SUMMARY
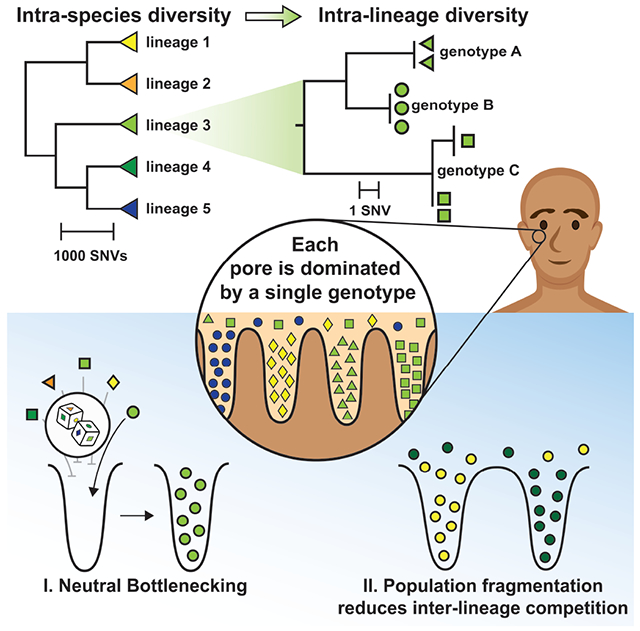
What enables strains of the same species to coexist in a microbiome? Here, we investigate if host anatomy can explain strain co-residence of Cutibacterium acnes, the most abundant species on human skin. We reconstruct on-person evolution and migration using whole-genome sequencing of C. acnes colonies acquired from healthy subjects, including from individual skin pores, and find considerable spatial structure at the level of pores. Although lineages (sets of colonies separated by <100 mutations) with in vitro fitness differences coexist within centimeter-scale regions, each pore is dominated by a single lineage. Moreover, colonies from a pore typically have identical genomes. An absence of adaptive signatures suggests a genotype-independent source of low within-pore diversity. We therefore propose that pore anatomy imposes random single-cell bottlenecks; the resulting population fragmentation reduces competition and promotes coexistence. Our findings suggest that therapeutic interventions involving pore-dwelling species might focus on removing resident populations over optimizing probiotic fitness.
Graphical Abstract
eTOC blurb:
Individual people typically harbor multiple lineages of Cutibacterium acnes, the most abundant species on human skin. Conwill et al. show that skin pores spatially segregate C. acnes genotypes via neutral bottlenecking (colonies isolated from the same pore typically differ by <1 mutation), thereby reducing competition and promoting coexistence among lineages.
INTRODUCTION
All host-associated microbiomes live in environments with spatially structured environmental variation generated by host anatomy and physiology. Spatial structure can be considered at multiple length scales—from location along the gastrointestinal tract down to the level of individual crypts and from distant regions on the skin down to the level of individual pores. Revealing the spatial structure of microbial communities is critical for interpreting the coexistence of diverse microbes (Chung et al., 2017; Welch et al., 2016), modeling community assembly and stability (Kerr et al., 2002; Ladau and Eloe-Fadrosh, 2019; Tropini et al., 2017), and predicting the response of microbiomes to therapeutics (Ferreiro et al., 2018; Koskella et al., 2017).
To date, microbiome biogeography studies have largely focused on taxonomic characterization at the species level or higher (Flowers and Grice, 2020; Grice and Segre, 2011; Oh et al., 2016), and intraspecies diversity has received relatively little attention, with some notable exceptions (Rossum et al., 2020; Zhou et al., 2020). Intraspecies diversity can emerge from both the migration of multiple strains to a host and the mutation of individual strains on the host. Sustained diversity arising from both processes has been observed within human microbiomes (Poyet et al., 2019; Zhao et al., 2019; Zhou et al., 2020).
Understanding the forces that generate and maintain intraspecies diversity at both of these levels is critical for the design of precision microbial therapeutics. For example, if adaptive forces like niche partitioning are critical to strain coexistence, then fine-scale manipulation of microbiomes will require understanding the genetic basis of strain success; however, if neutral forces (e.g. priority effects) determine strain composition (Koskella et al., 2017), then therapeutic approaches might depend instead on removal of extant strains.
Evolutionary reconstruction at the whole-genome level, when combined with fine-scaled sampling, provides an opportunity to reveal migration dynamics across a host and the forces maintaining intraspecies diversity (Chung et al., 2017; Jorth et al., 2015; Lieberman et al., 2016). While metagenomic sequencing provides a powerful approach for surveying microbiomes, it does not provide the resolution required for such evolutionary inference. Metagenomic approaches cannot distinguish whether a detected polymorphism reflects recent on-person mutation or the presence of homologous regions among co-colonizing strains. Moreover, metagenomics cannot determine whether a pair of de novo mutations occurred on the same or different genetic backgrounds (e.g. 2 mutations each at 20% frequency in the population). While single-cell sequencing can in theory provide information about genomic linkage, current technologies cover only a fraction of the genome and have high error rates. In contrast, culture-based approaches that profile bacterial colonies, each formed from a single cell in the original sample, enable true single-genotype resolution. We therefore use culture-based sequences to obtain the resolution needed for evolutionary reconstruction across an individual host.
The skin microbiome provides an excellent opportunity for studying how spatial structure shapes on-person diversity of commensal microbes due to the ease of acquiring samples across body sites and its tractability at multiple spatial scales (Byrd et al., 2018; Flowers and Grice, 2020). Here, we focus on Cutibacterium acnes, the dominant commensal of sebaceous skin (oily skin of the face and back), because: (1) it is prevalent and abundant across all healthy people; (2) multiple strains of this species stably coexist on each person (Oh et al., 2016); and (3) it can be sampled at multiple spatial scales. C. acnes is present on all healthy adults, comprising on average 92% of the bacterial community on sebaceous skin (computed from Table S3 in Oh et al., 2014). Despite its name, the role of C. acnes in acne vulgaris remains unclear (Dréno et al., 2018; Lomholt et al., 2017; McLaughlin et al., 2019; O’Neill and Gallo, 2018).
Each adult has a unique mix of C. acnes strains (Oh et al., 2014), which are found at substantially higher abundance within follicles of pilosebaceous units (skin pores) than on the skin surface (Acosta et al., 2021). C. acnes cells grow substantially faster in anaerobic conditions (Cove et al., 1983) and are thought to consume sebum (the oily substance produced by sebaceous glands at the bottoms of pores) (Brüggemann et al., 2004; Miskin et al., 1997), making pores their ideal environment (Fitz-Gibbon et al., 2013; Hall et al., 2018; Leeming et al., 1984). C. acnes’ residence in anatomical locations that differ greatly in oxygen concentration, nutrient availability, and exposure to the environment raises the possibility that strains display niche specificity. However, it is not yet clear if niche specialization contributes to strain coexistence or why these person-specific populations are resilient to invasion, particularly given the skin’s exposure to the environment.
Here, we report that the anatomy and physiology of human skin promotes substantial intraspecies diversity in part by segregating the C. acnes population across disconnected pores. Strikingly, we find that each skin pore is dominated by a single C. acnes lineage, despite coexistence of multiple lineages within the immediate vicinity. Reduced diversity persists down to the single-nucleotide variant (SNV) level, and phylogenetic reconstruction suggests the presence of single-cell bottlenecks within pores. These bottlenecks cannot be explained by adaptive sweeps, as we find no evidence of positive selection or parallel evolution among 2,445 on-person SNVs. We therefore propose a model in which pore anatomy and physiology gives rise to severe and genotype-agnostic population bottlenecking in skin pores, thereby reducing interstrain competition and promoting the maintenance of intraspecies diversity via non-selective means. More broadly, these findings present a framework for using SNV-level spatial biogeography to uncover migration dynamics at the subspecies level and highlight the capacity of anatomy to shape the ecology and evolution of commensal microbes.
RESULTS
C. acnes biogeography at unprecedented spatial and genetic resolution
To capture the biogeography of C. acnes on sebaceous skin of healthy people, we collected samples across multiple length scales (Figure 1A). At the finest scale, we collected material from inside single sebaceous follicles—where most C. acnes growth is thought to take place–using comedone extractors and pore strips (pore samples; Methods). We incidentally collected samples that included material from multiple adjacent follicles (multipore samples). In addition, we collected samples on a coarser spatial scale (forehead, nose, left/right cheek, chin, shoulder, back quadrants) by scraping a long toothpick back and forth over a large sebaceous skin region (scrape samples; Methods).
FIGURE 1: Cutibacterium acnes biogeography at high spatial resolution and high genetic resolution.
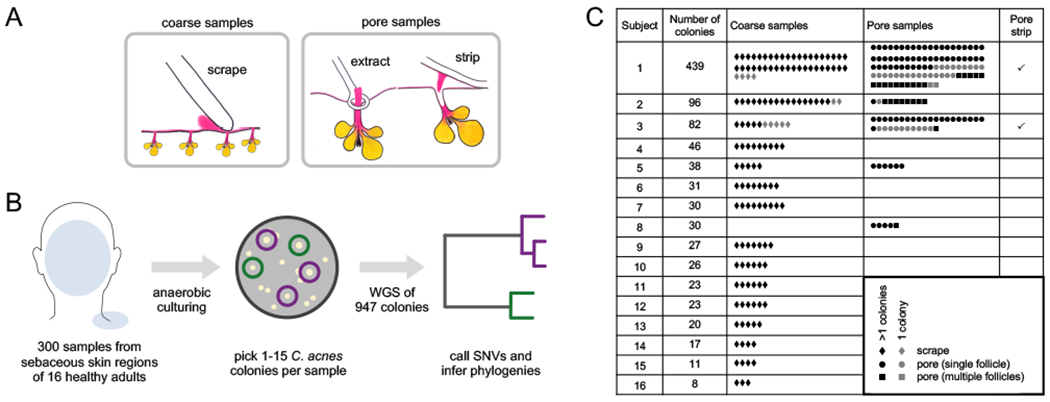
(A) We collected C. acnes colonies from sebaceous skin regions (forehead, nose, right cheek, left cheek, chin, shoulder, back) from 16 healthy adult subjects. Samples were acquired at coarse spatial resolution with a toothpick (scrapes; N=155) and fine spatial resolution (pore extracts and pore strips; N=145). This approach enabled us to examine C. acnes biogeography with spatial resolution down to a single pore (sebaceous follicle). (B) To understand how these samples were related to each other, we performed whole genome sequencing on 947 colonies (1-15 per sample), each of which represents the genetic content of a single cell that originated on the skin of one of our subjects. This approach enabled us to examine C. acnes biogeography with genetic resolution down to a single nucleotide variant (SNV). (C) Summary of samples studied, showing the number of colonies passing quality filters for each subject along with the breakdown of sample types (for more detail, see Methods and Table S1).
In total, we collected 300 samples from 16 healthy adults. This includes 145 pore samples from 5 of these subjects, two of whom were sampled in detail using pore strips (Figure 1B–C; Tables S1–S2).
Immediately after sampling, we streaked the collected material onto solid media and incubated it in an anaerobic environment that favors C. acnes growth (Methods). We randomly selected 1-15 colonies per sample with colony morphology consistent with C. acnes for whole genome sequencing (Figure 1B). All together, we obtained 947 high-quality genomes that passed purity and coverage filters (Methods).
C. acnes communities on individuals arise from multiple colonization events
We first classified colonies according to an established typing scheme (Scholz et al., 2014) (Methods); the six strain-types represented in our dataset cover the majority of known C. acnes diversity. Consistent with previous work (Lomholt et al., 2017; Oh et al., 2014), we find that multiple C. acnes strain-types typically reside on an individual person. Notably, the phylogenetic distribution of strain-types present varies considerably from person to person (Figure 2A).
FIGURE 2: C. acnes lineages from distinct colonization sources coexist on individuals.
