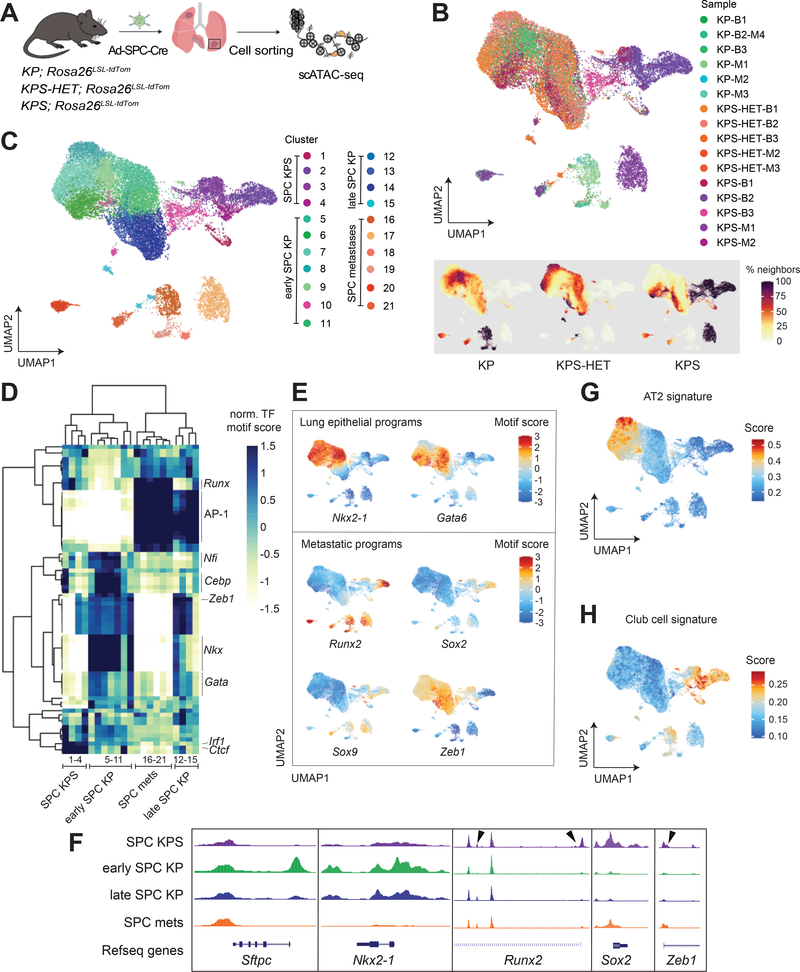
Figure 2. Epigenetic states in Smarca4-deficient primary tumors resemble metastases.
(A) Schematic diagram of the experimental strategy to isolate cancer cells from moribund animals for scATAC-seq. (B) UMAP visualization of single-cell chromatin accessibility profiles of 25,229 cells isolated from primary tumors (bulk) and metastases (met) of KP (bulk n=3; met n=4), KPS-HET (bulk n=3; met n=2), and KPS (bulk n=3; met n=2) animals colored by sample where B = bulk primary tumors and M = metastasis (upper panel), and by % of cell neighbors per genotype (lower panel). (C) UMAP visualization of single-cell chromatin accessibility profiles colored by cluster. (D) A heatmap showing the top variable accessible motifs across the dataset. Heatmap is colored by row-normalized mean motif scores of each cell cluster identified through the Louvain modularity method. (E) UMAP visualization of the scATAC-seq dataset colored by lung lineage and metastatic program motif scores. (F) Aggregated scATAC-seq tracks showing selected chromatin accessibility peaks per group. Arrows show gains of peaks in SPC KPS clusters. UMAP visualization of dataset colored by (G) AT2 and (H) club cell signature gene scores.
As shown in Fig. 2B and 2C, chromatin accessibility profiles from KP and KPS-HET primary tumors formed a continuum of states, while metastasis cells clustered separately as previously reported and characterized (20). By contrast, cancer cells isolated from the lungs of KPS animals generated strikingly distinct clusters composed almost exclusively of cells of this genotype, indicating unique epigenetic states in these cells (Fig. 2B and 2C). Importantly, KPS-specific clusters were clearly reproduced using two distinct dimensionality reduction methods (Fig. 2B and 2C; Supplementary Fig. S2D and S2E). A fraction of KPS cells belonged to clusters predominantly composed of KP and KPS-HET cells, likely reflecting the presence of SMARCA4-positive tumors in the lungs of KPS animals. A small number of KPS-HET cells were also found in KPS clusters, which we attribute to a fraction of KPS-HET tumors harboring cancer cells absent of SMARCA4 protein expression (Supplementary Fig. S2F). We identified TF motifs uniquely marking 21 clusters identified through the Louvain modularity method (21) and performed hierarchical clustering (Fig. 2D). We annotated the clusters based on their sample composition and chromatin accessibility profile in relation to features of early and advanced tumors previously described in this model (20,22): SPC KPS (composed almost exclusively of KPS cells; 1–4), early SPC KP (Nkx2–1 high; 5–11), late SPC KP (Nkx2–1 low; 12–15), and SPC metastases (composed of metastasis cells from all three genotypes; 16–21).
Tumor progression in the KP model is characterized by key epigenetic state transitions and loss of cell identity as cancer cells evolve towards an advanced state (20,22–24). Strikingly, SPC KPS clusters displayed characteristic features of SPC metastases clusters, despite being composed almost exclusively of primary tumor cells (Fig. 2D–2F). Activities for the lung lineage TFs, Nkx2–1/Ttf-1 and Gata6, were markedly absent in SPC KPS clusters, demonstrating a lack of lung lineage cell identity among these primary tumor cells. A fraction of cells from SPC KPS clusters were also highly enriched for peaks associated with TFs marking late tumors or metastatic cells, such as Runx2, Sox2, and Sox9 (20,25). Importantly, the activities of these TF programs were markedly higher in KPS cells compared to advanced KP or KPS-HET cells within late SPC KP clusters. Furthermore, decreased accessibility in motifs of the repressor Zeb1 indicated that a subset of KPS cells were undergoing epithelial-to-mesenchymal transition (26) – consistent with a metastatic-like phenotype. In contrast to cells isolated from KPS primary tumors, we did not detect clear TF activities that distinguished cancer cells isolated from metastases in KPS mice from those isolated from metastases in KP and KPS-HET animals (Fig. 2D). Instead, KPS metastasis cells generated multiple clusters and displayed variable accessibilities for pro-metastatic TF programs (Runx2, Onecut2, Sox2, Sox9), suggesting heterogeneous routes to metastases in the context of Smarca4 deficiency (Fig. 2D and 2E; Supplementary Fig. S2G).
While SPC KPS clusters had features consistent with an advanced cancer cell state, cells from these clusters were depleted for peaks associated with AP-1 TF family motifs (Fos, Jun, among others) in stark contrast to late SPC KP and SPC metastases clusters (Fig. 2D; Supplementary Fig. S2G). SWI/SNF complexes bind directly to AP-1 TF motifs, and members of this TF family have been shown to be important modulators of enhancer selection (27). Depletion of AP-1 motif accessibility may be attributed to the abrogation of a direct interaction between the AP-1 TF family member, JUNB, and SWI/SNF complexes upon Smarca4 deficiency, which we have previously shown through quantitative mass spectrometry (28). Interestingly, metastasis-derived cells isolated from KPS animals were enriched for peaks with AP-1 TF motifs, suggesting that a gain in the activities of these TF programs may be a key event selected for in the transition of KPS primary tumors to a fully metastatic cell state. Given that the absence of AP-1 activity distinguishes KPS clusters from SPC metastases clusters, the gain in AP-1 motif accessibility as cancer cells from KPS primary tumors transition into a pre-metastatic/metastatic cell state results in KPS metastases cells that cluster with those isolated from KP and KPS-HET animals (Figure 2D).
Altogether, these data show that Smarca4 inactivation in tumor-initiating cells leads to cancer cells with distinct cell states that largely recapitulate metastatic cell states. In contrast to cancer cells from KP and KPS-HET primary tumors, which undergo similar epigenetic state transitions and a gradual loss of lineage fidelity throughout tumor evolution, those from KPS primary tumors are characterized by a general lack of lung lineage specificity and the robust activation of TF programs associated with metastases in a subset of cells. Thus, we hypothesized that Smarca4 inactivation in certain tumor-initiating cells facilitates the rapid acquisition of a metastatic-like cell state.
Smarca4-deficient primary tumors exhibit a club cell state
We next sought to understand the heterogeneity of SMARCA4-deficient tumors in KPS animals. Notably, SMARCA4-negative tumors in this model tended to be either of low tumor grade or highly advanced. In particular, we sought to identify determinants of high grade tumors in this context. These tended to be located by the airways of the lungs of KPS animals – in sharp contrast to high grade tumors from KP and KPS-HET mice, which were predominantly located in the alveolar spaces (Supplementary Fig. S2H). We hypothesized that these highly advanced tumors arose from an atypical cell-of-origin within the bronchioles or the bronchioalveolar duct junction (BADJ) that is uniquely sensitive to transformation upon SMARCA4 loss.
Although independent studies have demonstrated that SPC+ AT2 cells are the predominant cell-of-origin in the Kras-driven GEMM of lung adenocarcinoma (19,29), other cell types, including the club cell secretory protein-positive (CCSP+) bronchiolar epithelial club cell (30) and the SPC+CCSP+ double positive bronchioalveolar stem cell (BASC) (31), may have this ability given the proper context. Importantly, BASCs are able to regenerate multiple cell lineages, including AT2 and club cells upon lung injury (32,33). Among human LUAD patients, both AT2 and club cells are hypothesized to be the cell-of-origin of LUAD as these tumors tend to express markers of these lineages (34).
To investigate the role of the cell-of-origin in our data, we scored each cell in the scATAC-seq dataset for AT2 and club cell identities using the promoter and gene body accessibilities (also referred to as gene scores (20)) of a set of marker genes for each cell type identified from single cell RNA-sequencing (scRNA-seq) of the developing lung (35). The AT2 signature was enriched in early SPC KP clusters and depleted in late SPC KP, SPC KPS, and SPC metastases clusters (p < 2.2 × 10−16, Fig. 2G). Consistent with a distinct cell-of-origin giving rise to advanced SPC KPS tumors, KPS clusters displayed a strikingly specific enrichment of the club cell signature, which was completely absent in clusters largely comprised of primary tumors from KP and KPS-HET animals (p < 2.2 × 10−16, Fig. 2H).
Cells within SPC KPS clusters, therefore, have a club cell state. These results point towards the club cell state potentially being highly sensitive to malignant transformation and rapid tumor progression in the absence of Smarca4. Such a state may be achieved through Kras-driven transformation of SPC+CCSP+ BASCs in this model, since these cells could adopt a club cell state (32,33) upon tumor initiation by SPC-Cre.
CCSP+ cells are highly sensitive to malignant transformation in the absence of Smarca4
To formally test whether the club cell state is sensitive to transformation in the setting of Smarca4 inactivation, we initiated tumors in the lungs of KP, KPS-HET, and KPS animals using adenoviruses in which Cre expression was driven by the Scgb1a1 (CCSP) promoter (Fig. 3A), which is predominantly active in the club cell population (18).
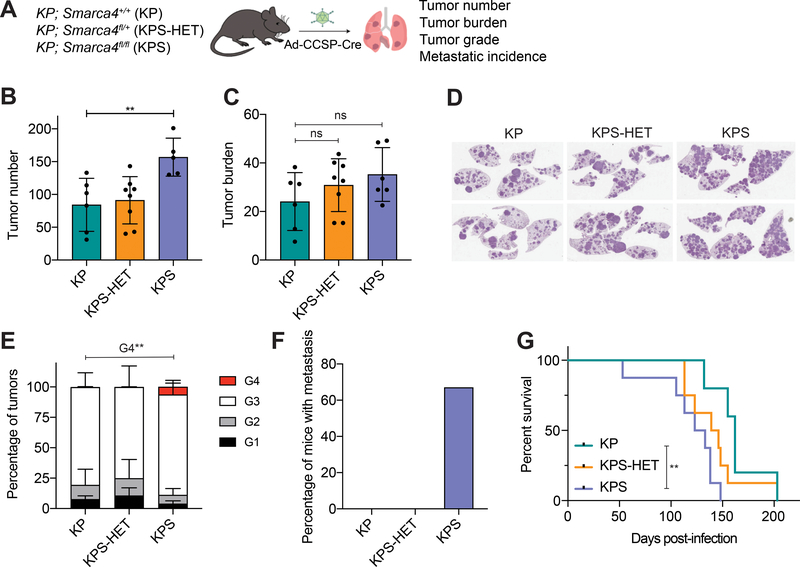
Figure 3. Smarca4 inactivation in tumor-initiating CCSP+ cells accelerates tumor progression.
