Abstract
The exact pathogenesis of inflammatory bowel disease (IBD), a chronic gastrointestinal inflammatory disease comprising Crohn’s disease and ulcerative colitis, remains unclear. Studies on ubiquitination, which regulates the degradation of inflammation signalling pathway molecules, and deubiquitination have provided novel insights. Targeting the ubiquitin-specific protease (USP) family of deubiquitinases elucidates IBD signalling pathway mechanisms and possibly, IBD therapeutic solutions. Here, we characterised USPs as chief regulators of pro-inflammatory signalling pathways, including nuclear factor-κB and transforming growth factor-β; analysed the relationship between USPs and IBD pathogenesis in terms of genetic susceptibility, intestinal epithelial barrier, immunity, and gut microbiota; and discussed future research prospects.
Subject terms: Inflammatory bowel disease, Ubiquitylation
Facts
Inflammatory bowel disease (IBD) is a chronic intestinal disorder. Various signalling pathways have a regulatory effect on IBD, but the specific activation and regulation mechanism of the signalling pathway is still unclear.
Deubiquitination implicates numerous biological processes and plays an essential role in various human disease generation.
Ubiquitin specific proteases (USPs) are largest family of deubiquitinating enzymes and involved in the signalling pathway regulation as well as pathogenesis of IBD.
The findings regarding the role of USPs in IBD may point out future researches and approaches to the therapeutic targets of IBD.
Open questions
What are the constituents and functions of USPs family?
How USPs regulate the inflammation signalling pathways?
What is the role of USPs in the pathogenesis of IBD?
Inflammatory bowel disease (IBD), a chronic gastrointestinal inflammatory disease comprising Crohn’s disease (CD) and ulcerative colitis (UC), has become prevalent worldwide [1]. Rising case incidence and pathological therapy challenges have recently led to extensive research on the pathology and treatment of IBD, a disease with precipitating factors such as impaired intestinal barrier, altered gut bacteria, and disordered immune responses [2]. Understanding the impact of these pathological alterations is key to discovering treatment targets, and multiple studies have focused on the signalling pathways involved in IBD pathogenesis to clarify the underlying mechanisms [3].
Despite recent studies on IBD mechanisms and pathways, a precise identification of signalling pathway targets has yet to be achieved. Previous researches have elucidated the function of enzymes in ubiquitination, a post-translational process that provides an important opportunity for the regulation of immune-inflammatory response signalling molecules [4]. However, deubiquitination, the process that reverses ubiquitination, has not been examined critically with respect to IBD regulation. Deubiquitinating enzymes (DUBs), a superfamily of proteases that can be categorized by catalytic mechanism into cysteine proteases (USPs, UCHs, OTUs, MJDs, MINDYs and ZUP1) and metalloproteases (JAMMs), are critical in regulating ubiquitin signalling [5, 6] and associated with human diseases such as cancer, centre nervous system disease, autoimmune disease and infections [7]. DUBs remove ubiquitin from specific substrate proteins and affect subsequent proteasomal degradation and sub-cellular localization, which maintains a balanced level of protein quality control and homoeostasis between ubiquitination and deubiquitination in physiological condition [7]. Ubiquitin-specific proteases (USPs) are the largest family of DUBs and play vital roles in deubiquitinating modification. The present study characterises USPs as the primary regulators of pro-inflammatory signalling pathways, including the nuclear factor-κB (NF-κB) and the transforming growth factor-beta (TGF-β) pathways. We analysed the relationship between USPs and IBD pathogenesis in terms of genetic susceptibility, immunity, intestinal epithelial barrier function, and gut microbiota and discussed prospects for future research on USPs and IBD.