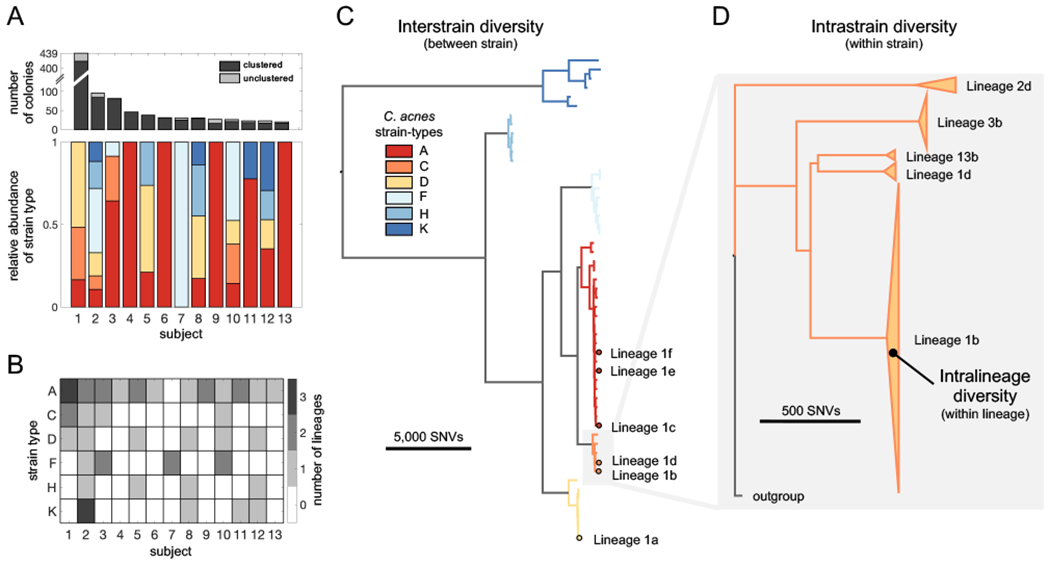
(A) Multiple coexisting strain-types of C. acnes typically reside on the skin of healthy adults, with the composition varying between individuals (lower panel). The number of C. acnes colonies per subject is shown in the top panel, with the number of unclustered colonies in light gray. Only subjects for whom at least 20 colonies passed quality filters are shown (Methods). The relative abundances of C. acnes strain-types on each subject is based on an established C. acnes strain typing scheme (Scholz et al., 2014). (B) Subjects often have multiple lineages belonging to the same strain-type. We note that the number of lineages detected on an individual is sensitive to sampling depth (Figure S1). (C) The phylogenetic relationship of the 53 C. acnes lineages detected across all subjects is shown and colored by C. acnes strain-type. All six distinct coexisting lineages found on Subject 1 are highlighted. (D) A zoom-in of strain-type C illustrates that lineages within a strain-type are separated by large genetic distances relative to intralineage diversity. The heights of triangles are proportional to the number of colonies in each lineage and their widths represent the extent of intralineage genetic divergence. Lineages are named by subject number and then indexed by size within each subject using lowercase letters.
To assess whether colonies of the same strain-type might originate from independent colonization events, we quantified genomic divergence using a reference-based approach and focused on single nucleotide variants (SNVs) (Methods; Figure 1). This approach captures the vast majority of the intraspecies variation because C. acnes has a small accessory genome (~10% variation between strain-types), primarily composed of genomic islands whose patterns of presence and absence correspond to the core genome phylogeny (Brzuszkiewicz et al., 2011; Scholz et al., 2016; Tomida et al., 2013) (Tables S3 and S4).
We found that colonies from the same individual, but not different individuals, were often very closely related, suggesting the presence of person-specific populations (Figure S1). This disparity suggests that closely related colonies emerge from on-person diversification from a recent ancestor on that individual (Zhao et al., 2019; Zhou et al., 2020). We therefore clustered colonies into lineages based on genetic distances, resulting in 53 lineages, each of which contains colonies from only one subject (Methods; Figure 2B–D; Table S5). Colonies from the same lineage are separated by fewer than 100 SNVs across their core genome. Because we imposed a minimum lineage size of 3 colonies, some colonies do not belong to any lineage; these represent either low-abundance genotypes or transient non-resident genotypes from the external environment.
The clustering of colonies into lineages allowed us to estimate the number of colonization events on each individual. Each lineage might represent a distinct colonization event (Zhao et al., 2019), or a lineage might reflect multiple colonization events if a person is colonized by multiple closely related genotypes (e.g. multiple genotypes from a parental lineage transferred to a child). Therefore, the number of C. acnes lineages detected on a person represents the minimum number of C. acnes genotypes that successfully colonized a person. We note that we underestimate lineage coexistence on many subjects, as most were not sampled exhaustively (Figure S1, Table S1). Intriguingly, we often detected multiple lineages of the same strain-type on an individual subject (Figure 2B), demonstrating that an individual host can be colonized by the same strain-type multiple times. In the most extreme case, Subject 2 has been colonized by at least 9 distinct lineages from 6 strain-types.
As expected from the literature, differences in mobile gene content between lineages correlated well with core-genome differences (Figure S2A) (Brzuszkiewicz et al., 2011; Scholz et al., 2016; Tomida et al., 2013). Consequently, we find many cases where lineages with similar gene content coexist on an individual--suggesting that differences in gene content cannot explain coexistence. Moreover, we do not find more aggregate gene content on an individual than would be expected from randomly drawing lineages (Figure S2B–D), suggesting that functional differences are not a strong factor in lineage coexistence. Within lineages, we find few cases of gene content variation (Table S3), indicating relatively slow rates of gene gain and loss. Plasmids make up most of the mobile gene content variation within lineages,with 23% colonies having evidence of a plasmid (Brüggemann et al., 2012; Kasimatis et al., 2013) (Methods; Table S2).
Coexistence of C. acnes strain-types does not arise from specificity to anatomical niches
To test if strain-types coexist because they are equally competitive, we measured their growth rates in vitro. Even in the simplest of laboratory conditions, we noticed substantial differences between colonies originating from the same person. We assessed growth rates for 25 colonies from the most abundant lineages on Subject 1 and Subject 2 (the most intensively sampled subjects), representing diverse strain-types and cultured from the same timepoint for each subject (Figure S3). We find that growth rates can vary substantially (P < 10−3 for both subjects, ANOVA), by as much as 80% across colonies and with variation apparent both within and across lineages. We therefore sought to identify what enables C. acnes strain-types with different competitive abilities to coexist in vivo.
The stable coexistence of diverse C. acnes strain-types might arise from niche specialization to anatomical features. In particular, the environment on the skin surface differs dramatically from that inside skin pores in terms of oxygen concentration, nutrient availability, and other factors (Adamson and Lipoff, 2021; Plewig et al., 2019). We therefore looked for differences in strain-types when sampling directly from the follicle of a pore (extract and pore strip samples) as compared with sampling across the skin surface (scrape samples). However, we did not observe strain-type exclusivity to the skin surface vs skin pores on Subject 1 (Figure 3A) or across subjects (Figure 3B). This suggests that C. acnes strain-types are not exclusively specialized to either the pore or the skin surface environment.
FIGURE 3: C. acnes lineages are spatially segregated into different pores despite coexistence on skin regions.
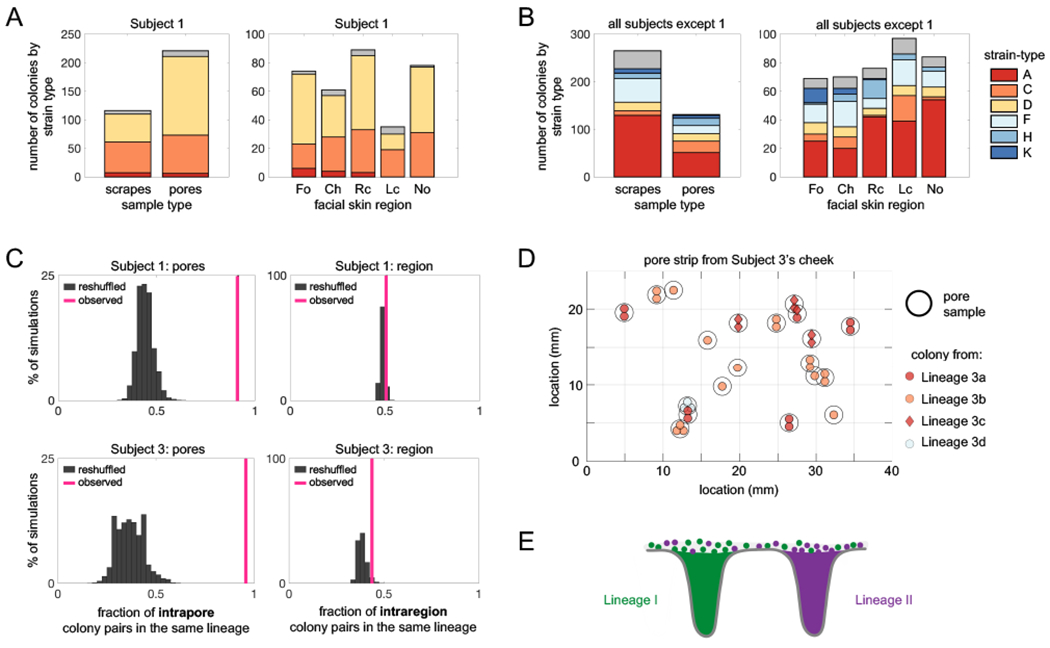
(A-B) Niche specificity does not explain coexistence of strain-types on an individual. (A) C. acnes strain-types on Subject 1 are not specific to sample type (coarse scrape samples vs pore samples) or to skin regions (Fo=forehead, Ch=chin, Rc= right cheek, Lc=left cheek, No=nose). (C) Multiple strain-types or lineages do not typically coexist within the same pore. Pairs of colonies from the same pore belong to the same lineage 91% of the time on Subject 1 and 96% of the time on Subject 3 (pink lines). In contrast, randomly reshuffled colonies from the same pore belong to the same lineage only 44% of the time for Subject 1 and 37% of the time for Subject 3 (gray histograms; P<0.0001 for both cases). In comparison, colonies originating from different pores within the same facial region are only slightly more likely to be from the same lineage than when compared to a random model (P=0.03 for Subject 1 and P=0.02 for Subject 3). This analysis excludes pore samples originating from multiple follicles. (D) A pore strip from Subject 3 (left cheek section) illustrates how pores housing different lineages can be in close vicinity to each other. Each black circle represents a single pore sample, and each interior symbol represents a colony from that follicle. Symbol colors indicate strain-type and symbol shapes indicate lineage membership. (E) These findings demonstrate that lineages are spatially segregated into different pores, despite lineage coexistence within skin regions.
We next explored the possibility that some strain-types are better adapted to particular skin regions (e.g. nose vs forehead). Diverse strain-types coexist in close proximity within facial skin regions on Subject 1 (Figure 3A). This pattern holds across subjects, where strain-types are broadly found across facial skin regions instead of being found only in certain regions (Figure 3B). This lack of exclusivity to facial regions is consistent with previous metagenomic and culture-based studies (Lomholt et al., 2017; Oh et al., 2014, 2016). Some subjects, however, harbor substantial compositional differences in their C. acnes strain-types between the face and back, a pattern also apparent in publicly available metagenomic data (Figure S4) (Oh et al., 2014). Interestingly, we find no consistency in which strain-types are enriched on faces and backs. This lack of consistency argues against a ‘back-adapted’ or ‘face-adapted’ strain-type and instead implicates neutral forces such as limited migration or priority effects (forces that favor early colonizers over new migrants). Together, these findings support a model in which C. acnes strain-types are not exclusively specialized to specific anatomical regions.
Each skin pore is dominated by only one lineage
The lack of niche specialization to anatomical features raises the question of how the skin environment prevents strain-types from outcompeting each other. We next investigated fine-scale spatial resolution, focusing on the lineage level and on samples obtained from pore follicles (pore strips and extracts).
At the level of individual pores, we observe a striking absence of diversity. This can be most clearly seen by close examination of Subjects 1 and 3, from whom we sampled the greatest numbers of pores (Figure 3C). Pairs of colonies originating from a pore belong to the same lineage at a significantly higher rate than would be expected if genotypes were randomly distributed across pores (P<0.0001 for both Subjects 1 and 3; Figure 3C). Notably, from the most densely sampled pore, 11 of 11 colonies are from the same lineage (Table S2). This segregation persists even when pores are closely spaced: in a 1 square cm section of a pore strip from Subject 3, we found 3 different lineages, despite each pore containing only a single lineage (Figure 3D). We occasionally detect minority lineages from pore samples (Figure S5); we were unable to determine whether they reflect true minority populations, partitioning within a sebaceous filament (Piewing et al., 2019), or surface contamination.
Although low within-pore diversity (Figure 3E) might arise from sampling methods that only capture representatives from a part of the follicle, we note that previous work using light microscopy to image skin biopsies after blackhead extraction suggests that extractions are capable of removing the majority of the follicular contents (Plewig, 1974).
Monocolonization of pores results from neutral bottlenecks
Spatial segregation of C. acnes lineages in skin pores could arise from priority effects or from pore-specific selection shaped by the host or other microbes. We reasoned that these mechanisms would result in different degrees of within-pore diversity when examined at the whole-genome level, as well as different signals of adaptive evolution. Exclusion via priority effect or adaptive sweep within a pore would result in a single genotype within each pore, while selection for members of a particular lineage would sometimes result in coexistence of distinct migrants of the same lineage.