(A) Schematic diagram of the experimental strategy of tumor initiation in CCSP-expressing cells in the lungs of KP (n=6), KPS-HET (n=8), and KPS (n=6) animals using adenoviral CCSP-Cre (Ad-CCSP-Cre), and subsequent analysis 16 weeks post-infection. (B) Quantification of tumor number (number of tumors per mouse) in KP (n=6), KPS-HET (n=8), and KPS (n=5) animals 16 weeks post-infection. Data are mean ± s.d. One-way ANOVA: F(2, 16) = 6.786, p = 0.0073; Dunnett’s multiple comparisons test: **adjusted p-value = 0.0078. (C) Quantification of tumor burden (% tumor area/lung area per mouse) in KP (n=6), KPS-HET (n=8), and KPS (n=6) animals 16 weeks post-infection. Data are mean ± s.d.; one-way ANOVA: not significant (ns). (D) Representative H&E staining of tumor-bearing lungs of KP, KPS-HET, and KPS animals 16 weeks post-infection. (E) Distribution of histological tumor grades (% tumor grade area/total tumor area where G1 = Grade 1, G2 = Grade 2, G3 = Grade 3 and G4 = Grade 4) in KP (n=6), KPS-HET (n=8), and KPS (n=6) animals 16 weeks post-infection. Data are mean ± s.d.; one-way ANOVA (G4): F(2, 17) = 9.102, p = 0.0021; Dunnett’s multiple comparisons test: **adjusted p-value = 0.0044. (F) Percentage of KP (n=6), KPS-HET (n=8), and KPS (n=6) mice with metastasis 16 weeks post-infection. Chi-square: p = 0.0029. (G) Survival curves of KP (n=5), KPS-HET (n=8), and KPS (n=8) animals infected with Ad-CCSP-Cre through intratracheal instillation. Log-rank test (KP vs KPS): **p = 0.0086.
In striking contrast to KPS animals in which tumors were initiated using SPC-Cre, these KPS animals displayed a significant increase in the number of tumors per mouse compared to KP (1.86X) and KPS-HET (1.72X) animals (Fig. 3B). They also exhibited a trend towards increased tumor burden compared to their Smarca4 wild-type and heterozygous counterparts (Fig. 3C and 3D). Histological examination of these tumors for SMARCA4 expression revealed that the majority of tumors (86–96%) within the lungs of these KPS animals were SMARCA4-negative (Supplementary Fig. S3A and S3B) in contrast to the tumors arising from SPC-expressing cells in KPS animals (Supplementary Fig. S1A and S1B). These KPS animals were also consistently enriched for higher grade tumors (Grades 3–4; Fig. 3E) and all metastatic lesions in this cohort were identified exclusively in KPS animals (Fig. 3F), some of which displayed multiple metastatic lesions within a single tissue (Supplementary Fig. S3C). Importantly, all high grade tumors and metastases identified in KPS animals lacked SMARCA4 protein expression (Supplementary Fig. S3D and S3E). Of note, we detected SMARCA4-negative cells within some tumors in the majority of KPS-HET animals infected with the CCSP-Cre virus (Supplementary Fig. S3F and S3G). Consistent with a potent tumor suppressive role for SMARCA4 in LUAD, these KPS animals rapidly succumbed to disease and displayed decreased overall survival compared to KP animals (Fig. 3G). Thus, Smarca4 inactivation in CCSP+ tumor-initiating cells promotes tumor progression at multiple stages of lung tumorigenesis.
Collectively, these data demonstrate a cell type-specific role for SMARCA4 in tumor initiation and progression in the lung. These models show that the tumor suppressive function of SMARCA4 is influenced by the tumor cell-of-origin. While SMARCA4 loss inhibits tumor progression in the vast majority of transformed SPC+ cells, predominantly AT2 cells, it promotes malignant transformation and consistently accelerates tumor progression in transformed CCSP+ cells, predominantly club cells.
Epigenetic states of Smarca4-deficient primary tumors are driven by SMARCA4 loss
We next sought to determine whether the epigenetic states of KPS clusters we previously identified by scATAC-seq are driven by an alternative cell-of-origin for KPS tumors in the SPC KP model or are a direct effect of Smarca4 loss-of-function. To address this, we performed scATAC-seq on sorted cancer cells arising from CCSP-expressing cells from KP, KPS-HET, and KPS animals (Fig. 4A).
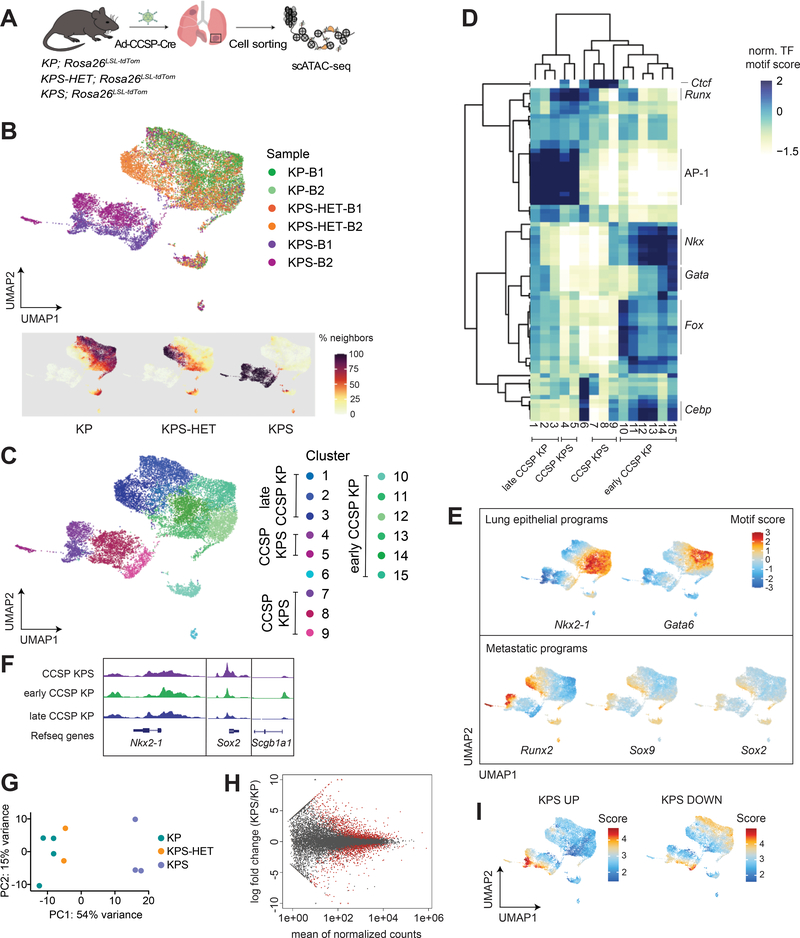
Figure 4. The epigenetic states of Smarca4-deficient primary tumors are driven by SMARCA4 loss.
(A) Schematic diagram of the experimental strategy to isolate cancer cells from moribund animals for scATAC-seq. (B) UMAP visualization of single-cell chromatin accessibility profiles of 16,321 cells isolated from primary tumors of KP (n=2), KPS-HET (n=2), and KPS (n=2) animals colored by sample where B = bulk primary tumors (upper panel) and by % of cell neighbors per genotype (lower panel). (C) UMAP visualization of scATAC-seq profiles colored by cluster. (D) A heatmap showing the top variable accessible motifs across the dataset. Heatmap is colored by row-normalized mean motif scores of each cell cluster identified through the Louvain modularity method. (E) UMAP visualization of the scATAC-seq dataset colored by lung lineage and metastatic program motif scores. (F) Aggregated scATAC-seq tracks showing selected chromatin accessibility peaks per group. (G) Principal component analysis (PCA) of RNA-seq data generated from KP (n=4), KPS-HET (n=2), and KPS (n=3) primary tumors. (H) MA plot showing differentially expressed genes between KP (n=4) and KPS (n=3) primary tumors. Red dots represent differentially enriched genes (adjusted p-value < 0.05). (I) UMAP visualization of the CCSP scATAC-seq dataset colored by the mean gene score of differential genes increased and decreased (adjusted p-value < 0.05, |FC|>1.5) in KPS primary tumors (n=3) compared to KP (n=4) primary tumors.
We generated chromatin accessibility profiles from 16,321 cells isolated from the tumor-bearing lungs of 2 animals per genotype (Supplementary Fig. S4A–S4C). Similar to SPC-Cre-initiated tumors, KP and KPS-HET primary tumors formed a continuum of epigenetic states, while KPS primary tumors displayed distinct states represented by clusters composed exclusively of cells from KPS animals (Fig. 4B and 4C).
We uncovered 15 distinct clusters, identified differential TF motifs across the dataset, and performed hierarchical clustering (Fig. 4C and 4D). We annotated these clusters as early CCSP KP (Nkx2–1 high; 10–15), late CCSP KP (Nkx2–1 low; 1–3), and CCSP KPS (composed almost exclusively of KPS cells; 4, 5, 7–9) based on their chromatin accessibility profile and sample composition (Fig. 4C and 4D). Of note, we could detect enrichment of Fox motifs (Foxa1, Foxc1, among others) in cluster 10, an early CCSP KP cluster, representing normal club cells and early transformed club cells. Fox motifs are among the most enriched motifs distinguishing normal club cells from normal AT2 cells (Supplementary Fig. S4D).
We also detected epigenetic state transitions and loss of cell identity as KP and KPS-HET tumor cells from this model progressed towards an advanced state, consistent with those observed in tumors arising from SPC-expressing cells in KP and KPS-HET animals. For example, a gain in the activity of the AP-1 TF family accompanied the loss of Nkx2–1 activity that demarcates early CCSP KP from late CCSP KP (Fig. 4D and 4E; Supplementary Fig. S4E). Additionally, we could detect reduced lung lineage TF activities (Nkx, Cebp, Gata families) in cells belonging to late CCSP KP clusters compared to those in early CCSP KP clusters.
By contrast, cells in CCSP KPS clusters were almost entirely depleted for peaks harboring these lung lineage TF motifs (Fig. 4D and 4E; Supplementary Fig. S4E). We detected low activity for Nkx2–1 and Cebpa in cluster 9, representing a KPS cell state early in tumor progression, while programs associated with more advanced tumors (Runx2, Sox2, Sox9) were activated in another subset of KPS cells and are reflective of a KPS cell state late in tumor progression (Fig. 4D–4F). Thus, the marked reduction of lung lineage TF activities and activation of pro-tumorigenic programs in KPS cells are consistent features of Smarca4-deficient primary tumors.
Similar to KPS primary tumors initiated from SPC-expressing cells, KPS cancer cells initiated from CCSP-expressing cells were also depleted for AP-1 TF family motifs (Fig. 4D; Supplementary Fig. S4E). However, a subset of cells in late CCSP KPS clusters had high AP-1 TF activity, and were also enriched for programs defining a metastatic cell state, most notably Runx2 (Fig. 4D and 4E; Supplementary Fig. S4E). Thus, a gain in AP-1 TF activity in Smarca4-deficient cells occurs in primary tumors transitioning into a metastatic cell state. Gain in AP-1 TF activity in Smarca4-deficient cells was also observed in metastases from SPC-Cre-infected KPS animals (Supplementary Fig. S2G). Of note, Ctcf and Irf/Stat activities were the most clearly enriched programs in KPS clusters (Fig. 2D and 4D; Supplementary Fig. S4E).
KPS primary tumors display distinct transcriptional profiles
Our results, thus far, indicate that complete loss of Smarca4 during tumor initiation results in distinct epigenetic states in KPS primary tumors throughout tumor evolution. To determine whether this distinction is maintained at the transcriptional level, we performed bulk RNA sequencing on sorted cancer cells from the lungs of moribund tumor-bearing KP, KPS-HET, and KPS animals. Both unsupervised hierarchical clustering and principal component analysis (PCA) revealed that KPS samples clearly separated from KP and KPS-HET samples (Supplementary Fig. S4F; Fig. 4G), indicating a unique transcriptional profile associated specifically with complete Smarca4 loss.
To understand the gene expression programs characterizing KPS primary tumors, we next performed pairwise analysis to determine the most significant differentially expressed genes between KPS and KP primary tumors (Fig. 4H). Top genes increased (adjusted p-value < 0.05, |FC| > 1.5) in KPS primary tumors were enriched for genes characterizing the inflammatory response, while those decreased (adjusted p-value < 0.05, |FC| > 1.5) were enriched for genes characterizing xenobiotic metabolism (Supplementary Fig. S4G). Genes constituting a classical epithelial-mesenchymal transition signature were represented in both directions (Supplementary Fig. S4G). Interestingly, the oxidative phosphorylation signature, which was previously found to be the most significant gene set increased in KPS primary tumors (12), was not enriched in our analysis of KPS primary tumors (p = 0.56; FDR = 0.67).
We next sought to understand the extent to which the transcriptional profiles of KPS primary tumors correlated with the chromatin states of KPS primary tumors. To this end, we scored both CCSP and SPC scATAC-seq datasets using the mean gene scores of the most significantly increased (KPS UP) and decreased (KPS DOWN) genes in KPS primary tumors compared to those from KP by RNA sequencing (Fig. 4I and Supplementary Fig. S4H). Subsets of cells within KPS clusters scored highly for the KPS UP signature compared to cells in other clusters. Likewise, cells within KPS clusters tended to have lower gene scores for genes significantly decreased in KPS primary tumors compared to KP cells in the dataset. Furthermore, top genes in either direction (p < 0.00001) displayed similar directionalities in mean gene scores in CCSP KPS samples (Supplementary Fig. S4I). Of note, we could detect one KPS-HET sample that displayed reduced gene scores in top genes decreased in KPS primary tumors. These analyses indicate a degree of correlation between bulk transcript levels and mean gene scores in these datasets.