USP role and mechanism
USP structure and catalytic mechanism
As the largest family of DUBs, USPs bear both similarities with, and critical differences from, the other cysteine protease DUBs. All DUB catalytic domains contain a primary ubiquitin-binding site that enables numerous interactions with ubiquitin in the distal region of a polyubiquitin chain [6]. USP catalytic domains, similar to those of classical cysteine proteases, rely on a catalytic dyad or triad of conserved amino acid residues; thus, the various isopeptide-bond hydrolysation processes of different USPs are quite similar to one another. In contrast to other cysteine protease DUBs, USPs contain specific USP domains exhibiting a functional catalytic mechanism and consisting of three conserved subdomains resembling the thumb, palm, and fingers of a right hand. The USP catalytic centre, which includes a C-terminal His Box and N-terminal Cys Box with catalytic His and Cys residues, respectively, is located at the interface between the thumb and palm subdomains, and the finger subdomain grips the distal ubiquitin in the ubiquitin chain during interactions [6]. USP family members have varying molecular structures, with most comprising a core catalytic domain with insertions and other N- and C-terminal extensions, which may consist of different domains or sequences and perform specific functions [8]. As examples, the USP5 N-terminus contains a ubiquitin-binding zinc-finger domain required for binding the ubiquitin C-terminus diglycine motif [9], while the USP7 N-terminal tumour necrosis factor receptor (TNFR)-associated factor domain recognizes various substrates, enhancing USP7 catalytic activity [10]. CYLD is the only USP family member that lacks the finger subdomain [11].
Ubiquitin code and USPs
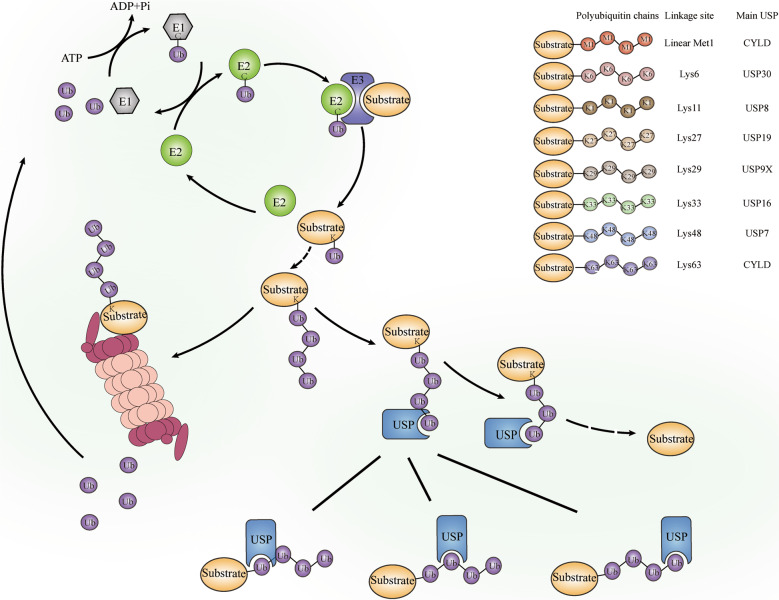
Ubiquitin, a small conserved regulatory protein composed of 76 amino acids, covalently attaches to substrates to act as a modular marker. During ubiquitination, substrate proteins can be attached to a monoubiquitin or to polyubiquitin chains, via multiple interaction modes. Four types of substrate ubiquitination provide additional diversity: monoubiquitination, multi-monoubiquitination, homotypic polyubiquitination, and heterotypic polyubiquitination. Ubiquitin can be linked by one of seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63), or by the first methionine (Met1) in a polyubiquitin chain [12]. As a regulatory post-translational modification, ubiquitination is reversible via DUBs, peptidases that can cleave ubiquitin from substrate proteins (Fig. 1). DUBs regulate eukaryotic ubiquitination dynamics by cleaving ubiquitin or ubiquitin-like proteins from target proteins or pro-proteins, preventing 26S proteasome degradation of the substrates [7]. As the largest subgroup of DUBs, USPs recognise a variety of substrates with Lys48-, Lys63- and Met1-linked ubiquitin chains, and regulate various biological processes, including cell migration, tumorigenesis, immunity response and inflammation activation [13]. Aberrant expressions, or blocked activity of USPs commonly cause abnormal tissue development and a series of disorders, such as abnormal expression of pro-inflammatory factors and impaired intestinal barrier function [14, 15].
Fig. 1. The role and mechanism of USPs family.