At the level of individual SNVs, we find a striking lack of C. acnes intrapore diversity, with colonies from the same pore clustering tightly together on the phylogeny (Figure 4; Figures S6–8; Table S6; Methods). Colonies from the same pore often form monophyletic clades, and in some cases share mutations not detected anywhere else or rare plasmid variants (Figure S9). Moreover, metrics of intrapore diversity are extremely low relative to each lineage’s total diversity, as assessed by genetic distances to various inferred most recent common ancestors (MRCAs). Colonies in Lineage 1a (the largest lineage from Subject 1) from single pore samples have on average less than 1 mutation since their intrapore MRCA, whereas pairs of pores from this lineage typically have 4-8.5 mutations (25%-75% percentiles) since their interpore MRCA (Figure 5A). This pattern of extremely low intrapore diversity, in both absolute and relative scales, is consistent across lineages and subjects (Figure 4B; Figure 5A; Figures S6–8).
FIGURE 4: Each pore harbors only a small fraction of intralineage diversity.
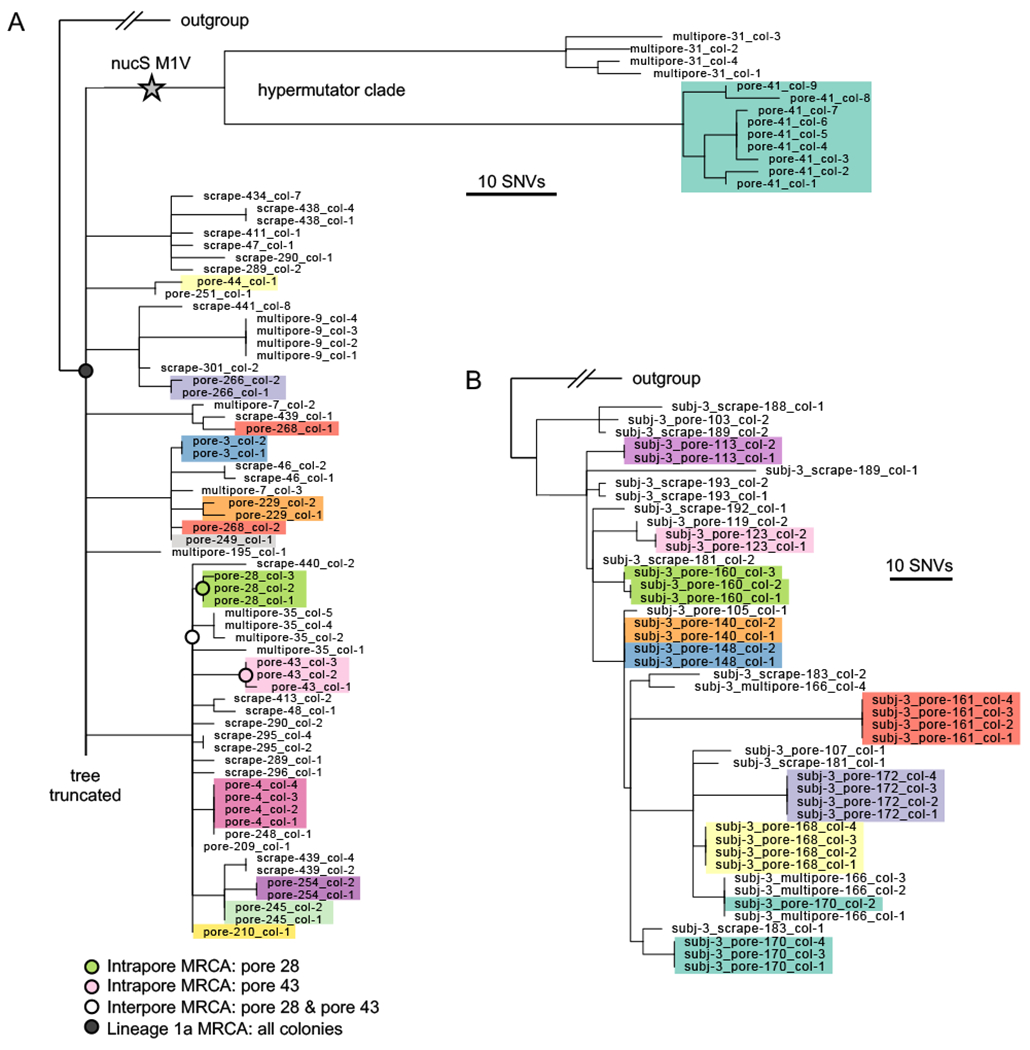
(A) Maximum parsimony tree of the most abundant lineage on Subject 1, Lineage 1a, in which each leaf represents a single colony. Colonies are colored by pore (excluding multipore samples and pore samples with only one colony), emphasizing low within-pore diversity. The long branches at the top of the phylogeny display a hypermutator phenotype (Figure S6). For any given non-hypermutator pore, the mean genetic distance of colonies to the pore’s inferred most recent common ancestor (MRCA) is usually less than 1 SNVs (median across pores: o SNVs; 25%-75% percentiles: 0 to 1.1 SNVs). Four example inferred ancestral genotypes are marked on the tree. Due to space limitations, the tree is truncated; see Figure S6 for the complete tree. (B) Maximum parsimony tree of the most abundant lineage on Subject 3, Lineage 3a. Additional lineage trees with individual pore samples highlighted are available in Figure S8.
FIGURE 5: Neutral forces give rise to population bottlenecks during pore colonization.
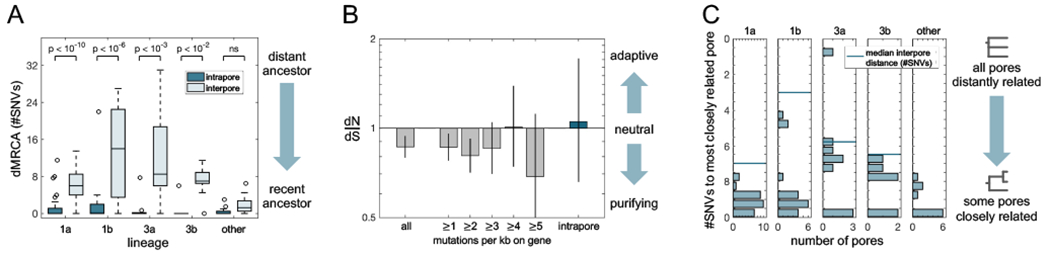
(A) The pattern of small intrapore dMRCAs (distance to the MRCA, averaged across colonies) as compared with interpore dMRCAs (average distance from the two pore MRCAs to their interpore MRCA) is consistent across subjects and lineages (Wilcoxon rank-sum test with Bonferroni correction). This indicates that pore populations are subject to recent strong bottlenecks and are ecologically isolated from each other. Analysis for (A) and (C) included all single pore samples (excluding hypermutators in Lineage 1a); pores from lineages containing fewer than five such samples are grouped as “other”. (B) Across all within-lineage SNVs, dN/dS (the ratio of nonsynonymous to synonymous mutations, relative to a neutral model; Methods) is slightly negative, indicative of purifying selection (P<0.0003). Values of dN/dS for genes with high mutational densities among within-lineage mutations are consistent with a neutral model, as is dN/dS for mutations inferred to have occurred inside pores. These findings suggest that non-adaptive evolution dominates C. acnes evolution on individuals and that adaptive sweeps are not responsible for low within-pore diversity. Error bars indicate 95% CIs. See Figure S12 for a more detailed analysis. (C) Pairs of pores on a person often share very recent common ancestry, suggesting that neutral bottlenecking occurred during a recent pore colonization or re-colonization event. The genetic distance between two pores is equal to twice the interpore dMRCA. Given that the number of pores sampled per subject was vastly smaller than the total number of pores on a person, these values underestimate the commonality of shared mutations between pore populations.
Although the molecular clock rate for C. acnes is not known and we were unable to accurately measure it (Figure S10), all reported bacterial molecular clocks from human infection or colonization range between 0.5 SNVs/genome/year and 30 SNVs/genome/year (Didelot et al., 2016; Zhao et al., 2019). Therefore, our observation of low intrapore diversity (median O SNVs since pore MRCA, 25%-75% percentiles: 0-0.6 SNVs; Methods) suggests that the population within each pore typically descended from a single cell about 1 year ago and hints that priority effects may be important to the exclusion of other strain-types.
There are two pore samples in Lineage 1a that have diverged further from the lineage MRCA (45 and 56 SNVs vs a mean of 9 SNVs; Grubbs outlier test) and harbor more intrapore diversity. We suspected that this excess diversity might be due to hypermutation, an accelerated mutation rate that is common in laboratory experiments (Sniegowski et al., 1997) and in vivo (LeClerc et al., 1996), usually caused by a defect in DNA repair (Oliver, 2010). Consistent with this hypothesis, these colonies share a mutation that eliminates the start codon of the nucS gene, which encodes for an endonuclease critical for the repair of transition mutations (Castañeda-García et al., 2020; Ishino et al., 2018). Indeed, we observe an enhancement in the ratio of transition to transversion mutations in the hypermutator clade (Figure S6). This finding suggests that these pores were physiologically similar to other pores, and that an increased mutation rate enabled C. acnes to accumulate more diversity between the most recent single-cell ancestor and sampling. Interestingly, we only recovered colonies with the nucS mutation at the first of 5 sampling timepoints from Subject 1, suggesting that this hypermutation is unlikely to be associated with long-term adaptation to this host. Similarly, we did not observe any other lineages across subjects with evidence of hypermutation.
The finding of a recent single-cell ancestor for each pore is particularly surprising given that single pores contain on average 50,000 colony-forming units of C. acnes (max 108 CFU; Figure S11)(Claesen et al., 2020). Such large population sizes generally limit the speed of neutral genetic drift (Hartl and Clark, 2006); classic models of neutral evolution predict that it would take over 100,000 bacterial generations (in this case, likely hundreds of years) for a neutral mutation to sweep a population of this size. Therefore, our observations suggest the presence of either conditions that enhance genetic drift or adaptive mutational sweeps that swiftly purge diversity.
To test if adaptive sweeps might be responsible for purging diversity inside pores, we examined within-lineage mutations for evidence of past adaptation. Parallel evolution is a common signature of adaptation in bacteria that manifests as an enrichment of mutations in genes or pathways under selection relative to a neutral model (Lieberman et al., 2011; Zhao et al., 2019). However, we detected no cases of parallel evolution across all 2,445 de novo mutations in coding regions, across mutations occurring on a subject, across mutations occurring within a lineage, or among intrapore mutations (Figure S12, Methods). Moreover, we identified a depletion of nonsynonymous (amino-acid changing) mutations among all de novo mutations (dN/dS < 1 with P < 0.0003, Figure 5B). Critically, ratios of dN/dS were invariant to the number of times a gene was mutated, the inferred age of a mutation, or the functional pathways considered (Figure S12). The absence of adaptive signals argues against selective sweeps as the driver of within-pore bottlenecks. Furthermore, rates of gene content changes were too slow for a model in which bottlenecks are driven by adaptive gene gains or losses (Table S3). Instead, we propose that low within-pore genetic diversity stems from frequent, neutral population bottlenecks induced by pore anatomy and physiology.
Pore anatomy and physiology are sufficient to create bottlenecks during colonization
We next asked if these recent population bottlenecks occurred long after pore populations were established, or, instead, during recent migration into a pore. If pore populations are segregated for long periods of time, the recent bottlenecks observed here would reflect only the most recent bottleneck in a series of in-pore bottlenecks; in this case, sequential intrapore sweeps would create large genetic distances between the MRCAs of each pore. Instead, we find that most pores have a closely related population in another pore, with many pairs of pores sharing SNVs inferred to have occurred recently (Figure 5C; Figures S6–8). These findings are consistent with recent transmission of genotypes between pores. Combined with our observations of young populations within pores (Figure 5A), the finding of recent common ancestors between pores supports a model in which neutral bottlenecking occurred during recent pore colonization or re-colonization events.
We propose that pore physiology can create such bottlenecks (Figure 6A; Figure S13). We modeled the process of pore colonization, using published values for relevant physiological parameters (Butcher and Coonin, 1949; Cove et al., 1983; Plewig, 1974) and the assumption that most C. acnes growth occurs in the favorable conditions at the bottom of pores. First, since C. acnes is not motile (Brüggemann et al., 2004), it must rely on growth and diffusion in order to reach the bottom of a pore. Estimations of the diffusion coefficient of a bacterial cell in sebum and of the sebum flow speed suggest that most potential colonizers are quickly pushed out of the follicle by the sebum flow (Butcher and Coonin, 1949; Plewig, 1974); it is rare for a cell to remain in a pore for more than one doubling-time. Second, C. acnes cells likely cannot proliferate rapidly until they reach lower depths in the pore, where the environment is anaerobic and nutrient rich due to sebum production (Cove et al., 1983; Flowers and Grice, 2020). Third, solid obstacles, including bacterial mass and dead human cells (Jahns and Alexeyev, 2014; Plewig et al., 2019), embedded in sebum will further slow diffusion, making it even more difficult for potential invaders to colonize. In this way, pore physiology could enable a lucky single cell to found a pore’s resident population, with abundant growth at the bottom of the pore blocking new migrants.