Smarca4 inactivation directly results in global SWI/SNF loss-of-function leading to reduced chromatin accessibility at lung lineage motifs
We next sought to determine the underlying mechanism behind the altered cell states observed in KPS primary tumors. We chose to focus on the absence of lung lineage TF activities in KPS clusters. Primary tumor cells within KPS clusters, including those representing a KPS cell state early in tumor progression, consistently showed reduced accessibilities of lung lineage TF motifs. Loss of lineage specificity in cancer cells can facilitate the acquisition of cell states that support tumor progression. This particular feature of KPS primary tumor cells may explain the acceleration of lung tumorigenesis resulting in the increased incidence of highly advanced tumors and metastases observed in KPS animals upon Smarca4 inactivation.
We hypothesized that the lack of lung lineage TF activities signified by the decrease of their motif accessibilities in Smarca4-deficient cells may be caused by loss of expression of the TFs themselves or altered SWI/SNF function at their binding sites as a consequence of Smarca4 loss. To discriminate between these possibilities, we first compared Nkx2–1 and Gata6 transcript levels of sorted cancer cells from KP, KPS-HET, and KPS primary tumors. We observed a significant decrease in the expression levels of both TFs in KPS primary tumors by bulk RNA sequencing (Fig. 5A and 5B). Reduced transcript levels may indicate a general reduction in expression of these TFs across all KPS tumors or may reflect the increased frequency of Grade 4 tumors – which typically lose expression of both proteins – in KPS animals. We next quantified NKX2–1 and GATA6 positive cells in KP, KPS-HET, and KPS primary tumors by immunohistochemistry. Among lung tumors initiated from SPC+ and CCSP+ cells in KPS animals, we could detect a striking reduction in NKX2–1 and GATA6 protein expression in SMARCA4-negative Grade 4 tumors as expected. By contrast, these proteins were readily detected in SMARCA4-negative tumors of lower grade (Fig. 5C and 5D; Supplementary Fig. S5A and S5B). Altogether, these data suggest that loss of NKX2–1 and GATA6 protein expression does not primarily drive the general absence of their activities in KPS clusters.
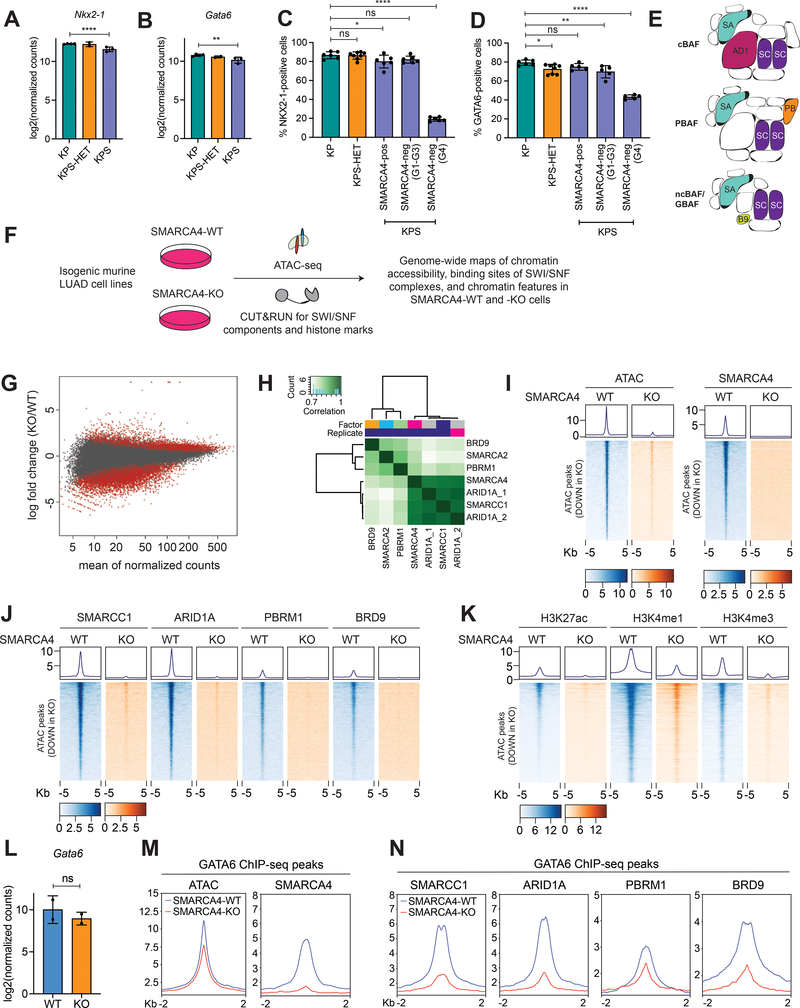
Figure 5. Reduced chromatin accessibility in Smarca4-deficient LUAD is a direct effect of SWI/SNF loss-of-function upon Smarca4 inactivation.
(A) Transcript levels of Nkx2–1 in KP (n=4), KPS-HET (n=2), and KPS (n=3) primary tumors (****pDE = 7.91e-05 by DESeq2). (B) Transcript levels of Gata6 in KP (n=4), KPS-HET (n=2), and KPS (n=3) primary tumors (***pDE = 0.00250 by DESeq2). (C) Quantification of NKX2–1 staining in lung tumors initiated from CCSP+ cells in KP (n=6), KPS-HET (n=8), and KPS (n=6) animals (grouped by SMARCA4 protein expression status and histological tumor grade). The average percentage of NKX2–1-positive cells per tumor in each mouse is shown. Data are mean ± s.d. One-way ANOVA: F(4, 27) = 286.9, p < 0.0001; Dunnett’s multiple comparisons test: *adjusted p-value = 0.0375; ****adjusted p-value < 0.0001. (D) Quantification of GATA6 staining in lung tumors initiated from CCSP+ cells in KP (n=6), KPS-HET (n=8), and KPS (n=5) animals (grouped by SMARCA4 protein expression status and histological tumor grade). The average percentage of GATA6-positive cells per tumor in each mouse is shown. Data are mean ± s.d. One-way ANOVA: F(4, 24) = 56.03, p < 0.0001; Dunnett’s multiple comparisons test: *adjusted p-value = 0.0269; **adjusted p-value = 0.0048; ****adjusted p-value < 0.0001. (E) Schematic drawing of the three major classes of mammalian SWI/SNF complexes. SA = SMARCA2/4, AD1 = ARID1A, SC = SMARCC1, PB = PBRM1, B9 = BRD9. (F) Schematic diagram of the approach to generate genome-wide maps of chromatin accessibility, SWI/SNF binding, and chromatin features in isogenic pairs of Smarca4 wild-type and knockout murine LUAD cell lines. (G) MA plot showing differential bulk ATAC-seq peaks between SMARCA4-WT (n=2) and –KO (n=2) cell lines. Red dots represent differentially enriched peaks (adjusted p-value < 0.05). (H) Representative normalized read-count correlation plot of SWI/SNF subunit CUT&RUN consensus peaks in Smarca4 wild-type murine LUAD cells (SMARCA4-WT). (I) Representative ATAC-seq and SMARCA4 CUT&RUN density metaplots and heatmaps showing chromatin accessibility and SMARCA4 occupancy at differential ATAC-seq peaks at distal intergenic regions significantly reduced in SMARCA4-KO LUAD. (J) Representative CUT&RUN density metaplots and heatmaps showing occupancies of the pan-SWI/SNF component SMARCC1 and SWI/SNF class-specific components ARID1A (cBAF), PBRM1 (PBAF), and BRD9 (ncBAF/GBAF) at differential ATAC-seq peaks at distal intergenic regions significantly reduced in SMARCA4-KO LUAD. (K) Representative CUT&RUN density metaplots and heatmaps showing chromatin features (H3K27ac, H3K4me1, and H3K4me3) at differential ATAC-seq peaks at distal intergenic regions significantly reduced in SMARCA4-KO LUAD. (L) Transcript levels of Gata6 in SMARCA4-WT (n=2) and –KO (n=2) cell lines in RNA-seq dataset. Gata6 is not differentially expressed (pDE = 0.10302 by DESeq2). (M) Representative metaplots showing chromatin accessibility and SMARCA4 occupancy at GATA6 binding sites in SMARCA4-WT and –KO LUAD. (N) Representative metaplots showing occupancies of the pan-SWI/SNF component SMARCC1 and class-specific components ARID1A (cBAF), PBRM1 (PBAF), and BRD9 (ncBAF/GBAF) at GATA6 binding sites in SMARCA4-WT and –KO LUAD.
We next sought to determine the consequences of SMARCA4 loss on the ability of SWI/SNF complexes to bind and open chromatin. SWI/SNF gene products assemble in a combinatorial fashion resulting in three classes of complexes in mammalian cells (36,37): canonical BAF (BAF), polybromo-associated BAF (PBAF), and noncanonical or GLTSCR1/GLTSCR1L-associated BAF (ncBAF/GBAF; Fig. 5E). These complexes have both overlapping and unique subunits, as well as binding sites on chromatin (37,38). As one of two mutually exclusive ATPases that can assemble into all three complexes, SMARCA4 has a central role in SWI/SNF function and directly regulating chromatin accessibility.
To determine the direct effects of Smarca4 loss on chromatin accessibility and SWI/SNF binding in Kras-driven lung adenocarcinoma, we took advantage of isogenic pairs of Smarca4 wild-type and knockout KP cell lines. These lines were generated by transiently expressing Cas9 and a guide RNA for Smarca4 (or control guide) in KP tumor-derived cell lines and screening single cell clones for complete loss of SMARCA4 expression (28). We performed bulk ATAC-seq and CUT&RUN (C&R) epigenomic profiling of SWI/SNF components in two pairs of isogenic Smarca4 wild-type (SMARCA4-WT, n=2) and knockout (SMARCA4-KO, n=2) cell lines generated from two independently derived parental KP cell lines (Fig. 5F). Additionally, we generated genome-wide maps of chromatin features characterizing promoters and enhancers in these cells, as these are the sites predominantly bound by SWI/SNF complexes.
We identified 9,497 differential bulk ATAC-seq peaks between SMARCA4-WT and SMARCA4-KO lines (q < 0.05, LFC > 1; Fig. 5G). Statistically significant differential peaks were over-represented (1.75X) in the down direction in SMARCA4-KO cells (p = 3.8e-199, hypergeometric test) indicating a general compaction of chromatin upon Smarca4 inactivation. Motifs enriched among differential bulk ATAC-seq peaks between SMARCA4-WT and SMARCA4-KO lines were reminiscent of the clearest motif changes we observed in vivo upon Smarca4 loss (AP-1, Ctcf, among others, Supplementary Fig. S5C). This suggests that these changes are direct effects of Smarca4 inactivation, rather than an indirect effect of transformation or tumor progression. To understand the extent to which these cell lines captured the chromatin states observed in vivo, we next scored each cell in both SPC and CCSP scATAC-seq datasets for correlation to the chromatin accessibility profiles of the four single cell clones (Supplementary Fig. S5D). Cells from late KP and metastases clusters generally scored highly for these profiles compared to early KP clusters, consistent with the notion that 2D cell lines resemble late stage tumors and metastases. Cells from KPS clusters also scored highly for these profiles, supporting the idea that Smarca4 loss results in an advanced tumor cell phenotype. Importantly, chromatin states of KPS clusters best correlated with the chromatin accessibility profiles of SMARCA4-KO cells, while those of late KP clusters best matched the chromatin accessibility profiles of SMARCA4-WT cells. These analyses suggest that these cell lines are reasonable models to study the direct impact of Smarca4 loss on SWI/SNF binding to chromatin.
We mapped the genome-wide binding profile of pan-SWI/SNF components SMARCA4 and SMARCC1, as well as subunits distinguishing the three complex classes: ARID1A (cBAF), PBRM1 (PBAF), and BRD9 (ncBAF/GBAF) in SMARCA4-WT and SMARCA4-KO cell lines. Additionally, we mapped the binding profile of SMARCA2, the only other catalytic subunit of SWI/SNF, in order to understand any compensatory effects mediated by SMARCA2 that may occur in the absence of SMARCA4. Peaks of the various SWI/SNF components correlated well with one another in varying degrees, suggesting overlapping binding sites among them in Kras-driven Smarca4-wild-type LUAD (Fig. 5H). Importantly, we detected robust ATAC-seq and SWI/SNF C&R peaks at transcription start sites (TSS) of stably expressed genes in both SMARCA4-WT and –KO cell lines except for SMARCA4 C&R peaks in SMARCA4-KO cells as expected (Supplementary Fig. S5E and S5F), indicating that these are high quality datasets to examine the direct consequences of Smarca4 loss on SWI/SNF function in chromatin regulation.