During the ubiquitination process, the cysteine group of the ubiquitin-activating enzyme (E1) conjugates the C-terminal carboxyl group of ubiquitin to initiate ubiquitination with assumption of an ATP. Subsequently, the ubiquitin-conjugating enzyme (E2) binds to E1 and activated ubiquitin complex, catalysing the ubiquitin transfer from E1 to the active site of E2 through transesterification reaction. Then the ubiquitin-protein ligase (E3) establishes an isopeptide bond with C-terminal glycine on ubiquitin and lysine on substrate to form a ubiquitin-substrate complex. Ubiquitin can be conjugated by another ubiquitin to form a polyubiquitin chain via seven Lys residues (Lys6, 11, 27, 29, 33, 48 and 63) or Met1 by E1/E2/E3 recruitment. Once marked by a ubiquitin chain especially Lys48-linked chain, substrate tends to be degraded by proteasome. However, this process can be reversed by DUBs including multiple USPs. For example, CYLD can deubiquitinates Met1- and Lys63-linked polyubiquitin chains for signalling [29]; USP30, the only human USP that inserted in the outer mitochondrial membrane, deubiquitinates Lys6-linked di-ubiquitin for mitophagy [112]; USP8 deubiquitinates Lys11 for autophagy regulation [113]; USP19 deubiquitinates Lys27-linked polyubiquitination for innate immune responses [114]; USP9X is implicated to deubiquitinate Lys29-linked polyubiquitination [115]; USP16 deubiquitinates Lys33-linked polyubiquitination for signalling [40]; USP7 deubiquitinates Lys48-linked polyubiquitin chains for suppression of proteasomal degradation [45]. With specific USP domain, USPs can recognized the ubiquitin and catalyse the cleavage of polyubiquitin chains in the proximal, middle or distal linkage.
USP regulation
USPs can be controlled at the synthesis and degradation levels. USP regulation during synthesis via mRNA expression is demonstrated by the antagonistic regulation of USP22 and USP44 mRNA expression, which ensures faithful stem cell differentiation [16, 17]. Moreover, DUB transcription is upregulated in specialised cell types or at various differentiation stages, providing the potential regulation of USP levels [18]. Finally, USP activation can be modulated by post-translational mechanisms, primarily via phosphorylation. For instance, AKT activation promotes the phosphorylation, and the subsequent subcellular localisation in the membrane and cytoplasm, of USP4 [19]. PINK1-induced phosphorylation of ubiquitin at Ser65 severely alters the ubiquitin electrostatic potential and surface properties via the addition of a negative charge, consequently inhibiting the function of USP2, USP8, USP15 and USP30 [20]. In addition, substrate binding induces conformational changes, thus regulating USP activation. Binding to ubiquitin produces rearrangements in the USP catalytic triad ubiquitin-binding site, such as the realignment of the catalytic triad in USP7 [21], the translocation of two surface loops, blocking loops 1 and 2, in USP14 [22], and the movement of the finger domain in USP8 [23]. USP activation can also be regulated by scaffold or adaptor binding, such as USP14 inactivation via blockage by two surface loops until it is properly localised and associated with the proteasome [8].
USPs and inflammation signalling pathways
The prominent role of USPs in the molecular mechanism of multiple pathways is demonstrated by their regulation of protein activation via the dissociation of monoubiquitin or polyubiquitin chains from ubiquitinated substrates. Here, we summarise relevant associations between USPs and inflammatory pathway signals, which may elucidate the regulatory role of USPs.
USPs and the NF-κB signalling pathways
NF-κB is a family of structurally related eukaryotic protein transcription factors that comprise p50/p105, p52/p100, RelA/p65, c-Rel and RelB [24]. Under basal conditions, NF-κBs interact with NF-κB inhibitory proteins (IκBs) in the cytoplasm, forming a deactivated trimer [25]. Multiple extracellular signals activate a cytoplasmic IκB kinase complex (IKK) which phosphorylates inhibitory IκBs, leading to the separation of IκBs from NF-κB and the dissociation of IκBs via the ubiquitin-proteasome dependent pathway [26]. Stimulated NF-κBs migrate into the nucleus, bind to the DNA consensus, and activate the expression of target genes [24]. Studies have demonstrated that the NF-κB signalling pathway is essential for inducing pro-inflammatory gene expression, regulating the inflammasome, and activating inflammatory T lymphocyte and innate immune cell differentiation in the inflammatory colon of IBD patients [27, 28].