FIGURE 6: Pore physiology may contribute to neutral bottlenecking and to lineage coexistence.
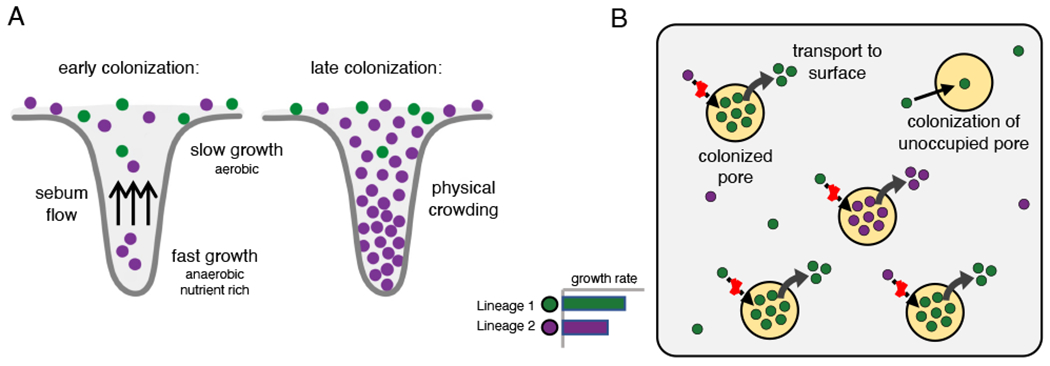
(A) Sebum flow out of the pore, the environmental gradient along the length of the pore, and physical crowding may make it more difficult for a cell to colonize a pore before a resident bacterium proliferates (see also Figure S13). (B) Bottlenecking at the level of individual pores reduces competition between lineages by spatially segregating populations with different genotypes. The skin surface allows for little growth and thus limits opportunities for competition (see also Figure S16).
Despite small distances between some pore MRCAs, the MRCA for each lineage as a whole is substantially older (Figure 5C). These data are consistent with a model in which pore populations studied here were established long after a given lineage initially migrated onto a subject’s skin. We therefore propose that these colonization events may represent pore re-colonization events following a disturbance to the underlying community, perhaps caused by the immune system, phage predation, or physical clearing.
Pores are colonized by C. acnes genotypes from distant locations
To understand migration dynamics across pores, we turned to pore strip data, where each pore sampled has defined spatial coordinates (Figure S14; Table S7). In the case that pores are colonized preferentially by their neighbors, we would expect to see spatial confinement of genetic variants that emerged recently. However, similar to our previous observation that lineages themselves are not specific to certain facial skin regions, we find that closely related pores can be separated by large physical distances (e.g., Figure S7). To assess this quantitatively, we created a neutral model in which spatial coordinates are randomly shuffled and assessed whether pores with closely related genotypes were more likely to be in the vicinity of each other than by random chance, and we find no evidence of spatial confinement at the SNV level (Figure S15). This finding suggests that the timescale for a new genotype spreading across facial skin regions is faster than the timescale for further genetic diversification. This is consistent with a model in which C. acnes cells primarily grow within pores and are transferred across the skin to newly opened pores via long-range dispersal mechanisms (e.g. washing or touching).
Skin pores promote coexistence and stability of extant C. acnes lineages
Altogether, our results support a model in which bottlenecking in skin pores and, therefore, skin anatomy and physiology, play a major role in C. acnes on-person ecology (Figure 6). As a consequence of severe spatial segregation into island-like units, C. acnes populations in different pores do not rely on the same resources for growth. Bacteria have little opportunity to compete elsewhere, as minimal growth occurs on the skin surface, and migration of bacteria to the surface is limited by sebum flow rather than intrinsic fitness (Figure 6B; Figure S16). Theoretical work has proposed that spatial segregation promotes neutral coexistence by reducing the strength of ecological interactions (Coyte et al., 2015). We propose that the reduction in competition promoted by isolated pores is an extreme version of ecological isolation, and that this promotes the coexistence of C. acnes lineages, even if they have fitness differences and distinct survival strategies.
In addition, the priority effects created by pores may help explain the surprising observations that an individual’s strain-types are stable over time despite the skin’s exposure to the outside world (Oh et al., 2016). First, the physiology of pores insulates their C. acnes populations from the external environment. Moreover, sebum flow ensures that C. acnes cells on the skin surface originating from pores outnumber those originating from the environment. Consequently, already established lineages will have a higher likelihood of colonizing a newly available pore. Longer timeseries data will be crucial to understanding the extent to which pores stabilize community dynamics over the host’s lifetime.
Taken together, our findings support a model in which skin pores play a critical role in C. acnes ecology. Skin pores provide an environment well-suited for C. acnes growth, but population bottlenecking limits the amount of genetic diversity each pore harbors. As a consequence, skin pores both reduce competition between strain-types via spatial segregation and favor the existing community via a priority effect. These forces work together to create a stable skin population.
DISCUSSION
Skin pores promote intraspecies diversity via neutral processes
In this work, we have shown that skin anatomy strongly influences intraspecies diversity in C. acnes, a prevalent and prominent commensal on human skin. Our culture-based approach and fine-scaled sampling methods enabled us to examine C. acnes biogeography with resolution down to single SNVs and single skin pores (Figures 1–2). This resolution was essential for uncovering that the C. acnes population in a single skin pore is extremely bottlenecked (Figures 3–5). As most growth happens within pores, we propose that this bottlenecking contributes to the stable coexistence of diverse C. acnes populations on individual adults (Oh et al., 2016), despite differences in fitness and despite the skin’s exposure to the environment (Figure 6).
We did not sample enough individuals in this study to characterize how different skincare regimens or history of treatment for acne might alter C. acnes biogeography. As we only studied adult subjects without active acne vulgaris, future studies will be needed to understand implications of these findings for acne (Dréno et al., 2018; Lomholt et al., 2017; McLaughlin et al., 2019; O’Neill and Gallo, 2018). However, we note that we found similar patterns across all subjects studied, suggesting that our observation of low within-pore C. acnes diversity is unlikely to be driven by a specific skincare regimen.
Future studies will be needed to understand if the findings we report for C. acnes are relevant to other skin commensals, and, more broadly, if crypt-like structures promote neutral bottlenecking and intraspecies diversity in other microbiomes. Intriguingly, our dataset includes 3 pore samples from which we cultured multiple clonal Cutibacterium granulosum colonies (Figure S17), hinting that the process leading to low within-pore C. acnes diversity may also apply to other related pore-dwelling species on human skin (Mak et al., 2013). However, we do not necessarily expect these patterns to hold for Staphylococcus epidermidis and related species, which are thought to grow primarily at the tops of pores and on the skin surface (Plewig et al., 2019). Regardless, S. epidermidis lineages have been shown to co-exist on individuals and within broad geographic regions (Zhou 2019). While the reason for this coexistence is not well understood, one possible factor is that the greater amount of variable gene content in S. epidermidis allows for more niche-specialization.
Beyond the skin, the crypts of the mouse large intestine have been shown to promote priority effects among Bacteroides (Whitaker et al., 2017) and crypts in the mouse stomach are thought to promote priority effects for Heliobacter pylori (Fung et al., 2019). However, at least for Bacteroides fragilis, toxin secretion is thought to be integral to exclusion of other strains (Hecht et al., 2016); this non-neutral filtering mechanism may explain why strain co-existence in this species is rare despite the presence of crypts and priority effects (Garud et al., 2019). We speculate that the importance of crypt-like structures in maintaining intraspecies diversity will depend both on microbial strategies and whether the particular anatomical and physiological conditions induce single-cell bottlenecks.
Role of skin pores in the balance of neutral and adaptive evolution
Our finding that SNV-driven adaptive evolution is exceedingly rare in C. acnes evolution—to the point where it is undetectable here—is surprising in light of recent reports of rapid adaptive evolution in other stable members of human microbiomes (Poyet et al., 2019; Zhao et al., 2019). While low population sizes can limit adaptive evolution (Hartl and Clark, 2006), C. acnes populations on individuals can reach up to 1010 cells, suggesting ample potential for on-person evolution. One possible explanation for our observation is that few beneficial mutations remain to be explored (Wielgoss et al., 2013; Wiser et al., 2013). For example, the skin microenvironment might be relatively stable compared to the variable environment of the human gut, selective pressure from bacteria might be limited by the relatively low complexity of the microbial community on skin (Oh et al., 2014), or follicle structure or sebum flow might limit phage predation (Lourenço et al., 2020)—all of which would result in fewer opportunities for adaptation for skin commensals.
Alternatively, it is possible that our observations of largely neutral evolution arise from the dominance of stochastic forces on the skin. To that end, we hypothesize that the physical structure of pores may create an environment in which luck and location—rather than genomically-encoded fitness—predict success, therefore limiting the adaptive potential of C. acnes on individual people. Bottlenecking suppresses selective forces by both reducing competition between cells with different genotypes and by introducing randomness in which cells get to proliferate (Barrick and Lenski, 2013; Lieberman et al., 2005; Tenaillon et al., 2016). In addition, genetic drift may be favored because the number of cells that are actually growing might be substantially smaller than the census population (e.g. if bacterial replication were restricted to the very bottom of the follicle) (Hartl and Clark, 2006). In the case of a narrow growth region, physical crowding of cells inside a pore (Jahns and Alexeyev, 2014; Plewig et al., 2019) may exclude beneficial mutants from the growth layer (Schreck et al., 2019; Karita et al., 2021). These proposed mechanisms emphasize how host anatomy has the potential to suppress selective forces and tip the balance toward more neutral outcomes. They also raise an interesting question of whether these structures have evolved because limiting commensal evolution is beneficial to the host (Foster et al., 2017).
Implications for microbial therapeutics
Understanding how host anatomy and physiology influence strain-level composition in microbiomes is critical to the design of precision microbiome therapeutics—particularly those that are intended to engraft into the existing community or remove a member of that community. This study of skin pores exemplifies how host anatomy can contribute to strain-level coexistence and stability via non-adaptive means, with implications for the development of microbiome-based therapeutics (Costello et al., 2009; Paetzold et al., 2019; Schmidt, 2020). In particular, these results suggest that the ability of a probiotic strain to engraft on sebaceous skin may hinge less on the probiotic strain’s competitive fitness and more on efficient removal or destabilization of the existing community prior to treatment.
Here, we have shown that evolutionary reconstruction of mutations—including neutral ones—at the SNV scale reveals migration dynamics in the microbiome and provides insight into the processes by which genetic diversity is maintained. We anticipate that future studies applying similar evolutionary approaches to other microbes will accelerate development of the mechanistic understanding needed for precision microbiome engineering.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tami Lieberman (tami@mit.edu).
Materials availability
Bacterial isolates generated in this study are available from the lead contact upon reasonable request. This study did not generate new unique reagents.
Data and code availability
All raw sequencing data have been deposited in NCBI-SRA and are publicly available as of the date of publication. Genome assemblies have been deposited on GitHub and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Additionally, processed data are available in Tables S3, S4, and S6.