Differential ATAC-seq peaks that were reduced upon SMARCA4 loss displayed robust SMARCA4 occupancy in SMARCA4-WT cells, demonstrating that these peaks are direct SMARCA4 binding sites (Fig. 5I). There was a clear loss of binding at these sites of the pan-SWI/SNF component, SMARCC1, as well as all three class-specific subunits ARID1A, PBRM1, and BRD9 in SMARCA4-KO cells (Fig. 5J). Of these, ARID1A occupancy displayed the greatest reduction. These changes were accompanied by a loss of primed sites (H3K4me1), as well as enhancer (H3K27ac, H3K4me1) and promoter (H3K27ac, H3K4me3) activities (Fig. 5K). These results are consistent with a model in which SMARCA4 loss results in a defect in the ability of all three major classes of SWI/SNF complexes to bind and open chromatin. Furthermore, the reduction in chromatin accessibility, primed sites, and active regulatory regions appear to be largely a direct effect of the loss of SWI/SNF binding.
We hypothesized that the reduction in chromatin accessibility caused by SMARCA4 inactivation and SWI/SNF loss-of-function occurs at lung lineage motifs, thereby rendering these sites inaccessible to their associated TFs. To test this, we chose to examine previously identified GATA6 binding sites in KP LUAD (39), as these tumor-derived cell lines maintain GATA6 expression, in contrast to NKX2–1 (Fig. 5L; Supplementary Fig. S5G). Consistent with this hypothesis, Smarca4 inactivation resulted in a decrease in chromatin accessibility at GATA6 binding sites, demonstrated by a reduction of ATAC-seq peak strength at these sites in SMARCA4-KO cells compared to SMARCA4-WT cells (Fig. 5M). We observed robust SMARCA4 occupancy at GATA6 binding sites in SMARCA4-WT cells, but not in SMARCA4-KO cells, in line with a direct role for SMARCA4 in remodeling chromatin at these sites (Fig. 5M). Furthermore, occupancies of the pan-SWI/SNF subunit SMARCC1 as well as class-specific subunits ARID1A, PBRM1, and BRD9 likewise were significantly reduced at GATA6 binding sites in the absence of SMARCA4 (Fig. 5N). Thus, we conclude that the loss of GATA6 activity in Smarca4-deficient cells is largely caused by the inability of SWI/SNF complexes to bind and open chromatin at GATA6 binding sites upon Smarca4 inactivation. The reduction in chromatin accessibility at these sites would, in turn, prohibit GATA6 from dictating transcriptional programs that maintain cell identity.
We next examined ATAC-seq peaks increased upon SMARCA4 loss to determine whether these changes are also a result of altered SWI/SNF binding (Supplementary Fig. S5H). Gain of SWI/SNF function has been described as a key driver of malignancy in other SWI/SNF-mutant cancers, most notably synovial sarcoma (40). However, in our datasets the majority of gained peaks did not display increased SMARCC1, ARID1A, PBRM1 or BRD9 occupancy (Supplementary Fig. S5I), indicating that increased accessibility in SMARCA4-KO cells is not a direct result of increased SWI/SNF activity at these sites. Instead, we observed a slight reduction in the occupancies of these components at these sites. Thus, increased accessibility upon SMARCA4 loss may be a secondary effect or due to loss of SWI/SNF-mediated chromatin compaction.
Next, we examined the binding profile of SMARCA2, a paralog of SMARCA4, and the only other SWI/SNF subunit with catalytic activity. SMARCA2 has been previously identified to be a synthetic lethal target in SMARCA4-deficient cancers (41,42), and SMARCA2 inhibitors have been developed as a potential targeted therapy for SMARCA4-mutant cancers (43). We detected modest changes in SMARCA2 occupancy among differential ATAC-seq peaks (Supplementary Fig. S5J). However, we did detect 138 differential SMARCA2 peaks between SMARCA4-WT and –KO cell lines, 137 of which were increased in SMARCA4-KO cells (Supplementary Fig. S5K). The increase in SMARCA2 peaks occurred at direct SMARCA4 binding sites, was accompanied by ATAC-seq peaks, and was strongly enriched for AP-1 TF family motifs (Supplementary Fig. S5L and S5M). SMARCA4 loss, therefore, results in some compensatory chromatin remodeling activity by SMARCA2 at certain TF binding sites.
SMARCA4-mutant LUAD show heterogeneous chromatin states
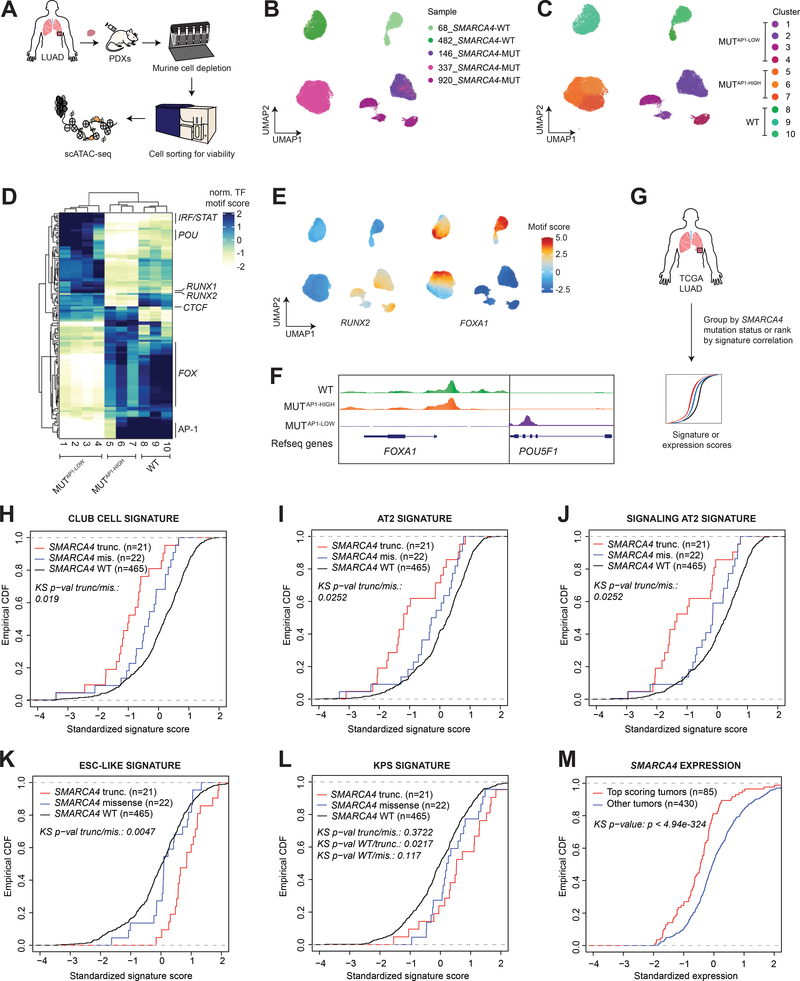
We next sought to determine whether the TF-directed programs we observed in our murine models recapitulated those in LUAD patients harboring loss-of-function SMARCA4 mutations. We profiled the chromatin accessibility of single cells from PDX models of human KRAS-mutant lung adenocarcinomas with intact SMARCA4 (n=2), and those with biallelic inactivating SMARCA4 alterations (n=3) identified through MSK-IMPACT (Supplementary Table S1) (44). We generated chromatin accessibility profiles from 30,992 single cells following murine cell depletion and cell sorting for viability (Fig. 6A and 6B; Supplementary Fig. S6A–S6C). Each PDX model clustered independently, suggesting that these patient samples have evolved and selected for distinct epigenetic states (Fig. 6C). Despite these differences among patients, we identified programs broadly characterizing these clusters that were associated with SMARCA4 status (Fig. 6D). We therefore grouped these clusters into three categories: SMARCA4-MUTAP−1-low (1–4), SMARCA4-MUTAP−1-high (5–7), and SMARCA4-WT (8–10, Fig. 6C and 6D). Importantly, all three SMARCA4-mutant samples were represented in the SMARCA4-MUTAP−1-low group, but not in the SMARCA4-MUTAP−1-high group.
Figure 6. Human SMARCA4-mutant LUAD recapitulates key features of the murine model.
(A) Workflow of the experimental strategy to isolate cancer cells from SMARCA4 wild-type (n=2) and mutant (n=3) LUAD PDX models for scATAC-seq. UMAP projection of scATAC-seq profiles of 30,992 cancer cells colored by (B) sample and (C) cluster. (D) A heatmap showing the top variable accessible motifs across the dataset. Heatmap is colored by row-normalized mean motif scores of each cell cluster identified through the Louvain modularity method. (E) UMAP projection of scATAC-seq dataset colored by RUNX2 and FOXA1 motif scores (F) Aggregated scATAC-seq tracks showing marker chromatin accessibility peaks per group. (G) Schematic diagram of TCGA LUAD analyses. Empirical cumulative distribution function (CDF) plot of standardized 10x club cell (H), 10x AT2 (I), 10x signaling AT2 (J), ESC-like (K), and KPS (L) signature scores in TCGA LUAD patients with intact SMARCA4 (WT), SMARCA4 truncating mutations, and SMARCA4 missense mutations. (M) Empirical CDF plot comparing standardized SMARCA4 expression between top-scoring TCGA LUAD correlated with the KPS signature (z score > 1) and the rest of the cohort.
Marker motifs for these clusters revealed that a subset of SMARCA4-mutant PDXs, in particular those with p53 pathway inactivation (SMARCA4-MUTAP−1-low), recapitulated key features of murine KPS clusters (Fig. 6D; Supplementary Fig. S6D). These had low activity for the AP-1 TF family, and increased accessibilities for RUNX2 and IRF1 motifs. They were also depleted for peaks harboring the FOX TF family motifs and enriched for activities of POU TFs, indicating a highly undifferentiated cell state (Fig. 6D and 6E) consistent with our findings in murine models of SMARCA4 loss. Changes in FOX and POU TF motif accessibilities in SMARCA4-MUTAP−1-low clusters were accompanied by a loss and gain, respectively, in peaks of the TFs themselves (Fig. 6F). Notably, individual SMARCA4-mutant samples belonging to the SMARCA4-MUT-AP-1low group tended to generate multiple clusters, demonstrating a substantial level of heterogeneity of epigenetic states within individual SMARCA4-deficient patient samples.
Transcriptional profiles of SMARCA4-mutant LUAD are poorly correlated with club cell and AT2 signatures and are enriched for an embryonic stem cell-like signature
We next turned to The Cancer Genome Atlas (TCGA) (6) to explore the relevance of our models to human LUAD by examining a larger set of SMARCA4-mutant tumors (Fig. 6G). We first investigated whether we could detect indications of a club cell-of-origin specifically in human SMARCA4-mutant LUAD. We examined the expression levels of SCGB1A1 and SFTPC, two frequently used markers that distinguish the club cell and AT2 lineages, respectively, in TCGA LUAD grouped by SMARCA4 mutation status (SMARCA4 wild-type, SMARCA4 missense mutant, and SMARCA4 truncating mutant), and observed no differences in their expression levels among the groups (Supplementary Fig. S7A and S7B). We next scored the transcriptional profiles of these tumors for club cell and AT2 signatures derived from an extensive single cell RNA-sequencing study of the human lung (45). In these analyses, we included signatures associated with canonical AT2s, as well as signaling AT2s, a distinct subset of AT2s that express genes involved in Wnt signaling defined by this study (45).
Transcriptional profiles of SMARCA4 truncating mutant tumors had significantly decreased correlation with all three cell type-specific signatures derived from both 10x Chromium and SmartSeq2 (SS2) platforms compared to the transcriptional profiles of tumors with intact SMARCA4 (Fig. 6H–J and Supplementary Fig. S7C–E). Importantly, these differences were maintained when comparing SMARCA4-wild-type and -mutant TCGA LUAD samples matched by tumor grade (Supplementary Fig. S7F–N). Altogether, these analyses indicate that SMARCA4-mutant LUAD, specifically those with SMARCA4 truncating mutations, correlate poorly with gene sets that are associated with either putative cell-of-origin.
We reasoned that these results may reflect SMARCA4-mutant tumors being more undifferentiated than their Smarca4-wild-type counterparts – in line with observations from phenotypic analyses and chromatin profiling of the murine models of Smarca4-deficient LUAD. We therefore scored these tumors for a core embryonic stem cell (ESC)-like gene module, which consists of genes upregulated in both mouse and human embryonic stem cells (46). Interestingly, the ESC-like gene expression signature exhibited strikingly increased correlation with the transcriptional profiles of SMARCA4 truncating mutant tumors compared to those with SMARCA4 missense mutations or intact SMARCA4 (Fig. 6K), a trend not observed when examining a cell proliferation signature (Supplementary Fig. S7O). When tumors were stratified by their correlation with the ESC-like gene module, a substantial fraction of top-scoring tumors (z > 1) had significantly reduced SMARCA4 expression (Supplementary Fig. S7P). Furthermore, top-scoring tumors were specifically enriched for SMARCA4 truncating mutations (p = 4.75e-03), but not missense mutations (p = 0.38). Collectively, these data show that the transcriptional profiles of SMARCA4 truncating mutant tumors are not only significantly less associated with lineage signatures, but also enriched for an embryonic signature – pointing towards a poorly differentiated state in SMARCA4-deficient LUAD.