All original code has been deposited at GitHub and is publicly available as of the date of publication. The GitHub repository is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| C. acnes isolates | This manuscript | N/A |
| C. granulosum isolates | This manuscript | N/A |
| Biological samples | ||
| Skin scrapes and skin pore samples from healthy people | This manuscript | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Brucella Blood Agar | Hardy Diagnostics | A30 |
| QuickExtract buffer | EpiCentre | QE09050 |
| Lysozyme | Millipore Sigma | 62971 |
| PCRClean-DX SPRI beads | Aline Biosciences | C-1003-250 |
| Polyethylene glycol (PEG) 8000 | Hampton Research | HR2-535 |
| Magnesium chloride (MgCl2) | Ambion | AM9530G |
| ReadyLyse Lysozyme Solution | EpiCentre | R1810M |
| KAPA HiFi HotStart ReadyMix | Roche | 7958927001 |
| Reinforced Clostridial Media (RCM) | Oxoid | CM0149 |
| Critical commercial assays | ||
| PureLink Genomic DNA Kit | Invitrogen | K182002 |
| Illumina Tagment DNA TDE1 Enzyme and Buffer Kits | Illumina | 20034198 |
| High-Molecular Weight Genomic DNA Kit | Qiagen | 67563 |
| Ligation Sequencing Kit and Native Barcoding Expansion 1-12 | Oxford Nanopore | SQK-LSK109 and EXP-NBD104 |
| Blackhead Removal Activated Carbon Mask | Mengkou | 4716872044078 |
| Deposited data | ||
| Raw sequencing data | This manuscript | NCBI-SRA BioProject: PRJNA771717 |
| Assembled genomes for each C. acnes lineage | This manuscript | GitHub: https://github.com/arolynconwill/cacnes_biogeo |
| Hybrid assemblies of C. acnes colonies with plasmids | This manuscript | GitHub: https://github.com/arolynconwill/cacnes_biogeo |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| 16S V1-V3 forward primer (27F-plex): TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATCMTGGCTCAG | Khadka et al., 2021 | N/A |
| 16S V1-V3 reverse primer (534R-plex): GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGATTACCGCGGCTGCTGG | Khadka et al., 2021 | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| All original code | This manuscript | https://github.com/arolynconwill/cacnes_biogeo |
| Snakemake (v6.4.1) | Mölder et al., 2021 | https://snakemake.readthedocs.io/en/stable/ |
| Matlab (v2015b, v2018a) | Mathworks | https://www.mathworks.com/products/matlab.html |
| Cutadapt (v1.18) | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| Sickle (v1.33) | Joshi and Fass, 2011 | https://github.com/najoshi/sickle |
| Bowtie 2 (v2.2.6) | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SAMtools (v1.5) and BCFtools (v1.2) | Li et al., 2009 | https://github.com/samtools/ |
| Kraken 2 (v2.0.7) | Wood et al., 2019 | https://github.com/DerrickWood/kraken2/wiki |
| Bracken (v2.5) | Lu et al., 2017 | https://github.com/jenniferlu717/Bracken |
| BLAST (v2.7.1) | NCBI | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| PHYLIP (v3.69) | Fenselstein, 2005 | https://evolution.genetics.washington.edu/phylip.html |
| FigTree (v1.4.4) | Andrew Rambaut | https://github.com/rambaut/figtree |
| SPAdes (v3.13) | Bankevich et al., 2012 | https://github.com/ablab/spades |
| Prokka (v4.8.1) | Seemann, 2014 | https://github.com/tseemann/prokka |
| CD-HIT (v4.8) | Li et al., 2006; Fu et al., 2012 | http://weizhong-lab.ucsd.edu/cd-hit/ |
| Filtlong (v0.2.0) | Wick, 2018 | https://github.com/rrwick/Filtlong |
| Unicycler (v0.4.8) | Wick et al., 2017 | https://github.com/rrwick/Unicycler |
| SATIVA | Kozlov et al., 2016 | https://github.com/amkozlov/sativa |
| QIIME2 (v2020.11) | Bolyen et al., 2018 | https://qiime2.org |
| DADA2 (v1.18.0) | Callahan et al., 2016 | https://benjjneb.github.io/dada2/index.html |
| Other | ||
| Public C. acnes genomes | NCBI GenBank | Accession: NC_018707.1 |
| Public C. acnes plasmid sequences | NCBI GenBank | CP003294 and CP017041 |
| Public C. granulosum genomes | NCBI GenBank | NZ_LT906441.1 |
| SILVA database (version 132) | Quast et al., 2013 | https://www.arb-silva.de |
EXPERIMANTAL MODEL AND SUBJECT DETAILS
Sixteen healthy adult subjects who had not taken antibiotics in the past 3 months were recruited under a protocol approved by MIT’s Institutional Review Board. Subjects included individuals from different ethnic and geographic backgrounds and had different histories of antibiotic treatment; as this study was designed to identify general trends and was not powered to identify associations, and to maintain subject anonymity, these histories are not reported. Subjects were asked to wash their face with gentle soap prior to sampling to enrich for resident bacteria. To sample from diverse anatomical features—including skin pores (sebaceous follicles) and the skin surface—three sampling methods were employed (Figure 1A). A single member of the research team collected all the samples, including from themselves (Subject 1).
Scrape samples were collected using a long sterile toothpick to survey bacteria from both the surface and the tops of pores within a given facial region. Scrape samples were collected from each subject at 8 standardized regions: forehead, left cheek, right cheek, chin, upper right back, upper left back, lower left back, and lower right back (from all but Subject 8, from whom only pore samples were taken). From some subjects, additional scrape samples were collected (Tables S1 and S2). Each toothpick was dragged at an angle using 1-2 inch strokes about 10 times over the region to be sampled, turning occasionally to maximize biomass collection. Each toothpick was then used to immediately inoculate Brucella Blood Agar plates (Hardy Diagnostics) and spread for single colonies using fresh inoculator loops.
For select subjects, samples from inside pore follicles were collected using a comedone extractor or pore strips. Pilosebaceous units (pores) to be sampled via comedone extraction were identified visually as blackheads or whiteheads, and a sterilized comedone extractor was used to apply pressure to the surrounding area of skin. Most extracts removed contents from a single pore as a semi-solid plug. However, some attempts resulted in the extraction of contents from multiple follicles; these samples were labeled as ‘multipore’ samples. For single-pore and multipore samples, a sterile plastic inoculator loop was then used to transfer the pore contents to a Brucella Blood Agar plate. This first inoculator loop was struck multiple times on the plate to disturb the follicular plug, which was then struck out for single colonies as above. When multiple pores were extracted simultaneously, contents from all extracted pores were processed together and the sample was labeled as containing contents from multiple pores. Some extracts (indicated in Table S1) were processed like pore strip samples (below) in order to conduct amplicon sequencing as well.
For pore strips, a commercially available product (Blackhead Removal Activated Carbon Mask, Mengkou) was applied to the cheeks, nose, and forehead and allowed to dry. The dried film was carefully peeled off and segments were placed into sterile petri dishes for processing. Spatial coordinates for pore strip samples are available in Table S7. Under a dissection microscope, individual extracts were plucked off using sterilized forceps and placed into individual wells of a microplate containing 50 μl of QuickExtract DNA Extraction Solution (Epicentre). Extracts were disturbed by pipetting up and down (samples did not completely dissolve even after mixing). A 5 uL aliquot was used to inoculate a Brucella Blood Agar plate and struck for single colonies. The remainder was used for amplicon sequencing as described below (see 16S amplicon sequencing). Sampling across subjects is summarized in Figure 1C and Table S1, which include all samples from which at least one colony passed quality filters (see below).
METHOD DETAILS
Culturing and single-colony sequencing
Culture plates were incubated in an anaerobic environment at 37°C for 5-7 days to enrich for C. acnes. Random colonies suspected to be C. acnes based on colony morphology were selected for further profiling. From most samples, up to 4 colonies were chosen for further processing; additional colonies were chosen on a few samples for further depth (see Table S2 for details on colonies that passed all filters). Selected colonies were resuspended in 200 μL of PBS, and 150 μL of the material was used for gDNA extraction. To obtain more pure freezer stocks, a small subset of colonies was restreaked prior to making freezer stocks; these colonies were used for growth-rate analysis and long-read sequencing and are indicated in Figure S3 and below (see C. acnes plasmid analysis), respectively. The remainder of the colony was mixed with glycerol to reach a final concentration of 20% and frozen at −80°C. DNA was extracted in 96-well plates using the PureLink Genomic DNA Kit (Invitrogen), using instructions for gram-positive bacteria, with the exception of longer incubations times (12 hours lysozyme step; 3 hours proteinase K step) and elution into a smaller volume (20 ul). Genomic libraries for Illumina sequencing were prepared using the Illumina Tagment DNA TDE1 Enzyme and Buffer Kits with previously described protocol modifications (Baym et al., 2015). Libraries were pooled and sequenced on Illumina NextSeq and HiSeq using 75-bp paired end reads to an average depth of 76 reads for colonies passing eventual filters (Table S2).
Clustering colonies into lineages
Colonies were clustered into lineages using SNV calls from an alignment-based approach with pipelines implemented in Snakemake (V6.4.1) (Mölder et al., 2021) and Matlab (v2015b for pre-processing steps executed in Snakemake pipelines). Adapters were removed using Cutadapt (v1.18) (Martin, 2011) and reads were trimmed using Sickle (v1.33; -q 20 -l 50 -x -n) (Joshi and Fass, 2011). Next, reads were aligned using Bowtie 2 (v2.2.6; -X 2000 --no-mixed --dovetail) against Cutibacterium acnes C1 (RefSeq NC_018707) (Langmead et al., 2009; Minegishi et al., 2013). Candidate single nucleotide variants were called using SAMtools (V1.5) mpileup (-q30 -t SP -d3000), bcftools call (-c), and bcftools view (-v snps -q .75) (Li et al., 2009). For each candidate variant, information for all reads aligning to that position (e.g. base call, quality, coverage), across all samples, were aggregated into a data structure for local filtering and analysis. Colonies were omitted from further analysis if less than 90% of their reads were assigned to Cutibacterium acnes according to Bracken (v2.5) (Lu et al., 2017; Wood et al., 2019) with the standard Univec database including all RefSeq genomes (153 of an initial 1546 colonies), if they had a median coverage below 10 across candidate variant positions (283 of 1393 colonies remaining), or if they had a major allele frequency below 0.65 for over 1% of variant positions with coverage greater or equal to 4 reads (50 colonies of 1110 remaining). In all, these filters retained 1080 colonies.
We filtered candidate SNVs using publicly available code implemented in Matlab (v2018a for analyses performed locally) (see Data Availability) similar to that previously published (Lieberman et al., 2014). Basecalls were marked as ambiguous if the FQ score produced by SAMtools was above −30, the coverage per strand was below 3, the major allele frequency was below 0.9, or more than 50% of reads supported indels. Remaining variant positions were discarded for clustering analysis if no unmasked polymorphisms remained. In addition, all SNVs in regions of the reference genome with homology to C. acnes plasmids were removed (see section on C. acnes plasmids). These SNV calls were used to calculate pairwise distances between colonies, equal to the number of positions where both colonies had non-ambiguous base calls and where the base calls differed. This distance matrix was used as input to clustering algorithm DBSCAN, using a distance threshold of 35 SNVs and a minimum cluster size of 3 (Figure S1). Clusters with a mean pairwise distance of below 80 SNVs were allowed to merge together (this allowed the hypermutator colonies to be part of Lineage 1a; see Figure S6).
Some colonies showed evidence of non-purity at this step, with mixed alleles at positions that distinguished colonies within the same initial cluster from each other. This nonpurity could have emerged during initial sample collection (no attempt was made to purify colonies into isolates before sequencing), during sample processing (all samples were processed in 96 well plates), or due to index hopping. Thus, after performing initial clustering, we removed colonies with a mean major allele frequency below 0.95 across within-cluster SNVs (variant positions that had base calls in at least 67% of colonies and with a median coverage of at least 10) for which the colony had sufficient coverage (greater or equal to 8 reads). Clustering and SNV identification were then repeated iteratively until no colonies with evidence of impurity remained, first by restricting clustering to colonies from the same subject only, and then by allowing clustering across subjects and allowing cluster merging (106 colonies were removed during this step). Finally, there were 7 colonies that clustered with Subject 1 despite originating from other subjects; these were removed due to suspected contamination, since Subject 1 was involved in sample acquisition and processing. In all, 947 high quality colonies and 53 clusters--termed lineages--remained and were used in subsequent analysis. Detailed information about all subjects, colonies, and lineages can be found in Tables S1, S2, and S5.
Classification of lineages into strains types
We used lineage-specific assemblies (see Mobile element analysis) to identify the global strain-types, using the previously described SLST scheme (Scholz et al., 2014). We used BLAST to compare known SLSTs to custom BLAST databases created from lineage assemblies. Some lineages had no exact matches, indicating a new SLST. In this case, we classified the lineage by the super-SLST level (e.g. “A” for SLST “A1”), based on SLST with the best alignment (blastn with default parameters; highest bit score for alignment lengths greater or equal to 480 bp) (Altschul et al., 1990; Camacho et al., 2009). The super-SLST for each lineage is available in Table S5.
SNV calling and evolutionary inference
To determine SNV positions within each lineage, basecalling was repeated using the following process: first, basecalls were marked as ambiguous if the FQ score produced by SAMtools was above −30, the coverage per strand was below 3, the major allele frequency was below 0.75, or more than 25% of reads supported indels; second, genomic positions with a median coverage below 12 reads across samples or where at least 34% of basecalls were ambiguous across samples were omitted. In addition, to remove variants that emerged from recombination or other complex events, we identified SNVs that were less then 500 bases apart and for which the correlation of non-ancestral allele frequencies (see below) across colonies within a lineage exceeded 0.75 (Table S8); these positions, as well as regions on the reference genome with homology to plasmids (see C. acnes plasmid analysis), were removed from downstream analysis.
All remaining genomic positions that passed these strict filters and retained two non-ambiguous alleles were considered SNV positions and were investigated across samples. To call genotypes for as many colonies as possible at these SNV positions, including ones with low coverage, basecalls were repopulated from the raw data and only marked as ambiguous only if the coverage per strand was below 1, the total coverage below 3, the major allele frequency below 0.67, or more than 25% of reads supported a deletion. Details on SNVs detected in each lineage are available in Table S6.
Phylogenetic reconstruction was done using dnapars from PHYLIP (V3.69) (Fenselstein, 2005). Trees were rooted using the ancestral allele as determined below. Example trees are shown in Figure 4 and Figures S6–8. Ancestral alleles were determined by using the most closely related lineage from a different subject (as measured by mean pairwise distance between colonies belonging to different lineages) as an outgroup: the ancestral allele was taken as the most common allele across 10 random colonies from the outgroup (or fewer colonies if the outgroup lineage contained less than 10 colonies). If outgroup colonies did not have any calls at that position, then the reference genome was used as the ancestral allele. All trees figures were generated using FigTree (v1.4.4).
Phylogenetic reconstruction across lineages (Figure 2C) was performed using the inferred ancestral genotype of each lineage (for positions that did not vary within the lineage, the ancestral genotype was taken as the basecall across non-ambiguous samples; for positions that did vary within the lineage, the ancestral genotype was determined from an outgroup as described above). A parsimony tree was generated using dnapars as above, using variable positions with basecalls in greater than 10% of lineage ancestors. The tree is midpoint-rooted.
Calculation of distances to MRCAs
To understand the evolutionary history of bacteria within and between pores, we calculated values of dMRCA (distance to most recent common ancestor) for sets of colonies (Figures 4 and 5). For vertically evolving organisms, this value has more interpretability than other metrics of diversity (e.g. average pairwise difference), representing the relative time since the set of organisms under consideration had a single-celled ancestor. In addition, dMRCA is robust to unequal sampling depth between clades on a phylogeny.
For each calculation of dMRCA, we inferred the genotype of the MRCA by assuming that, for each variable genomic position within the set of colonies, the ancestral allele was equal to that defined for the lineage ancestor (see SNV calling and evolutionary inference). We define the dMRCA for each pore as the mean of the number of SNVs distinguishing each colony from the pore MRCA. We exclude multipore samples as well as pore samples with only a single colony from calculations of intrapore dMRCAs. We define the interpore dMRCA for a pair of pores as the mean number of SNVs distinguishing the MRCA of each of the two pores and interpore MRCA. The genetic distances between pores reported in Figure 5C refer to the number of SNVs differing between the inferred ancestors of a given pair of pores.
Parallel evolution analysis
To search for genes with evidence of mutational enrichment, we first counted how many times each gene was mutated (mi). We then computed the probability of observing ≥ mi mutations according to a Poisson distribution with λ = Mpi, where M is the total number of mutations observed on coding regions and pi is the expected probability that a random mutation lands on that gene, taking into account gene length, codon distribution, and observed mutational spectrum (the relative rates of nucleotide conversion; for instance, the numbers of A:T->T:A vs A:T->C:G mutations observed). This analysis masked all regions of the reference genome with homology to C. acnes plasmids (see C. acnes plasmid analysis). To account for multiple hypotheses, we performed the Benjamini-Hochberg procedure (treating each unmasked gene on the genome as a hypothesis). We find no compelling evidence of parallel evolution when considering all de novo mutations or mutations at the intrasubject or intralineage levels (Figure S12).
To look for signs of positive selection, we computed dN/dS, the ratio of nonsynonymous mutations to synonymous mutations relative to a neutral model. Observed mutations were called as nonsynonymous (N) or synonymous (S) according to the reading frames in the annotated reference genome; in the event that there was an ancestral mutation (fixed in all colonies in a lineage) that differed from the reference genome, the basecall at that position was considered when determining if a SNV on that codon was N or S. Our neutral model was used to assess the expected N/S ratio, based on the observed mutational spectrum and the codon distribution of each gene. Figure 5B shows a summary of this analysis, and Figure S12 shows an extended version that considers mutations by subject, lineage, mutational age, and gene function. All results were consistent with neutrality or with weak purifying selection. We note that one limitation of this study is that it focused on ongoing evolution and would not capture any potential adaptive sweeps that occurred in the past, for example, immediately after a strain colonized an individual.
Mobile element and gene content analysis
To systematically identify gains and losses within each lineage, we constructed a pan-genome for all colonies from each lineage. For each lineage, we concatenated up to 250,000 reads from each member colony with ≥ 99% of reads assigned to C. acnes at the species level by Bracken (since no such colonies existed in Lineage 2i, we used a purity threshold of 95%; see Clustering colonies into lineages). We then assembled each lineage pangenome with SPAdes (V3.13; careful mode) (Bankevich et al., 2012) using minimum contig length of 500 bp. Lastly, we aligned reads from each member colony to its assembled pangenome (see SNV calling and evolutionary inference).
We then looked for genomic regions that were missing from some, but not all, colonies in a lineage. We identified candidate mobile elements as continuous regions over 500 bp with a copy number (relative to the rest of the genome) below 0.25x in a given colony. We also considered each contig from the assembled genome as a candidate mobile element region. We then filtered these candidate regions, requiring a mean copy number (relative to the rest of the genome) less than or equal to 0.15x and mean coverage of less than or equal to 2.5 reads; we also required that the region to have strong support in at least one other colony (mean copy number greater or equal to 0.85x). Regions with homology to C. acnes plasmids were masked (see next section). We merged all overlapping regions found in colonies from the same lineage, and these regions are reported in Table S3.
For comparison of gene-content across lineages, we considered all pan-genome contigs with an average copy number of greater than 0.5x and all protein-coding genes annotated by Prokka (V4.8.1) with at least 50 amino acids (Seemann 2014). We clustered genes across all lineages using CD-HIT with a 95% identity clustering cutoff (V4.8.1; cd-hit -i lineages_all.fasta -O lineages_all_db_95 -c 0.95 -n 5 -M 64000 -d 0) (Li et al., 2006; Fu et al., 2012). This resulted in 3825 gene clusters (Table S4). Analysis of gene presence/absence in each lineage is presented in Figure S2.
C. acnes plasmid analysis
During the mobile element analysis, we noted the presence of gain/loss regions with homology to known C. acnes plasmids (NCBI CP003294 and CP017041) (Brüggemann et al., 2012; Kasimatis et al., 2013). We also noted that additional gain/loss regions had similar coverage patterns across samples to known plasmid regions. To better understand plasmid gene content, we performed long read sequencing for five colonies, which cover diverse genotypes on Subject 1: subj-1_scrape-439_col-5, subj-1_scrape-440_col-3, subj-1_scrape-441_col-3, subj-1_scrape-442_col-6, and subj-1_scrape-443_col-6. We used the Qiagen High-Molecular Weight Genomic DNA Kit (Catalog #67563) following the protocol recommended for gram-positive bacteria, with increased lysozyme as above (see Culturing and single-colony sequencing). MIT’s BioMicroCenter performed size selection with SPRI beads (using 1-2 ug input gDNA shear with 50 ul SPRI beads and 01x MgCl500-Peg5 buffer, as described Stortchevoi et al., 2020), library preparation with Oxford Nanopore kits EXP-NBD104 and SQK-LSK109, and sequencing on a R9 PromethION flow cell over 72 hours. Long reads were filtered using Filtlong (v0.2.0, -- min_length 20000 --keep_percent 99 --target_bases 500000000) (Wick, 2018), and hybrid assemblies were generated using Unicycler (v0.4.8, default parameters) (Wick et al., 2017). Scaffolds with homology (blastn, default parameters, total alignment lengths > 2,000 bp) to known C. acnes plasmids were designated as plasmid scaffolds. We note that the scaffold originating from colony subj-1_scrape-443_col-6 is actually a transposon that can be found on some C. acnes plasmids (e.g. NCBI CP017041 and our scaffold originating from colony subj-1_scrape-439_col-5) or in the absence of the plasmid (e.g. colony subj-1_scrape-443_col-6); we therefore do not treat it as a plasmid in downstream analyses.
To determine which colonies in our dataset had evidence of plasmid presence, we aligned short reads to these five plasmid scaffolds using the same procedure for alignments to the C. acnes reference genome. A colony was deemed as having a plasmid if it had a copy number over 0.33x (relative to the rest of the genome) across at least 75% of at least one of the plasmid scaffolds. Plasmid presence/absence is available in Table S2 (along with transposon presence/absence which was determined in the same manner, with a threshold of 90% instead of 75%) and indicated on lineage trees in Figures S6–8.
To see how the plasmids in our dataset were related to each other, we generated a phylogenetic tree comparing a region common to as many plasmids as possible. We used alignments to the plasmid scaffold generated from isolate subj-1_scrape-441_col-3 (this scaffold had the most lineages with at least one positive colony). We then masked positions where fewer than 85% of plasmid-positive colonies had a copy number over 0.75 and removed colonies that had a copy number of less than 0.75 over fewer than 75% of these positions (this removed 1 of 216 plasmid-positive colonies). Basecalls were marked as ambiguous if the quality was below 30, the coverage per strand was below 3, or the major allele frequency was below 0.67. This retained 867 variable positions, which were used to generate a parsimony tree using the same procedure as for lineage trees (Figure S9).
To avoid calling SNVs on mobile elements for genome focused analyses, we masked regions on the reference genome where there was an alignment to one of our plasmid or transposon scaffolds or to known plasmid genotypes CP003294 and CP017041 (blastn using default parameters with a minimum alignment length of 200 bp and a maximum e-value of 0.001). In our analysis of gain/loss regions, we additionally masked any contig for which these alignments covered over half of the contig positions.
Cutibacterium granulosum analysis
There were 50 colonies for which greater or equal to 75% of reads were assigned as Cutibacterium granulosum according to bracken (see Clustering colonies into lineages). To characterize the within-species C. granulosum diversity, we used an alignment-based approach following the same procedure as above, but with C. granulosum NCTC 11865 (RefSeq NZ_LT906441.1) as the reference genome. Colonies were removed from the analysis if less than 72% of reads aligned to the reference genome (7 colonies) or if they had a mean coverage of 5x or below across candidate variant positions (1 colony). Basecalls were marked as ambiguous if the FQ score produced by SAMtools was above −30, the coverage per strand was below 3, or the major allele frequency was below .75. Remaining variant positions were discarded if 34% or more of all colonies were called as ambiguous, if the median coverage across all colonies at that position was below 12, or if no polymorphisms remained. Any colonies for which greater than 30% of variant positions were marked as ambiguous at this stage were removed (this removed 3 colonies). In all, these filters retained ~90,000 variable positions across 39 colonies. Basecalls were repopulated from the raw data and only marked as ambiguous only if the FQ score was above −30, the coverage was below 5, or the major allele frequency was below .67. A parsimony tree (Figure S17, left panel) was generated using the same process as for C. acnes.
We identified three pores (Subject 1, pore 17; Subject 1, pore 18; Subject 2, pore 87) for which there were multiple C. granulosum colonies and for which these colonies were monophyletic on the tree constructed above. For each case, we assembled a genome using the same procedure as for C. acnes lineages, using reads from all colonies from that pore. We then aligned reads from each colony (including all colonies from that pore and any additional colonies within 100 SNVs according to the above tree) onto its pore-specific assembled genome and called SNVs using the same filters as above in order to generate parsimony trees (Figure S17, right panels).
16S amplicon sequencing
Samples collected for community profiling were collected in QuickExtract buffer (see Human subjects and sample collection). After streaking for single colonies, the remainder of samples were lysed by adding 1 ul of ReadyLyse (Epicentre) and incubating at room temperature for 12 hours. A 1 uL aliquot was used to amplify the V1-V3 region using HiFi HotStart ReadyMix (KAPA BioSystems) and the Illumina PCR protocol. A spike of genomic DNA from Caulobacter crecentus, a species typically found in freshwater, was included in each PCR reaction to estimate the number of unique sequencing reads. Samples were cleaned and pooled as in (Baym et al., 2015). Samples were sequenced on an Illumina MiSeq (300 PE) to an average read depth of ~16,500.