Murine-derived KPS signature is enriched in human LUAD harboring SMARCA4 truncating mutations
Finally, we sought to determine whether the KPS transcriptional signature (q < 0.05, |FC| > 1.5) derived from our murine model captured the transcriptional profiles of human SMARCA4-mutant LUAD. The KPS signature was strongly associated with the transcriptional profiles of TCGA LUAD harboring SMARCA4 truncating mutations (Fig. 6L), indicating that this signature consistently characterizes both SMARCA4-deficient murine and human LUAD. When we stratified patients according to the correlation of the transcriptional profiles of their tumors to the KPS signature, top-scoring patients (z > 1) showed no differences in 5-year or overall survival compared to the rest of the cohort (Supplementary Fig. S7Q and S7R). However, these patients had significantly reduced SMARCA4 expression (Fig. 6M), and were strikingly enriched for SMARCA4 truncating mutations (p = 3.18e-03), but not missense mutations (p = 0.29). These analyses demonstrate that the murine models and datasets we have generated are relevant to human SMARCA4-deficient LUAD.
Discussion
SMARCA4, a catalytic component of the SWI/SNF chromatin remodeling complex, is among the top mutated genes in lung adenocarcinoma and its mutation is a major predictor of poor patient survival. However, the functional impact of SMARCA4 loss-of-function on tumor initiation, progression, and the chromatin landscape in lung cancer has been unclear to date.
Our experiments in autochthonous mouse models of LUAD identify CCSP+ lung cells to be uniquely sensitive to malignant transformation upon Smarca4 loss. SMARCA4-negative tumors initiated from SPC+ lung cells in KPS animals were either low grade or highly advanced, which can be explained by a heterogeneous population of SPC-expressing cells – including AT2 and BASCs – transformed at tumor initiation. Our results suggest that these cell types have differential inherent sensitivities to Smarca4 perturbation. Analysis of our scATAC-seq dataset supports a club cell state for high grade SMARCA4-deficient tumors in this model. Indeed, when we initiated tumors in KPS animals from CCSP-expressing cells, which are predominantly club cells, we observed a consistent increase in tumor number, grade and metastatic incidence, as well as a shorter overall survival for KPS animals. Collectively, these results show that the club cell state is uniquely sensitive to malignant transformation and tumor progression upon Kras activation and Trp53 loss in the absence of Smarca4 function.
This particular cell state can be adopted by BASCs upon differentiation. These cells have been shown through elegant lineage tracing experiments to have the ability to repopulate the club cell population upon injury (32,33). Alternatively, transformed AT2 cells in KPS animals may also transdifferentiate into a club cell state following tumor initiation in the context of Smarca4 loss. This possibility can be directly addressed by lineage tracing experiments; however, our data suggest that this is predominantly not the case. SMARCA4-negative high grade tumors arising from SPC-expressing cells in KPS animals were typically found by the airways where BASCs reside, while SMARCA4-negative low-grade tumors were located in the alveolar space. This supports a cell-of-origin switch and not an AT2 to club cell transdifferentiation event that gives rise to SMARCA4-negative high grade tumors initiated from SPC+ cells in KPS animals.
The phenotypes we describe in the SPC and CCSP models corroborate aspects of previous studies that investigated the effects of Smarca4 inactivation on tumor progression in Kras-driven GEMMs of LUAD (11,12). Cell-type specificity in the tumor suppressive function of Smarca4 in the lung provides a potential explanation for seemingly contradictory results in these studies, which used ubiquitous promoters to drive Cre expression and initiate tumors. The effects of Smarca4 loss on overall tumor progression would be dependent on the fraction of transformed cells that were singly SPC+ or CCSP+. These likely vary depending on the relative efficiencies of the method of viral transduction in these cell types, as well as the relative activities of the promoter used to drive Cre expression in SPC+ and CCSP+ cells. Our results in the CCSP model are consistent with the accelerated tumorigenesis observed by others in KPS animals in which tumors were initiated using adenoviral delivery of Cre (12). While we do not observe an enrichment for the oxidative phosphorylation signature described in this study in KPS tumors from our model, we do observe an association between this signature and SMARCA4 truncating mutant tumors in TCGA LUAD (Supplementary Fig. S7S). Interestingly, our results in the SPC model are also in line with the restrained tumor progression observed in Kras-driven GEMMs in which Smarca4 loss-of-function mutations were generated by CRISPR-Cas9 (11). We speculate that the majority of transformed cells harboring Smarca4 mutations in this case were AT2 cells, which would be consistent with previous work demonstrating this cell type to be the predominant cell-of-origin in this model (19,29).
The mutation spectrum of SWI/SNF subunits is distinct across cancer types, indicating that the requirements for subunit function are highly context-specific. Our results indicate that within murine LUAD, a distinct cell-of-origin underlies Smarca4 mutants. Our findings demonstrate cell-type specificity in the tumor suppressive function of Smarca4 and identify the requirements for cell state that are permissive for transformation upon Smarca4 mutation in the lung. Investigation of Smarca4 and SWI/SNF function in AT2 and club cells under normal physiologic conditions and in response to oncogenic stress will be key to understanding the differences underlying the sensitivities of these cell types to undergo malignant transformation upon Smarca4 inactivation. Interestingly, Kras-driven lung adenocarcinomas harboring Keap1 loss also display a bronchiolar cell-of-origin (47). KEAP1 mutations are strongly associated with SMARCA4 mutations in lung adenocarcinoma patients (8), leading us to speculate that most human tumors harboring both SMARCA4 and KEAP1 mutations arise from cells of the club cell lineage. Though we were unable to detect a specific enrichment of a club cell signature in SMARCA4-deficient human LUAD – presumably due to these tumors being highly undifferentiated – we anticipate that determining the cell-of-origin of distinct molecular subtypes of the human disease will be increasingly addressable as more extensive and sophisticated molecular profiling and analyses of both normal cell types and tumors throughout tumor evolution are performed.
Importantly, our studies also provide mechanistic insights into the tumor suppressive functions of Smarca4 in the lung, particularly in chromatin regulation. We show that Smarca4-deficient cancer cells have a chromatin state resembling that of metastatic cells. In contrast to Smarca4-intact primary tumors, which undergo a gradual loss of lung epithelial cell identity and lineage fidelity (20,22–24), Smarca4-deficient primary tumors largely lack activities of lung lineage TFs, similar to metastatic cells. SMARCA4 loss directly results in a defect in the ability of all three classes of SWI/SNF complexes (cBAF, PBAF, ncBAF/GBAF) to bind and open chromatin in regulatory regions, including target sites of lung lineage TFs. Among these, the most profound change occurs at cBAF binding sites. Inaccessible chromatin as a direct consequence of Smarca4 inactivation and SWI/SNF loss-of-function would directly hamper the ability of lung lineage TFs, such as GATA6, to maintain cell identity. Additionally, TF programs known to be active in metastatic-like and metastatic cells (20,25) are highly enriched in subsets of primary Smarca4-deficient tumor cells.
Primary lung adenocarcinomas display aberrant expression of transcription factors specifying epithelial lineages, while metastases exhibit transcriptional programs characteristic of a stem-like or progenitor state (25). Loss of the final differentiation state of the tumor cell-of-origin and the acquisition of plasticity in cancer cells enables the adoption of progenitor-like states or alternative differentiation states. Such phenotypic plasticity is thought to support malignancy throughout tumor evolution (48). That Smarca4-deficient primary tumors efficiently lose cell identity is a potential explanation for the rapid acquisition and selection of a cell state associated with highly advanced tumors and metastases and the acceleration of tumor progression observed in KPS animals.
Taken together, these data demonstrate a direct role for SMARCA4 loss in driving an aggressive malignant phenotype that underlies the poor prognosis of this molecular subtype of LUAD. More broadly, these data are in line with other highly undifferentiated SMARCA4-mutant malignancies observed in patients including small-cell-carcinomas of the ovary, hypercalcemic type (SCCOHT) (49–51) and thoracic sarcomatoid tumors, which are thought to represent undifferentiated or de-differentiated lung carcinomas (52,53).
Our work has focused on complete Smarca4 loss in conjunction with Kras activation and Trp53 loss in LUAD. Indeed, the KPS signature derived from our model best captures the tumor expression profiles of TCGA LUAD patients harboring SMARCA4 truncating mutations. Future studies modeling recurrent SMARCA4 missense mutations, which have been shown to have dominant negative and gain-of-function effects (54,55), will be crucial to understand their specific effects on lung cancer evolution. Additionally, modeling Smarca4 mutations in combination with other frequently co-occurring genetic alterations, such as Keap1 and Stk11 mutations, will be critical to assess the impact of SMARCA4 mutations in other contexts and to expand our repertoire of relevant Smarca4-mutant preclinical models that can be used to test therapeutic strategies.
While we have focused on the consequences of Smarca4 loss on tumor cell state and SWI/SNF function in chromatin regulation in this work, these models and datasets are also poised to assess other functions of SWI/SNF, such as maintenance of genomic stability upon Smarca4 loss and to examine potentially altered interactions among Smarca4-deficient lung cancers and the tumor microenvironment during tumor evolution. Furthermore, the CCSP KPS model is a useful preclinical platform to evaluate various therapeutic approaches that have been proposed for SMARCA4-mutant LUAD.
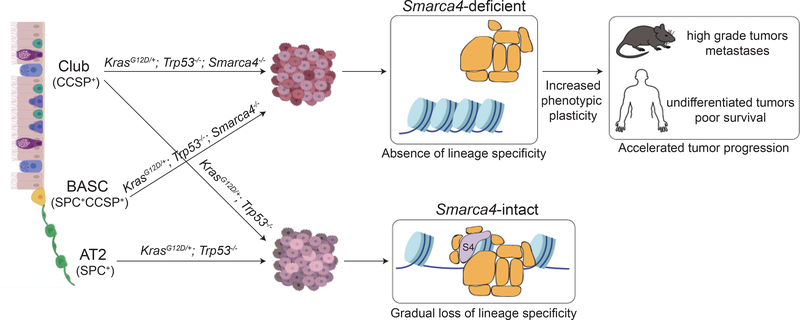
In sum, this work puts forth a model wherein SMARCA4 loss in transformed CCSP+ cells directly results in the inability of SWI/SNF complexes to bind to chromatin, and eject and mobilize nucleosomes, which prohibits lung lineage TFs from exerting lineage-specifying gene expression programs (Fig. 7). Absence of lineage specificity in turn promotes phenotypic plasticity of Smarca4-deficient cells, and accelerates the sampling and selection of pro-tumorigenic states throughout tumor evolution. Ultimately, this drives the increased incidence of high-grade tumors and metastases in Smarca4-deficient murine LUAD, and highly undifferentiated tumors and poor overall survival in NSCLC patients harboring SMARCA4 inactivating alterations. Collectively, this work provides a global view of Smarca4-mediated tumor suppression in the lung.
Figure 7. A model for Smarca4-mediated tumor suppression in the lung.
A club cell state is highly sensitive to malignant transformation upon oncogenic Kras activation and Trp53 deletion in the absence of Smarca4. Apart from club cells (blue), BASCs (yellow) can also adopt this state. Absence of Smarca4 in transformed cells results in the inability of SWI/SNF complexes to bind to chromatin, and mobilize and eject nucleosomes. Loss of SWI/SNF function prohibits lung lineage TFs from activating lung lineage gene expression programs. Absence of lineage specificity leads to increased phenotypic plasticity throughout tumor evolution, enables rapid selection of pro-tumorigenic programs, and accelerates tumor progression in the absence of Smarca4.
Methods
Mice
Mouse strains used in this study were previously published: KrasLSL-G12D (56), Trp53fl (57), Smarca4fl (16), Rosa26LSL-tdTomato (58). Mice were maintained in a mixed Sv129/C57BL/6 genetic background. Tumors were initiated using 1.0 or 2.5 X 108 plaque-forming units (PFU) of Ad-SPC-Cre or 1.0 X 108 PFU of Ad-CCSP-Cre (18) from the Viral Vector Core of the University of Iowa through intratracheal instillation as previously described (59) in age-matched (~8–12 weeks of age), sex-matched littermate cohorts. Animal health was monitored daily by the investigators and/or veterinary staff at the Department of Comparative Medicine at MIT. Cohorts of mice were euthanized by CO2 inhalation or cervical dislocation. Mice were euthanized at defined time points (17 weeks post-infection for Ad-SPC-Cre-infected animals in which 1.0 X 108 PFU per mouse was used, 14 weeks post-infection for Ad-SPC-Cre-infected animals in which 2.5 X 108 PFU per mouse was used, and 16 weeks post-infection for Ad-CCSP-Cre-infected animals in which 1.0 X 108 PFU per mouse was used) or upon reaching a body condition score under 2 for long-term studies. Animal studies were approved by the MIT Committee for Animal Care.