To classify amplicon sequence variants (ASV) on a species level, a classifier was built using the V1-V3 region of the raw sequences and taxonomy of the SILVA database (version 132) (Quast et al., 2013), with taxonomically-mislabeled sequences identified by the phylogeny-aware pipeline SATIVA (Kozlov et al., 2016) either corrected or removed. Staphylococcus species were specifically filtered by the methods presented in (Khadka et al., 2021). The genuses Cutibacterium, Acidipropionibacterium, Pseudopropionibacterium, Propionibacterium, and the Corynebacteriaceae and chronically-mislabelled Neisseriaceae families were also cleaned by the following filters: (i) sequences with incorrect higher taxonomic classes (ex. a species with the family Corynebacteriaceae but the genus Cutibacterium) were removed, (ii) sequences missing a species classification or assigned to non-species taxa (ex. Corynebacterium sp.) were removed, (iii) species with >60% similarity with other taxa were relabeled as a specific “taxa cluster”, (iv) taxonomically mislabeled sequences identified using SATIVA with greater than 90% confidence were relabeled and sequences with below 90% confidence removed. To reduce computational load, each family or genuses within the same family were grouped together in independent SATIVA runs. This removed about 2% of sequences from each group. This database was then used to train a naive Bayes classifier in QIIME2 (v2020.11).
QIIME2 was used to process and classify 16S reads, using Cutadapt and DADA2 (v1.18.0) (Bolyen et al., 2018; Callahan et al., 2016; Martin, 2011). To visualize species diversity, all spike-in sequences, unclassified reads, and reads with only a domain-level classification were removed from further analysis (Figure S11).
Growth curve assays
We measured growth rates for two sets of isolates: 9 isolates from three Subject 1 lineages collected at the same timepoint and 16 isolates from four Subject 2 lineages collected at the same timepoint. Frozen stocks of isolates were revived on RCM plates (Oxoid CM0149) that were reduced overnight anaerobically at 33°C. For each isolate, independent replicates were started from distinct colonies on the plate and grown to saturation over 4 days in deep-well plates in 400 uL of RCM in anaerobic conditions at 33°C. From this, cultures for growth curves were inoculated by diluting 2 μl of saturated culture into 198 μl of RCM in microtiter plates. Growth curves were obtained inside a Tecan M Nano, taking readings of OD595 nm every 15 minutes. Growth rates were obtained for each replicate by fitting a linear regression of ln(OD) versus time for every 3 hour and 45 minute interval (15 timepoints) during the period of exponential growth, and taking the highest slope with R2 >0.99 (Figure S3).
Competition model
We developed a mathematical model to explore how possible features of pores influence strain competition and coexistence. We consider a scenario in which two strains have the same initial relative abundance, and one strain has a fitness advantage of 50%. Our model tracks the relative abundances of the two competing strains on the skin surface and in 10,000 pores, and we evaluate the competition outcome after a simulation time of 10 years.
All simulations start with a 50/50 mix of the two strains on the skin surface. For simulations that do not allow coexistence inside pores, 50% of pores start filled with one strain and 50% of pores start filled with the other. For simulations that allow coexistence inside pores, each pore initially contains a 50/50 mix of the two strains.
Using a time step of one day, we simulate the following 3 processes sequentially each time step:
Cell growth (deterministic): We compute the change in relative abundance of the two strains on the surface and inside each pore, assuming exponential growth with the population doubling times shown in Figure S16. In some simulations, we assume that strains cannot compete inside pores.
Migration (deterministic): Migration from the pores to the surface is fixed at a constant rate, where 1/3 of the cells on the surface are replaced by cells from pores each day, mimicking the transport of cells to the skin surface via sebum flow. Only some simulations allow migration from the surface to pores (see Figure S16).
Pore re-population (stochastic): Each day, we randomly select a subset of pores to be reinitialized, using Poisson statistics with an average pore lifetime of 1 year. Pores are recolonized by the indicated number of cells randomly selected from the surface population using binomial statistics.
A table of all parameter values used in simulations is available in Figure S16. This model was implemented in Matlab (v2018a).
QUANTIFICATION AND STATISTICAL ANALYSIS
Information on the statistical tests and simulations can be found in the figure legends and in the corresponding Method Details. Statistics for all genomic analyses were computed in Matlab and statistics for growth curve analyses were computed in R.
Supplementary Material
Table ST1: Summary of samples taken from and colonies culture from each subject, related to Figure 1
Table ST2: Colony metadata, related to Figure 1
Table ST3: All regions detected as gained or lost within a lineage, related to Figure 2
Table ST4: Gene cluster analysis, related to Figure 2
Table ST5: Summary of lineages detected on each subject, related to Figure 2
Table ST6: All SNVs detected in all lineages, related to Figure 4
Table ST8: All potential recombination events removed from de novo mutation analysis, related to Figure 4
Highlights:
C. acnes lineages coexist across an individual’s skin but not within the same pore.
Colonies isolated from the same skin pore are nearly clonal (<1 mutation apart).
Neutral bottlenecking rather than selection drives low within-pore diversity.
Population fragmentation limits competition between C. acnes genotypes.
ACKNOWLEDGMENTS
We thank Sarah Bi, Sean Kearney, Sean Gibbons, Allison Perrotta, Claire Duvallet, Tucker Lynn, and all Lieberman Lab members for experimental assistance and helpful discussions, the MIT BioMicroCenter for assistance in sequencing, and Oskar Hallatschek for helpful discussions. We thank Vicki Mountain and Scott Olesen for assistance with IRB protocols. We thank Sayeh Gorjifard for assistance in creating the graphical abstract. This work was funded by grants from the Broad Institute (to E.J.A.), the Smith Family Foundation (to T.D.L.), the MIT Center for Microbiome Informatics and Therapeutics (to T.D.L.), and the National Institutes of Health (1DP2GM140922-01 to T.D.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Acosta EM, Little KA, Bratton BP, Mao X, Payne A, Davenport D, Gitai Z (2021). Bacterial DNA on the skin surface overrepresents the viable skin microbiome. Biorxiv 2021.08.16.455933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson AS, and Lipoff JB (2021). Reconsidering Named Honorifics in Medicine—the Troubling Legacy of Dermatologist Albert Kligman. Jama Dermatol 157, 153–155. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. (2012). SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, and Lenski RE (2013). Genome dynamics during experimental evolution. Nat Rev Genet 14, 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, and Kishony R (2015). Inexpensive Multiplexed Library Preparation for Megabase-Sized Genomes. Plos One 10, e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2018). QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. Peerj Prepr 6, e27295v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, Hujer S, Dürre P, and Gottschalk G (2004). The Complete Genome Sequence of Propionibacterium Acnes, a Commensal of Human Skin. Science 305, 671. [DOI] [PubMed] [Google Scholar]
- Brüggemann H, Lomholt HB, Tettelin H, and Kilian M (2012). CRISPR/cas Loci of Type II Propionibacterium acnes Confer Immunity against Acquisition of Mobile Elements Present in Type I P. acnes. Plos One 7, e34171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Weiner J, Wollherr A, Thürmer A, Hüpeden J, Lomholt HB, Kilian M, Gottschalk G, Daniel R, Mollenkopf H-J, et al. (2011). Comparative Genomics and Transcriptomics of Propionibacterium acnes. Plos One 6, e21581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher E, and Coonin A (1949). The physical properties of human sebum. J Investigative Dermatology 12, 249–254. [PubMed] [Google Scholar]
- Byrd AL, Belkaid Y, and Segre JA (2018). The human skin microbiome. Nat Rev Microbiol 16, 143–155. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, and Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: architecture and applications. Bmc Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda-García A, Martín-Blecua I, Cebrían-Sastre E, Chiner-Oms A, Torres-Puente M, Comas I, and Blázquez J (2020). Specificity and mutagenesis bias of the mycobacterial alternative mismatch repair analyzed by mutation accumulation studies. Sci Adv 6, eaay4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Lieberman TD, Vargas SO, Flett KB, McAdam AJ, Priebe GP, and Kishony R (2017). Global and local selection acting on the pathogen Stenotrophomonas maltophilia in the human lung. Nat Commun 8, 14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesen J, Spagnolo JB, Ramos SF, Kurita KL, Byrd AL, Aksenov AA, Melnik AV, Wong WR, Wang S, Hernandez RD, et al. (2020). A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci Transl Med 12, eaay5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, and Knight R (2009). Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 326, 1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove JH, Holland KT, and Cunliffe WJ (1983). Effects of Oxygen Concentration on Biomass Production, Maximum Specific Growth Rate and Extracellular Enzyme Production by Three Species of Cutaneous Propionibacteria Grown in Continuous Culture. Microbiology 129, 3327–3334. [DOI] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, and Foster KR (2015). The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–666. [DOI] [PubMed] [Google Scholar]
- Didelot X, Walker AS, Peto TE, Crook DW, and Wilson DJ (2016). Within-host evolution of bacterial pathogens. Nat Rev Microbiol 14, 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dréno B, Pécastaings S, Corvec S, Veraldi S, Khammari A, and Roques C (2018). Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol 32, 5–14. [DOI] [PubMed] [Google Scholar]
- Fenselstein J (2005). PHYLIP (Phylogeny Inference Package). Distributed by the author: Department of Genome Sciences, University of Washington, Seattle. Available at https://evolution.genetics.washington.edu/phylip.html. [software] [Google Scholar]
- Ferreiro A, Crook N, Gasparrini AJ, and Dantas G (2018). Multiscale Evolutionary Dynamics of Host-Associated Microbiomes. Cell 172, 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz-Gibbon S, Tomida S, Chiu B-H, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, et al. (2013). Propionibacterium acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J Invest Dermatol 133, 2152–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers L, and Grice EA (2020). The Skin Microbiota: Balancing Risk and Reward. Cell Host Microbe 28, 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Schluter J, Coyte KZ, and Rakoff-Nahoum S (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, and Li W (2012). CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics 28 (23): 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud NR, Good BH, Hallatschek O, and Pollard KS (2019). Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. Plos Biol 17, e3000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, and Segre JA (2011). The skin microbiome. Nat Rev Microbiol 9, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JB, Cong Z, Imamura-Kawasawa Y, Kidd BA, Dudley JT, Thiboutot DM, and Nelson AM (2018). Isolation and Identification of the Follicular Microbiome: Implications for Acne Research. J Invest Dermatol 138, 2033–2040. [DOI] [PubMed] [Google Scholar]
- Hartl DL, and Clark AG (2006). Principles of Population Genetics (Oxford University Press; ). [Google Scholar]
- Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, and Wardenburg JB (2016). Strain competition restricts colonization of an enteric pathogen and prevents colitis. Embo Rep 17, 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino S, Skouloubris S, Kudo H, l’Hermitte-Stead C, Es-Sadik A, Lambry J-C, Ishino Y, and Myllykallio H (2018). Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res 46, gky460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns AC, and Alexeyev OA (2014). Three dimensional distribution of Propionibacterium acnes biofilms in human skin. Exp Dermatol 23, 687–689. [DOI] [PubMed] [Google Scholar]
- Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, et al. (2015). Regional Isolation Drives Bacterial Diversification within Cystic Fibrosis Lungs. Cell Host Microbe 18, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, and Fass J (2011). Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. Available at https://github.com/najoshi/sickle. [software]
- Karita Y, Limmer DT, and Hallatschek O (2021). Scale-dependent tipping points of bacterial colonization resistance. Biorxiv 13.444017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimatis G, Fitz-Gibbon S, Tomida S, Wong M, and Li H (2013). Analysis of Complete Genomes of Propionibacterium acnes Reveals a Novel Plasmid and Increased Pseudogenes in an Acne Associated Strain. Biomed Res Int 2013, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, and Bohannan BJM (2002). Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature 418, 171–174. [DOI] [PubMed] [Google Scholar]