Immunohistochemistry and histological analyses
Lung tissues were perfused with PBS through the heart and inflated with zinc formalin through the trachea. Tissues were then fixed overnight in zinc formalin, transferred to 70% ethanol, and embedded in paraffin. Tissues in the chest cavity of each mouse were also fixed and paraffin-embedded to identify micrometastases. Sections were cut at 4 μm thickness and stained for H&E for histological examination. For immunohistochemistry (IHC), slides were dewaxed and antigen retrieval was performed using citrate buffer (pH 6.0). Endogenous peroxidase was blocked using DAKO Dual Endogenous Enzyme Block and endogenous species protein was blocked using the appropriate species serum depending on the secondary antibody. Tissues were incubated with primary antibodies overnight. Primary antibodies used are: anti-SMARCA4 (Abcam Cat# ab110641, RRID:AB_10861578, 1:500), anti-Ki67 (Cell Signaling Technology, #12202, 1:200), anti-NKX2–1 (Abcam Cat# ab76013, RRID:AB_1310784, 1:1000), anti-GATA6 (Cell Signaling Technology, #5851, 1:400). ImmPRESS HRP secondary antibodies and the DAB Peroxidase Substrate Kit (Vector Laboratories) were used for signal detection. Tissues were counterstained with hematoxylin. Histological quantification of tumor area, tumor grade, and lung area was performed in H&E-stained sections using an automated deep neural network developed by Aiforia (nsclc_V25 or nsclc_V37) in collaboration with the Jacks and Tammela labs (20) under the guidance of Dr. Roderick T. Bronson. Quantification of the number of tumors per mouse was performed in H&E-stained sections in a blinded manner using QuPath (60). Identification of metastatic lesions in animals was performed by microscopic examination of H&E-stained sections of paraffin-embedded tissues of the chest cavity including thymus and lymph node. All H&E slides were independently examined by an expert in mouse pathology (R.T.B), who identified slides with Grade 4 tumors and metastases in a blinded manner. Classification of SMARCA4 protein expression status in tumors in KPS animals, measurements of tumor size, and percentages of Ki67-positive cells in Ad-SPC-Cre-infected KPS animals were determined using Aperio ImageScope (v12.3.2.8013). Measurements of the percentages of NKX2–1 and GATA6 positive cells were performed using QuPath (60).
Laser capture microdissection, DNA extraction, and genotyping PCR of tumors
SMARCA4-positive tumors in KPS animals identified by IHC were laser-capture microdissected from paraffin sections using the Veritas Laser Capture Microdissection microscope. DNA was extracted from individual tumors using the Arcturus PicoPure DNA Extraction Kit (Applied Biosystems). Dissected tumors sections were incubated in extraction solution containing Proteinase K at 65°C overnight, spun down, incubated at 95°C for ten minutes, and cooled. The samples were directly subjected to published genotyping protocols to identify Smarca4 recombined and floxed alleles (16).
Isolation of primary murine lung adenocarcinoma cells and metastases
Tumor-bearing lungs and macrometastases from moribund KP; Rosa26LSL-tdTomato, KPS-HET; Rosa26LSL-tdTomato, and KPS; Rosa26LSL-tdTomato animals were dissociated using the Miltenyi Biotec Lung Dissociation Kit (130–095-927). Tissues were submerged in Enzymes A and D diluted in 1X Buffer S, mechanically dissociated using dissecting scissors, and incubated at 37°C for 25 minutes with rotation. The dissociated cells were then filtered using a 100 μm strainer. Red blood cells were lysed using ACK (Thermo Scientific), and stained with APC-conjugated anti-CD31 (BioLegend Cat# 102510, RRID:AB_312917, 1:500), anti-CD45 (BD Biosciences Cat# 559864, RRID:AB_398672, 1:500), anti-CD11b (eBioscience, 17–0112-82, 1:500), and anti-TER119 (BD Biosciences Cat# 557909, RRID:AB_398635, 1:500). DAPI was used for live/dead staining. Fluorescence-activated cell sorting (FACS) was performed using a FACSAria sorter (BD) to isolate DAPI−/tdTomato+/APC− cancer cells. Isolated cancer cells were then subjected to sciATAC or Chromium Single Cell ATAC protocols. Alternatively, single cell suspensions were frozen in freezing media (DMEM supplemented with Penicillin-Streptomycin, 20% fetal bovine serum, and 10% dimethyl sulfoxide) prior to antibody staining, sorting, and scATAC-seq or RNA extraction for bulk RNA-seq at a later date.
Isolation of PDXs
SMARCA4-intact and –mutant LUAD PDXs, identified through MSK-IMPACT (44), were maintained in NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (Jackson Laboratories Stock #5557) (61). Tumor cells were thawed from frozen stocks and washed with PBS. Human cells were enriched using the Miltenyi Biotec Mouse Cell Depletion Kit (130–104-694). Cells were washed with column buffer, incubated with mouse cell depletion cocktail, and subjected to magnetic separation using an LS column. Human cells from the flow through were collected and stained with DAPI. Live cells were sorted using the a FACSAria sorter (BD), and subjected to the Chromium Single Cell ATAC protocol.
Chromium Single Cell ATAC of murine primary tumor-derived cancer cells and PDXs
Nuclei from sorted cells were isolated using the low cell input nuclei isolation protocol from 10x Genomics. Cells were spun at 300 rcf for 5 mins at 4°C and resuspended in 50 μl PBS + 0.04% BSA. Cells were spun at 300 rcf for 5 mins at 4°C, and 45 μl supernatant was removed prior to addition of 45 μl chilled Lysis Buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% NP-40, 0.01% Digitonin, 1% BSA), gentle pipetting, and incubation on ice for 6 mins. 50 μl of chilled Wash Buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% BSA, 0.1% Tween-20) was added, and the sample was spun at 500 rcf for 5 mins at 4°C prior to removal of 95 μl of supernatant. Nuclei were washed with 45 μl chilled Diluted Nuclei Buffer, spun at 500 rcf for 5 mins at 4°C, and resuspended in 7 μl chilled Diluted Nuclei Buffer prior to counting. After counting, nuclei were diluted to capture 2000–6000 nuclei per sample. 5 μl of nuclei was mixed with the ATAC Buffer and the Tn5 transposase and incubated for 60 mins at 37°C. Further processing of the sample and library generation was performed as described in the 10x Genomics Single Cell ATAC Regent Kits User Guide.
Single-cell ATAC-seq with combinatorial indexing of murine metastasis-derived and primary tumor-derived cancer cells
Fixation:
Sorted cancer cells were transferred to Eppendorf tubes precoated with 7.5% BSA and pelleted by centrifugation at 300 rcf for 3 minutes. The cell pellets were resuspended in 100 μl cold PBS and counted. Samples with a cell number over 60,000 were performed with fixation, and samples with a lower cell number were not fixed and immediately proceeded to the transposition step. Cells were fixed by adding 6.7 μl 1.6% formaldehyde to a final concentration of 0.1% and incubated at room temperature for 5 minutes. Fixation was stopped by adding 5.6 μl 2.5M glycine, 5 μl 1M pH 8.0 Tris, and 1.3 μl 7.5% BSA followed by incubation on ice for 10 minutes. The cells were pelleted by centrifugation at 500 rcf for 3 minutes at room temperature. Cells were gently washed twice with 0.5 mL PBS by pipetting against the side of the tube without resuspending the pellet followed by centrifugation at 500 rcf for 3 minutes.
Transposition:
1 μl fixed cells were distributed across a 96 well plate and combined with 7 μl transposition buffer (41.25 mM Tris-acetate, 82.5 mM K-acetate, 12.5 mM Mg-acetate, 20% DMF, 0.125% NP-40, 0.5% Protease Inhibitor Cocktail) and incubated at room temperature for 10 minutes. The assembled Tn5 was diluted 1:1 by adding 8 μl transposition buffer to 8 μl of the assembled oligo containing Tn5 as previously described (20). 1 μL of the Tn5 containing Ad1 and 1 μL of the Tn5 containing Ad2 were added to each well. The transposition reaction was carried out at 37°C at 300 rpm for 30 minutes. The reaction was stopped by adding 1 μl 0.5M EDTA to each well, mixing well, followed by incubation at 37°C for 15 minutes at 300 rpm. All 96 reactions were pooled and combined with 38.4 μl MgCl2. The sample was transferred to an Eppendorf tube precoated with 7.5% BSA. The sample was pelleted by centrifugation at 500 rcf for 2 minutes, then washed in 1mL Nuclei Isolation Buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40). The pellet was resuspended in 0.5 mL Nuclei Isolation Buffer and passed through a 40 μm filter then diluted to 13.3 cells/μl.
Reverse crosslinking and PCR:
1.5 μl of the sample was distributed across a 96-well plate and was combined with 2.5 μl reverse crosslink buffer (100 mM Tris pH 8.0, 400 mM NaCl, 2 mM EDTA pH 8.0, 2% SDS, and 40 mg/ml proteinase K), 0.5 μl of 10 μM P1 oligo and 0.5 μl of 10μM P2 oligo. Reverse crosslinking was carried out at 55°C for 16 hours in a thermal cycler. 5 μl of 10% Tween20 was added to quench SDS. PCR reactions were carried out by adding 12.5 μl 2x NEBNext PCR mix and 2.5 μl water to each well, then using the following conditions: 72°C for 5 min (extension), 98°C for 5 min, and thermocycling at 98°C for 10 sec, 70°C for 30 sec, and 72°C for 1 min. After thermocycling for 5 cycles, 5 μl from 4 randomly chosen wells was used to perform qPCR. 10 μl PCR mix with 0.6x SYBRgreen was added and cycled through the following conditions: 98°C for 30 sec (initial extension) and thermocycling at 98°C for 10 sec, 70°C for 30 sec, and 72°C for 1 minute for 25 cycles to determine the number of additional cycles required for the remaining samples on the plate. Libraries were amplified for 13–14 cycles in total. The libraries from each plate were pooled and purified using the Qiagen MinElute PCR purification column. Libraries were quantified using the KAPA library quantification kit and then sequenced on the Next-seq platform (Illumina) using a 150-cycle kit.
scATAC-seq data analysis
Single cell ATAC sequencing data were pre-processed using Cell Ranger ATAC to generate fragment files after removing duplicates. The reads were aligned to either mm10 or hg19 genome. The fragment files for each sample were used as input for peak calling with MACS v2.1.2 (62). All default options were used, with the following flags explicitly set: --nomodel, --nolambda, --keep-dup all, --call-summits. The peak summits were merged and padded with 150 base pairs (bp) at either end to obtain fixed-width peak windows. Only peaks with smallest p values were kept if peaks overlapped. Using the generated peak region list, the number of reads overlapping a given peak window was determined for each unique cell barcode tag. This generated a peak by cell counts matrix corresponding to ATAC reads in peaks for each cell profiled. The peak X cell count matrix was used to generate TF motif score X cell matrix using chromVAR (63).
Single-cell clustering and visualization
The dimension reduction of scATAC data was performed using cisTopics (64). The Uniform Manifold Approximation and Projection (UMAP) algorithm (65) was then applied to project single cells in two dimensions using the topics from cisTopics. To further cluster cell populations, the Louvain method for network community detection, a heuristic method based on modularity optimization (21) was then applied on a k-nearest neighbor (KNN) graph built using the topics and visualized in the original UMAP space.
Murine AT2 and club cell signature scores
We calculated the gene scores by summing the reads intersecting the gene body and promoter region (2 kb upstream of TSSs). To reduce sequencing depth bias, we then normalized gene scores by the total gene score per cell. We then selected the top 30 marker genes of each cell type based on a previous single-cell RNA-seq dataset (35) and defined the gene module score by averaging gene scores of selected genes for each single cell. P values are from Student’s t-test.
Matching scATAC-seq profiles to cellular identity
Previously identified normal lung scATAC-seq profiles were utilized to identify club and AT2-specific gene scores and motif signatures (20). To match each cell to a meta-cluster, scATAC-seq profiles were filtered for highly variable gene scores. The coefficient of variation (CV) for each gene was determined and filtered for genes with a CV > 1. Then, the most correlated (Pearson) meta-cell was used for cell type matching. Absolute differential gene scores were computed between club and AT2 cell types and determined as significant with a p-value < 0.01 and an absolute difference on > 1.5 between club and AT2 genes scores or motifs.
Scoring scATAC-seq datasets with bulk ATAC-seq profiles from cell lines
Peaks were first called on each bulk ATAC sample using MACS v2.1.2 (62) with the following flags explicitly set: --nomodel, --nolambda, --keep-dup all. We then used the getAnnotations function from chromVAR package (66) to find the overlap between bulk ATAC peaks and scATAC peaks. Next, we derived z score for each bulk ATAC peak set using the computeDeviations function from chromVAR, which compares the peak counts to GC matched background peak counts. The z score was further smoothed over 50 nearest neighbors and painted on UMAP.
Scoring scATAC-seq datasets with a KPS signature derived from bulk RNA sequencing
We scored scATAC cells with gene scores (see Signature Score section) of differential genes from bulk RNA-seq. To do this, we first identified significantly differential genes (DE genes) between KP and KPS using DESeq2 (adjusted p-value < 0.05, |FC| > 1.5) (67). Then we calculated the mean gene scores for each cell for the DE genes that are either up in KP or KPS. To visualize the score on scATAC-seq UMAP, we further smoothed the mean gene score over 50 nearest neighbors and painted on UMAP.
Cell lines and tissue culture
Isogenic pairs of Smarca4-deficient and –intact cell lines were generated in the Jacks laboratory from two independent KP tumor-derived cell lines and were previously described (28). T2 (SMARCA4-WT) and M (SMARCA4-KO) were generated from parental line LG1233. 36 (SMARCA4-WT) and 23 (SMARCA4-KO) were generated from parental line LG1234. Cell lines were authenticated by performing Western blots for SMARCA4 expression using both N- and C-terminal antibodies for SMARCA4 and by RNA-sequencing. Cells were maintained in DMEM (Corning 10–013-CV) supplemented with 10% fetal bovine serum and 1X Penicillin:Streptomycin solution (VWR 45000–652). All experiments were performed within 4–5 days from the time of thawing. Cell lines tested negative for Mycoplasma by the MycoAltertTM Kit from Lonza (10/17/17).
RNA extraction
RNA from isogenic cell lines was extracted from cell lines using TRIzol as per manufacturer’s instructions. RNA from sorted primary tumors was extracted using the Qiagen RNeasy Micro Kit (Cat # 74004) according to manufacturer’s instructions.
Bulk ATAC-seq
Bulk ATAC-seq was performed on cell lines as previously published (68).
CUT&RUN epigenomic profiling
CUT&RUN for SWI/SNF components and histone marks was performed as previously described (69).
Bead activation:
Concanavalin A-coated magnetic beads (Polysciences 86057–3) were activated by washing twice with 1 ml binding buffer (20 mM HEPES pH 7.9, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2). 10 μl of bead suspension was used per condition.
Binding of cells to activated beads:
Cells (5 X 105 per condition) were washed twice with 1.5 ml of wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 1X protease inhibitor), and resuspended in 1 ml of wash buffer. Activated bead suspension was added to each sample. Samples were then rotated at room temperature for 10 minutes.
Cell permeabilization and primary antibody binding:
Samples were subjected to a quick spin and placed on a magnet stand. The liquid was removed and discarded. 50 μl of the antibody solution (1:50 dilution of antibody in 0.1% digitonin-wash buffer and 2 mM EDTA) was then added to each sample and mixed. Samples were then rotated at 4°C overnight. The following day, the tubes were subjected to a quick spin and placed on a magnet stand. The liquid was removed and discarded, and the beads were washed twice with 1 ml of 0.1% digitonin-wash buffer (wash buffer + 0.1% digitonin) and resuspended in 50 ul of 0.1% digitonin-wash buffer. The following antibodies were used: anti-SMARCA4 (Abcam Cat# ab110641, RRID:AB_10861578), anti-SMARCA2 (Abcam Cat# ab15597, RRID:AB_443214), anti-ARID1A (Abcam, ab217154 and Cell Signaling Technology Cat# 12354, RRID:AB_2637010), anti-SMARCC1 (Cell Signaling Technology Cat# 11956, RRID:AB_2797776), anti-BRD9 (Abcam, ab155039), anti-PBRM1 (Active Motif Cat# 61381, RRID:AB_2793612), anti-H3K27ac (Active Motif Cat# 39133, RRID:AB_2561016), anti-H3K4me1 (Abcam Cat# ab8895, RRID:AB_306847), anti-H3K4me3 (Millipore Cat# 07–473, RRID:AB_1977252).
Binding of protein A-MNase:
2.5 μl of EpiCypher CUTANA pAG-MNase was added to each sample. Samples were then rotated at 4°C for 1 hour, subjected to a quick spin, and placed on a magnet stand. The liquid was removed and discarded. The samples were washed then with twice with 1 ml of 0.1% digitonin-wash buffer, and resuspended in 150 μl of 0.1% digitonin-wash buffer.
Targeted digestion and target chromatin release:
3 ul of 100 mM CaCl2 was then added to each tube with gentle vortexing and placed on a chilled metal block (4°C) for 2 hours with periodic gentle shaking throughout the incubation period. 100 μl 2X stop buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.02% digitonin, 50 μg/ml RNAse A, 50 μg/ml glycogen, 2 pg/ml heterologous spike-in DNA) was then added to each sample and mixed. The tubes were incubated at 37°C for 10 mins at 500 rpm, and then spun at 16,000 xg at 4°C for 5 mins. The samples were then placed on a magnet stand and the liquid was transferred to DNA low-bind tubes.
DNA extraction:
2 μl of 10% (w/v) SDS and 1.5 μl of proteinase K (20 mg/ml) was added to each tube and mixed by inversion. Samples were incubated at 70°C for 10 mins, after which 200 μl of phenol-chloroform-isoamyl alcohol 25:24:1 was added. Samples were vortexed and then transferred to a phase-lock tube, and centrifuged at 16,000 xg at room temperature for 5 mins. 200 μl of chloroform was then added and tubes were inverted to mix the solution and centrifuged at 16,000 xg at room temperature for 5 mins. The sample was then transferred to a 1.5 ml tube containing 2 μl of 2 mg/ml glycogen. 500 μl of 100% ethanol was added to each sample and mixed by inversion. Samples were chilled on ice for 10 mins, and subsequently centrifuged at 16,000 xg at 4°C for 10 mins. The liquid was decanted and the pellets were washed with 1 ml of 100% ethanol. Samples were then centrifuged at 16,000 xg at 4°C for 1 min. The liquid was decanted and drained on a paper towel. The pellets were air-dried for 5 mins and dissolved in 40 μl of 1 mM Tris-HCl at pH 8 with 0.1 mM EDTA.
Library preparation and sequencing:
Libraries were prepared by NEB UltraII. Paired-end Illumina sequencing was then performed.
RNA-seq data analysis
Single-ended 50mer RNA-seq reads for SMARCA4-WT (T2, 36) and SMARCA4-KO (M,23) samples were mapped to the UCSC mm9 mouse genome build (genome.ucsc.edu) using Bowtie (70) v1.0.1 and gene counts were quantified using RSEM (71) v1.2.12. Estimated expression counts generated by RSEM were upper-quartile normalized to a count of 1000 (72). Genes with low expression across all samples (upper-quartile of normalized counts < 10) were dropped from downstream analyses. Genes with normalized expression standard deviation less than 50 across all four samples were classified as stably expressed genes. These were used in downstream bulk ATAC-seq and CUT&RUN analyses where intervals of ATAC peaks overlapping transcription start sites of stably expressed genes were analyzed for chromatin accessibility and occupancy profiles across SMARCA4-WT and -KO conditions. Raw counts from RSEM were used to detect differentially expressed (DE) genes between SMARCA4-WT and -KO conditions using DESeq2 (67) v1.26.0 with WT as the baseline condition. These results were used to assess DE status of Gata6 and Nkx2–1. Processed data for RNA-seq are included in Supplementary Information (RNAseq_supp_material.xlsx).
Single-ended 50mer RNA-seq reads for KP, KPS-HET, and KPS primary tumor samples were trimmed to 35mers in order to drop lower quality 3’ read positions. 35mer reads were then mapped to the USCC mm9 mouse genome build (genome.ucsc.edu) using Bowtie (73) v1.2.3 and gene counts were quantified using RSEM (71) v1.3.1. Estimated expression counts generated by RSEM were used to detect differentially expressed (DE) genes between pairwise conditions using DESeq2 (67) v1.26.0. Processed data for RNA-seq are included in Supplementary Information (RNAseq_invivo_supp_tbl.xlsx).
CUT&RUN data analysis
Paired-end 25mer CUT&RUN reads were mapped to the UCSC mm9 mouse genome build (genome.ucsc.edu) using Bowtie2 (73) v2.2.6. Read-alignment BAM files were processed with Picard MarkDuplicates v2.17.0 (broadinstitute.github.io/picard/) to drop duplicate alignments and sorted by read-name using Samtools (74) v1.5. Alignments were converted to BED format using BEDTools (75) bamtobed v2.29.2 and subsequently to bedgraph format using genomecov. Peaks were called with the SEACR (76) v1.2 pipeline by selecting the top 0.5% of regions enriched using area under the curve (AUC) statistic. Peaks were annotated by genomic feature (promoter, distal-intergenic etc.) using ChIPseeker v1.22.1 with UCSC mm9 genome annotation. Differential peak analyses between WT and KO conditions per factor were conducted using DiffBind (77) v2.4.8. DiffBind normalized read-count correlation plots were generated using the default normalization scheme (sequencing depth based) and reads in consensus peaks across samples (peaks that overlapped in at least two samples) were used. Diffbind results for SMARCA2 peaks (WT: T2,36 vs KO: M,23 samples) were visualized using a volcano plot. Motif analyses for differentially represented SMARCA2 peaks (FDR < 0.05) were conducted using HOMER (78) v4.10 with a background set consisting of non-differentially enriched peaks (FDR = 1; |log2 fold-change| < 0.01). The known-motif result set from HOMER was reviewed for significantly enriched motifs. Read density metaplots and heatmaps were generated using deepTools (79) v3.0.1 on 1x normalized BAM files with +/− 5kb flanks upstream and downstream of peaks. Representative heatmaps and metaplots shown in Figure 5 and Supplementary Figure S5 show data generated from the T2 (SMARCA4-WT) and M (SMARCA4-KO) isogenic pair.
Gata6 binding sites were obtained from supplementary data for ChIP-seq peaks in GEO accession GSE124601 (39). Mouse genome build mm10 coordinates were translated to mm9 coordinates using the UCSC-tools liftOver utility. Occupancy metaplot profiles were generated using deepTools (79) v3.0.1 with +/− 2kb upstream and downstream flanks. Processed data for CUT&RUN are included in Supplementary Information (CnR_supp_material.xlsx).
Bulk ATAC-seq data analysis
Paired-end 40mer bulk ATAC-seq reads for SMARCA4-WT (T2, 36) and SMARCA4-KO (M,23) samples (two replicates each for T2, 36, M, 23) were mapped to the UCSC mm9 mouse genome build (genome.ucsc.edu) using Bowtie (70) v1.0.1. Read-alignment BAM files were processed with Picard MarkDuplicates v2.17.0 (broadinstitute.github.io/picard/) to drop duplicate alignments. BAMs for all samples were merged using Samtools (74) v0.1.13. Peaks were called on the merged BAM using MACS2 (80) v2.2.1. Read counts per peak per sample were quantified using BEDTools multicov (75) v2.26. Differential analysis for peaks was performed using DESeq2 (67) v1.16.1 using default “median of ratios” normalization. Significant peaks (q < 0.05, |log2 fold-change| > 1) were selected for downstream analyses. Peaks were annotated by genomic feature using ChIPseeker v1.22.1 with UCSC mm9 genome annotation. Per-feature motif analyses for differentially enriched peaks were conducted using HOMER (78) v4.10 against a background set of common non-enriched peaks for a given genomic feature (FDR > 0.5, |log2 fold-change| < 1.1). The known-motif result set from HOMER was reviewed for significantly enriched motifs. Read density metaplots and heatmaps were generated using deepTools (79) v3.0.1 on 1x normalized BAM files with +/− 5kb flanks upstream and downstream of peaks. Metaplots of chromatin accessibility for Gata6 binding sites were generated as described earlier (see CUT&RUN data analysis). Processed data for bulk ATAC-seq are included in Supplementary Information (ATACseq_supp_material.xlsx).
TCGA clinical data analyses
RNA-seq gene expression profiles of primary tumors and relevant clinical data of 515 lung adenocarcinoma (LUAD) patients were obtained from The Cancer Genome Atlas (6) (TCGA, gdac.broadinstitute.org). SMARCA4 and KRAS mutational status of TCGA tumor samples was retrieved from cBioPortal (81,82) using the TCGA PanCancer Atlas collection (gdc.cancer.gov/about-data/publications/pancanatlas) wherein 510 of 515 TCGA tumors had mutational status available. Within this dataset of 510 samples, counts are as follows: 465 SMARCA4 WT; 45 SMARCA4 mutant (21 truncating mutations; 22 missense mutations; 2 fusions); 154 KRAS mutant (9 SMARCA4 mutant, 145 SMARCA4 WT). Individual tumor transcriptomes were scored with signatures (marker genes with adjusted p-value < 0.05 for club, AT2 and signaling AT2 cells from published scRNA-seq of the lung (45)) using ssGSEA (83). Patients were stratified based on standardized ssGSEA scores and Kaplan-Meier 5-year and overall survival analyses were conducted to compare high-scoring patients with the rest of the cohort, and significance was assessed using the log-rank test. All survival analyses were conducted using the survival package in R. Patients were also grouped by mutational status, as described in relevant Figure legends, and the distribution of standardized signature ssGSEA scores across groups were illustrated using an Empirical Cumulative Distribution Function plot (ECDF) where significance was assessed using a Kolmogorov-Smirnov test. Standardized SMARCA4 expression counts were similarly illustrated using an ECDF plot. All statistical analyses were conducted in the R statistical programming language (R-project.org).