- Khadka VD, Key FM, Romo-González C, Martínez-Gayosso A, Campos-Cabrera BL, Gerónimo-Gallegos A, Lynn TC, Durán-McKinster C, Coria-Jiménez R, Lieberman TD, and García-Romero MT. (2021). The skin microbiome of patients with atopic dermatitis normalizes gradually during treatment. Front Cell Molec Microbiol 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Hall LJ, and Metcalf CJE (2017). The microbiome beyond the horizon of ecological and evolutionary theory. Nat Ecol Evol 1, 1606–1615. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Zhang J, Yilmaz P, Glöckner FO, and Stamatakis A (2016). Phylogeny-aware identification and correction of taxonomically mislabeled sequences. Nucleic Acids Res 44, 5022–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladau J, and Eloe-Fadrosh EA (2019). Spatial, Temporal, and Phylogenetic Scales of Microbial Ecology. Trends Microbiol 27, 662–669. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc JE, Li B, Payne WL, and Cebula TA (1996). High Mutation Frequencies Among Escherichia coli and Salmonella Pathogens. Science 274, 1208–1211. [DOI] [PubMed] [Google Scholar]
- Leeming JP, Holland KT, and Cunliffe WJ (1984). The Microbial Ecology of Pilosebaceous Units Isolated from Human Skin. Microbiology+ 130, 803–807. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Subgroup, 1000 Genome Project Data Processing (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W and Godzik A (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22:1658–9. [DOI] [PubMed] [Google Scholar]
- Lieberman E, Hauert C, and Nowak MA (2005). Evolutionary dynamics on graphs. Nature 433, 312–316. [DOI] [PubMed] [Google Scholar]
- Lieberman TD, Michel J-B, Aingaran M, Potter-Bynoe G, Roux D, Davis MR, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, et al. (2011). Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 43, 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, and Kishony R (2014). Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet 46, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman TD, Wilson D, Misra R, Xiong LL, Moodley P, Cohen T, and Kishony R (2016). Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat Med 22, 1470–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomholt HB, Scholz CFP, Brüggemann H, Tettelin H, and Kilian M (2017). A comparative study of Cutibacterium (Propionibacterium) acnes clones from acne patients and healthy controls. Anaerobe 47, 57–63. [DOI] [PubMed] [Google Scholar]
- Lourenço M, Chaffringeon L, Lamy-Besnier Q, Pédron T, Campagne P, Eberl C, Bérard M, Stecher B, Debarbieux L, and Sordi LD (2020). The Spatial Heterogeneity of the Gut Limits Predation and Fosters Coexistence of Bacteria and Bacteriophages. Cell Host Microbe 28, 390–401.e5. [DOI] [PubMed] [Google Scholar]
- Lu J, Breitwieser FP, Thielen P, and Salzberg SL (2017). Bracken: estimating species abundance in metagenomics data. Peerj Comput Sci 3, e104. [Google Scholar]
- Mak TN, Schmid M, Brzuszkiewicz E, Zeng G, Meyer R, Sfanos KS, Brinkmann V, Meyer TF, and Brüggemann H (2013). Comparative genomics reveals distinct host-interacting traits of three major human-associated propionibacteria. Bmc Genomics 14, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17, 10–12. [Google Scholar]
- McLaughlin J, Watterson S, Layton AM, Bjourson AJ, Barnard E, and McDowell A (2019). Propionibacterium acnes and Acne Vulgaris: New Insights from the Integration of Population Genetic, Multi-Omic, Biochemical and Host-Microbe Studies. Microorg 7, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi K, Aikawa C, Furukawa A, Watanabe T, Nakano T, Ogura Y, Ohtsubo Y, Kurokawa K, Hayashi T, Maruyama F, et al. (2013). Complete Genome Sequence of a Propionibacterium acnes Isolate from a Sarcoidosis Patient. Genome Announc 1, e00016–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskin JE, Farrell AM, Cunliffe WJ, and Holland KT (1997). Propionibacterium acnes, a resident of lipid-rich human skin, produces a 33 kDa extracellular lipase encoded by gehA. Microbiology+ 143, 1745–1755. [DOI] [PubMed] [Google Scholar]
- Mölder F, Jablonski KP, Letcher B, Hall MB, Tomkins-Tinch CH, Sochat V, Forster J, Lee S, Twardziok SO, Kanitz A, et al. (2021). Sustainable data analysis with Snakemake. F1000research 10, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, Barnabas B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, et al. (2014). Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Park M, Program NCS, Kong HH, and Segre JA (2016). Temporal Stability of the Human Skin Microbiome. Cell 165, 854–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A (2010). Mutators in cystic fibrosis chronic lung infection: Prevalence, mechanisms, and consequences for antimicrobial therapy. International Journal of Medical Microbiology 300, 563–572. [DOI] [PubMed] [Google Scholar]
- O’Neill AM, and Gallo RL (2018). Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 6, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzold B, Willis JR, Lima J.P. de, Knödlseder N, Brüggemann H, Quist SR, Gabaldón T, and Güell M (2019). Skin microbiome modulation induced by probiotic solutions. Microbiome 7, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plewig G (1974). Follicular Keratinization. J Invest Dermatol 62, 308–315. [DOI] [PubMed] [Google Scholar]
- Plewig G, Melnik B, and Chen W (2019). Plewig and Kligman’s Acne and Rosacea (Springer; ). [Google Scholar]
- Poyet M, Groussin M, Gibbons SM, Avila-Pacheco J, Jiang X, Kearney SM, Perrotta AR, Berdy B, Zhao S, Lieberman TD, et al. (2019). A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat Med 25, 1442–1452. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glöckner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossum TV, Ferretti P, Maistrenko OM, and Bork P (2020). Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol 18, 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C (2020). Out of your skin. Nat Biotechnol 38, 392–397. [DOI] [PubMed] [Google Scholar]
- Scholz CFP, Jensen A, Lomholt HB, Brüggemann H, and Kilian M (2014). A Novel High-Resolution Single Locus Sequence Typing Scheme for Mixed Populations of Propionibacterium acnes In Vivo. Plos One 9, e104199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz CFP, Brüggemann H, Lomholt HB, Tettelin H, and Kilian M (2016). Genome stability of Propionibacterium acnes: a comprehensive study of indels and homopolymeric tracts. Sci Rep-Uk 6, 20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreck CF, Fusco D, Karita Y, Martis S, Kayser J, Duvernoy M-C, and Hallatschek O (2019). Impact of crowding on the diversity of expanding populations. Biorxiv 743534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman T (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–9. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, and Lenski RE (1997). Evolution of high mutation rates in experimental populations of E. coli. Nature 387, 703–705. [DOI] [PubMed] [Google Scholar]
- Stortchevoi A, Kamelamela N, and Levine SS (2020). SPRI Beads-based Size Selection in the Range of 2-10kb. J Biomol Tech 31(1): 7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Barrick JE, Ribeck N, Deatherage DE, Blanchard JL, Dasgupta A, Wu GC, Wielgoss S, Cruveiller S, Médigue C, et al. (2016). Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida S, Nguyen L, Chiu B-H, Liu J, Sodergren E, Weinstock GM, and Li H (2013). Pan-Genome and Comparative Genome Analyses of Propionibacterium acnes Reveal Its Genomic Diversity in the Healthy and Diseased Human Skin Microbiome. Mbio 4, e00003–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C, Earle KA, Huang KC, and Sonnenburg JL (2017). The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host Microbe 21, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JLM, Rossetti BJ, Rieken CW, Dewhirst FE, and Borisy GG (2016). Biogeography of a human oral microbiome at the micron scale. Proc National Acad Sci 113, E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker WR, Shepherd ES, and Sonnenburg JL (2017). Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome. Cell 169, 538–546.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R (2018). Filtlong. Available at https://github.com/rrwick/Filtlong. [software]
- Wick RR, Judd LM, Gorrie CL, and Holt KE (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. Plos Comput Biol 13, e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgoss S, Barrick JE, Tenaillon O, Wiser MJ, Dittmar WJ, Cruveiller S, Chane-Woon-Ming B, Médigue C, Lenski RE, and Schneider D (2013). Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc National Acad Sci 110, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser MJ, Ribeck N, and Lenski RE (2013). Long-Term Dynamics of Adaptation in Asexual Populations. Science 342, 1364–1367. [DOI] [PubMed] [Google Scholar]
- Wood DE, Lu J, and Langmead B (2019). Improved metagenomic analysis with Kraken 2. Genome Biol 20, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lieberman TD, Poyet M, Kauffman KM, Gibbons SM, Groussin M, Xavier RJ, and Alm EJ (2019). Adaptive Evolution within Gut Microbiomes of Healthy People. Cell Host Microbe 25, 656–667.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Spoto M, Hardy R, Guan C, Fleming E, Larson PJ, Brown JS, and Oh J (2020). Host-Specific Evolutionary and Transmission Dynamics Shape the Functional Diversification of Staphylococcus epidermidis in Human Skin. Cell 180, 454–470.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table ST1: Summary of samples taken from and colonies culture from each subject, related to Figure 1
Table ST2: Colony metadata, related to Figure 1
Table ST3: All regions detected as gained or lost within a lineage, related to Figure 2
Table ST4: Gene cluster analysis, related to Figure 2
Table ST5: Summary of lineages detected on each subject, related to Figure 2
Table ST6: All SNVs detected in all lineages, related to Figure 4
Table ST8: All potential recombination events removed from de novo mutation analysis, related to Figure 4
Data Availability Statement
All raw sequencing data have been deposited in NCBI-SRA and are publicly available as of the date of publication. Genome assemblies have been deposited on GitHub and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Additionally, processed data are available in Tables S3, S4, and S6.
All original code has been deposited at GitHub and is publicly available as of the date of publication. The GitHub repository is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| C. acnes isolates | This manuscript | N/A |
| C. granulosum isolates | This manuscript | N/A |
| Biological samples | ||
| Skin scrapes and skin pore samples from healthy people | This manuscript | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Brucella Blood Agar | Hardy Diagnostics | A30 |
| QuickExtract buffer | EpiCentre | QE09050 |
| Lysozyme | Millipore Sigma | 62971 |
| PCRClean-DX SPRI beads | Aline Biosciences | C-1003-250 |
| Polyethylene glycol (PEG) 8000 | Hampton Research | HR2-535 |
| Magnesium chloride (MgCl2) | Ambion | AM9530G |
| ReadyLyse Lysozyme Solution | EpiCentre | R1810M |
| KAPA HiFi HotStart ReadyMix | Roche | 7958927001 |
| Reinforced Clostridial Media (RCM) | Oxoid | CM0149 |
| Critical commercial assays | ||
| PureLink Genomic DNA Kit | Invitrogen | K182002 |
| Illumina Tagment DNA TDE1 Enzyme and Buffer Kits | Illumina | 20034198 |
| High-Molecular Weight Genomic DNA Kit | Qiagen | 67563 |
| Ligation Sequencing Kit and Native Barcoding Expansion 1-12 | Oxford Nanopore | SQK-LSK109 and EXP-NBD104 |
| Blackhead Removal Activated Carbon Mask | Mengkou | 4716872044078 |
| Deposited data | ||
| Raw sequencing data | This manuscript | NCBI-SRA BioProject: PRJNA771717 |
| Assembled genomes for each C. acnes lineage | This manuscript | GitHub: https://github.com/arolynconwill/cacnes_biogeo |
| Hybrid assemblies of C. acnes colonies with plasmids | This manuscript | GitHub: https://github.com/arolynconwill/cacnes_biogeo |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| 16S V1-V3 forward primer (27F-plex): TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATCMTGGCTCAG | Khadka et al., 2021 | N/A |
| 16S V1-V3 reverse primer (534R-plex): GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGATTACCGCGGCTGCTGG | Khadka et al., 2021 | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| All original code | This manuscript | https://github.com/arolynconwill/cacnes_biogeo |
| Snakemake (v6.4.1) | Mölder et al., 2021 | https://snakemake.readthedocs.io/en/stable/ |
| Matlab (v2015b, v2018a) | Mathworks | https://www.mathworks.com/products/matlab.html |
| Cutadapt (v1.18) | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| Sickle (v1.33) | Joshi and Fass, 2011 | https://github.com/najoshi/sickle |
| Bowtie 2 (v2.2.6) | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SAMtools (v1.5) and BCFtools (v1.2) | Li et al., 2009 | https://github.com/samtools/ |
| Kraken 2 (v2.0.7) | Wood et al., 2019 | https://github.com/DerrickWood/kraken2/wiki |
| Bracken (v2.5) | Lu et al., 2017 | https://github.com/jenniferlu717/Bracken |
| BLAST (v2.7.1) | NCBI | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| PHYLIP (v3.69) | Fenselstein, 2005 | https://evolution.genetics.washington.edu/phylip.html |
| FigTree (v1.4.4) | Andrew Rambaut | https://github.com/rambaut/figtree |
| SPAdes (v3.13) | Bankevich et al., 2012 | https://github.com/ablab/spades |
| Prokka (v4.8.1) | Seemann, 2014 | https://github.com/tseemann/prokka |
| CD-HIT (v4.8) | Li et al., 2006; Fu et al., 2012 | http://weizhong-lab.ucsd.edu/cd-hit/ |
| Filtlong (v0.2.0) | Wick, 2018 | https://github.com/rrwick/Filtlong |
| Unicycler (v0.4.8) | Wick et al., 2017 | https://github.com/rrwick/Unicycler |
| SATIVA | Kozlov et al., 2016 | https://github.com/amkozlov/sativa |
| QIIME2 (v2020.11) | Bolyen et al., 2018 | https://qiime2.org |
| DADA2 (v1.18.0) | Callahan et al., 2016 | https://benjjneb.github.io/dada2/index.html |
| Other | ||
| Public C. acnes genomes | NCBI GenBank | Accession: NC_018707.1 |
| Public C. acnes plasmid sequences | NCBI GenBank | CP003294 and CP017041 |
| Public C. granulosum genomes | NCBI GenBank | NZ_LT906441.1 |
| SILVA database (version 132) | Quast et al., 2013 | https://www.arb-silva.de |