Gene set enrichment analysis for KPS signature
KPS signature genes were derived by taking the top differential genes (adjusted p value < 0.05; |FC| > 1.5) in KPS primary tumors vs KP primary tumors for both directions. Mouse identifiers were then mapped to human orthologues using the biomaRt R package. These mapped gene signatures were then tested for enrichment of pathways and annotated genesets derived from the Molecular Signature Database (MSigDB) (84) using the hypeR R package (85).
Statistics and reproducibility
Statistical analyses were performed as indicated in the Figure legends, Supplementary Figure legends and Methods for each experiment. GraphPad Prism software Version 8.3.0 or R (R-project.org) was used. No statistical method was used to determine sample size prior to experimentation. Mice with no detectable tumor burden at end point were excluded (SPC cohort from Figure 1: 1 KPS; CCSP cohort from Figure 3: 1 KP, 2 KPS-HET).
Data availability
The data discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus (86) and are accessible through GEO Series accession number GSE164867.
Supplementary Material
Statement of Significance.
We demonstrate cell-type specificity in the tumor suppressive functions of SMARCA4 in the lung, pointing towards a critical role of the cell-of-origin in driving SWI/SNF-mutant lung adenocarcinoma. We further show the direct effects of SMARCA4 loss on SWI/SNF function and chromatin regulation that cause aggressive malignancy during lung cancer evolution.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute, the Virginia and D.K. Ludwig Center at MIT, NIH P01 Jacks P01-CA42063, The Bridge Project, a partnership between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center (DF/HCC), a Koch Institute Frontier grant, and in part by a Koch Institute Cancer Center Support Grant P30-CA14051 from the National Cancer Institute. C.P.C. was supported by a Koch Institute Quinquennial Postdoctoral Fellowship and an American Cancer Society Postdoctoral Fellowship (PF-17-009-01-CDD). This work was supported, in part, by a grant from John and Georgia DallePezze to Memorial Sloan Kettering Cancer Center, Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30-CA008748), and the Druckenmiller Center for Lung Cancer Research. C.F.K. was supported by a Mission Boost Grant, MBG-18-204-01-COUN from the American Cancer Society and by the National Cancer Institute of the National Institutes of Health under Award Number R01CA216188. P.M.K.W. is a Damon Runyon Fellow. We thank the Jacks lab, particularly S. Naranjo and R. Romero for helpful discussions; A. Berns from the Netherlands Cancer Institute for Ad-SPC-Cre and Ad-CCSP-Cre; S. Henikoff for CUT&RUN reagents; T. Tammela and T. Westerling for development of the Aiforia deep neural network for murine NSCLC; J. Teixeira, K. Yee, and K. Anderson for administrative support; K. Mercer and M. Magendantz for laboratory and technical support; the Swanson Biotechnology Center, particularly the Flow Cytometry and Histology core facilities for technical support; the MIT BioMicro Center, particularly S. Levine, and the Harvard Bauer Core Facility, particularly N. El-Ali, for scATAC and sequencing support.
Conflict of Interest Statement:
T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific. He is also a co-Founder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. T.J. laboratory currently also receives funding from the Johnson & Johnson Lung Cancer Initiative and The Lustgarten Foundation for Pancreatic Cancer Research, but this funding did not support the research described in this manuscript. A.R. is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas, and was a scientific advisory board member of ThermoFisher Scientific, Syros Pharmaceuticals, Neogene Therapeutics and Asimov until 31 July 2020. From 1 August 2020, A.R. has been an employee of Genentech. G.J.R. has been an uncompensated consultant to Daiichi, Pfizer, and Mirati; he has institutional research support from Mirati, Takeda, Merck, Roche, Pfizer, and Novartis. C.F.K. reports consortium associations with Bristol-Myers Squibb (formerly known as Celgene Corporation) and Longfonds Stichting. The projects of those consortiums are not related to the work in this publication. A.J.S. reports consulting/advising role to Iovance Biotherapeutics, J&J, and Perceptive Advisors. C.M.R. has consulted regarding oncology drug development with AbbVie, Amgen, Astra Zeneca, Epizyme, Genentech/Roche, Ipsen, Jazz, Lilly, and Syros; he serves on the scientific advisory boards of Bridge Medicines, Earli, and Harpoon Therapeutics. J.D.B. holds patents related to ATAC-seq and scATAC-seq and serves on the Scientific Advisory Board of CAMP4 Therapeutics, seqWell, and CelSee.
References
- 1.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodges C, Kirkland JG, Crabtree GR. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy N, Malik S, Villanueva KE, Urano A, Lu X, Von Figura G, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015;29:658–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, et al. Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer. Cancer Cell. Elsevier; 2017;32:574–589.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–4. [DOI] [PubMed] [Google Scholar]
- 6.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. Nature Publishing Group; 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagogo-Jack I, Schrock AB, Kem M, Jessop N, Lee J, Ali SM, et al. Clinicopathologic Characteristics of BRG1-Deficient NSCLC. Journal of Thoracic Oncology. Elsevier; 2020;15:766–76. [DOI] [PubMed] [Google Scholar]
- 8.Schoenfeld AJ, Bandlamudi C, Lavery JA, Montecalvo J, Namakydoust A, Rizvi H, et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin Cancer Res. American Association for Cancer Research; 2020;26:5701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernando TM, Piskol R, Bainer R, Sokol ES, Trabucco SE, Zhang Q, et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat Commun. 2020;11:5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008;68:3689–96. [DOI] [PubMed] [Google Scholar]
- 11.Walter DM, Venancio OS, Buza EL, Tobias JW, Deshpande C, Gudiel AA, et al. Systematic In Vivo Inactivation of Chromatin-Regulating Enzymes Identifies Setd2 as a Potent Tumor Suppressor in Lung Adenocarcinoma. Cancer Res. American Association for Cancer Research; 2017;77:1719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lissanu Deribe Y, Sun Y, Terranova C, Khan F, Martinez-Ledesma J, Gay J, et al. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. Nat Med. 2018;24:1047–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orvis T, Hepperla A, Walter V, Song S, Simon J, Parker J, et al. BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res. 2014;74:6486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue Y, Meehan B, Fu Z, Wang XQD, Fiset PO, Rieker R, et al. SMARCA4 loss is synthetic lethal with CDK4/6 inhibition in non-small cell lung cancer. Nat Commun. 2019;10:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazar JE, Stehling-Sun S, Nandakumar V, Wang H, Chee DR, Howard NP, et al. Global Regulatory DNA Potentiation by SMARCA4 Propagates to Selective Gene Expression Programs via Domain-Level Remodeling. Cell Rep. 2020;31:107676. [DOI] [PubMed] [Google Scholar]
- 16.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17:5976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–8. [DOI] [PubMed] [Google Scholar]
- 18.Ferone G, Song J-Y, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij J-P, et al. SOX2 Is the Determining Oncogenic Switch in Promoting Lung Squamous Cell Carcinoma from Different Cells of Origin. Cancer Cell. 2016;30:519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Rock JR, Lu Y, Futtner C, Schwab B, Guinney J, et al. Evidence for type II cells as cells of origin of K-Ras–induced distal lung adenocarcinoma. PNAS. National Academy of Sciences; 2012;109:4910–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaFave LM, Kartha VK, Ma S, Meli K, Priore ID, Lareau C, et al. Epigenomic State Transitions Characterize Tumor Progression in Mouse Lung Adenocarcinoma. Cancer Cell Elsevier; 2020;38:212–228.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. IOP Publishing; 2008;2008:P10008. [Google Scholar]
- 22.Winslow MM, Dayton TL, Verhaak RGW, Kim-Kiselak C, Snyder EL, Feldser DM, et al. Suppression of lung adenocarcinoma progression by Nkx2–1. Nature. 2011;473:101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dost AFM, Moye AL, Vedaie M, Tran LM, Fung E, Heinze D, et al. Organoids Model Transcriptional Hallmarks of Oncogenic KRAS Activation in Lung Epithelial Progenitor Cells. Cell Stem Cell. 2020;27:663–678.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marjanovic ND, Hofree M, Chan JE, Canner D, Wu K, Trakala M, et al. Emergence of a High-Plasticity Cell State during Lung Cancer Evolution. Cancer Cell. 2020;38:229–246.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallée V-P, et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020;26:259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nature Reviews Molecular Cell Biology. 2019;20:69–84. [DOI] [PubMed] [Google Scholar]
- 27.Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, et al. AP-1 transcription factors and the SWI/SNF complex mediate signal-dependent enhancer selection. Mol Cell. 2017;68:1067–1082.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta M, Concepcion CP, Fahey CG, Keshishian H, Bhutkar A, Brainson CF, et al. BRG1 Loss Predisposes Lung Cancers to Replicative Stress and ATR Dependency. Cancer Res. American Association for Cancer Research; 2020;80:3841–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mainardi S, Mijimolle N, Francoz S, Vicente-Dueñas C, Sánchez-García I, Barbacid M. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. PNAS. 2014;111:255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutherland KD, Song J-Y, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111:4952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim CFB, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q, Liu K, Cui G, Huang X, Yao S, Guo W, et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet. 2019;51:728–38. [DOI] [PubMed] [Google Scholar]
- 33.Salwig I, Spitznagel B, Vazquez-Armendariz AI, Khalooghi K, Guenther S, Herold S, et al. Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. EMBO J. 2019;38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wistuba II, Gazdar AF. Lung Cancer Preneoplasia. Annual Review of Pathology: Mechanisms of Disease. 2006;1:331–48. [DOI] [PubMed] [Google Scholar]
- 35.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. Nature Publishing Group; 2014;509:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alpsoy A, Dykhuizen EC. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol Chem. 2018;293:3892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell. Elsevier; 2018;175:1272–1288.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michel BC, D’Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJ, et al. A noncanonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol. 2018;20:1410–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnal-Estapé A, Cai WL, Albert AE, Zhao M, Stevens LE, López-Giráldez F, et al. Tumor progression and chromatin landscape of lung cancer are regulated by the lineage factor GATA6. Oncogene. Nature Publishing Group; 2020;39:3726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson BG, Helming KC, Wang X, Kim Y, Vazquez F, Jagani Z, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol. 2014;34:1136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111:3128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papillon JPN, Nakajima K, Adair CD, Hempel J, Jouk AO, Karki RG, et al. Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. J Med Chem. American Chemical Society; 2018;61:10155–72. [DOI] [PubMed] [Google Scholar]
- 44.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of Molecular Diagnostics. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. Nature Publishing Group; 2020;587:619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Best SA, Ding S, Kersbergen A, Dong X, Song J-Y, Xie Y, et al. Distinct initiating events underpin the immune and metabolic heterogeneity of KRAS-mutant lung adenocarcinoma. Nat Commun. 2019;10:4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy N, Hebrok M. Regulation of Cellular Identity in Cancer. Developmental Cell. Elsevier; 2015;35:674–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witkowski L, Carrot-Zhang J, Albrecht S, Fahiminiya S, Hamel N, Tomiak E, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–43. [DOI] [PubMed] [Google Scholar]
- 50.Jelinic P, Mueller JJ, Olvera N, Dao F, Scott SN, Shah R, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46:424–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramos P, Karnezis AN, Craig DW, Sekulic A, Russell ML, Hendricks WPD, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 2014;46:427–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Loarer F, Watson S, Pierron G, de Montpreville VT, Ballet S, Firmin N, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47:1200–5. [DOI] [PubMed] [Google Scholar]
- 53.Rekhtman N, Montecalvo J, Chang JC, Alex D, Ptashkin RN, Ai N, et al. SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. J Thorac Oncol. 2020;15:231–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodges HC, Stanton BZ, Cermakova K, Chang C-Y, Miller EL, Kirkland JG, et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol. 2018;25:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clapier CR, Verma N, Parnell TJ, Cairns BR. Cancer-Associated Gain-of-Function Mutations Activate a SWI/SNF-Family Regulatory Hub. Mol Cell. 2020;80:712–725.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marino S, Vooijs M, Gulden H van der, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 58.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature Protocols. Nature Publishing Group; 2009;4:1064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174:6477–89. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biology. 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nature Methods. 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.González-Blas CB, Minnoye L, Papasokrati D, Aibar S, Hulselmans G, Christiaens V, et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nature Methods. 2019;16:397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnology. 2019;37:38–44. [DOI] [PubMed] [Google Scholar]
- 66.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nature Methods. 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current Protocols in Molecular Biology. 2015;109:21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nature Protocols. Nature Publishing Group; 2018;13:1006–19. [DOI] [PubMed] [Google Scholar]
- 70.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. Nature Publishing Group; 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meers MP, Tenenbaum D, Henikoff S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics & Chromatin. 2019;12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. Nature Publishing Group; 2012;481:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research. 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature Protocols. Nature Publishing Group; 2012;7:1728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. American Association for Cancer Research; 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS -driven cancers require TBK1. Nature. Nature Publishing Group; 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Federico A, Monti S. hypeR: an R package for geneset enrichment workflows. Bioinformatics. 2020;36:1307–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus (86) and are accessible through GEO Series accession number GSE164867.