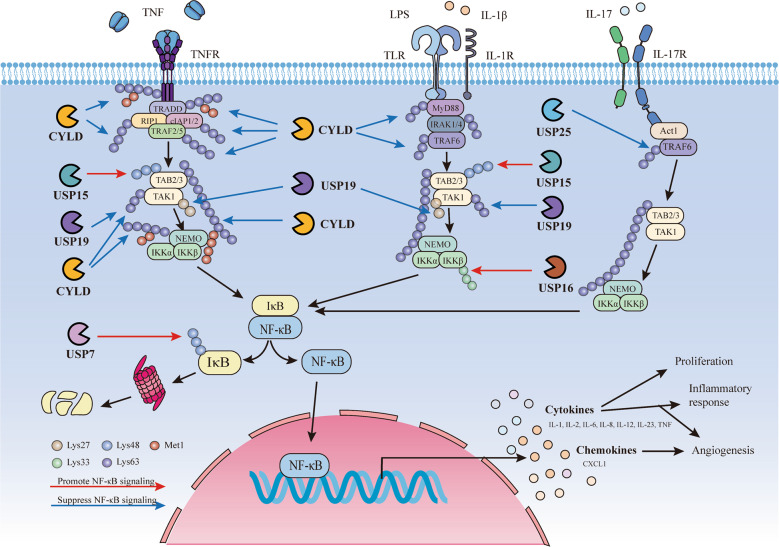
The USP family exerts crucial functions in regulating the NF-κB signalling pathway (Fig. 2). CYLD specifically diverts the Lys63- and Met1-linked polyubiquitin chains through two steps of cleavage reaction: initial binding and subsequent hydrolysis [29]. CYLD deubiquitinates various key negative regulators in the NF-κB pathway, including the NF-κB essential modulator (NEMO), receptor-interacting protein 1 (RIP1), TNFR-associated factor (TRAF)2, TRAF6, TRAF7, TNF receptor type 1-associated death domain protein (TRADD), RIP kinase(RIPK)1 and RIPK2 [29, 30]. Previous studies have demonstrated that CYLD causes TRAF2 ubiquitination reduction, and that a CYLD active site increases ubiquitinated TRAF2 levels, leading to TRAF2-mediated NF-κB activation, suggesting that CYLD inhibits NF-κB signalling activation through deubiquitination in TRAF2. A similar inhibitory effect has also been observed in NF-κB activation mediated by TRAF6 [31]. TNF-stimulated CYLD-deficient A549 cells exhibit increased Met1- and Lys63-linked ubiquitination of TNFR1, RIP1 and TRADD, suggesting that, when present in the TNFR signalling complex, these proteins carry Met1- and Lys63-linked ubiquitin chains, which can be antagonized by CYLD [32]. The T cells of CYLD-deficient mice overexpress p50, RelA, RelB and p100, which may be attributed to the absent inhibitory role of CYLD in TAK1 ubiquitination [33]. Upon anti-CD3 stimulation, CYLD-deficient mice further presented enhanced NEMO ubiquitination and elevated NF-κB activation, revealing the role of CYLD as a NEMO deubiquitinase and NF-κB negative regulator [34]. Containing the coil coiled 2-leucine zipper-zinc finger domain at the C-terminus, NEMO binds preferentially to Lys63-linked polyubiquitin chains, which can then be disconnected by CYLD. The Lys63-specific CYLD deubiquitinating activity removes the Lys63-linked ubiquitin chain from NEMO on RIP1, maintaining the IKKα-IKKβ-NEMO trimer structure and forming the RIP1–Fas-associated protein with the death domain (FADD)–caspase-8 complex, which participates in the negative feedback loop of NF-κB activation [35]. The loss of CYLD protects against TNF-α-induced apoptosis [36]. In addition, CYLD targets Lys63-linked ubiquitin chains on MyD88 during inflammation induced by Haemophilus influenzae infection [37].
Fig. 2. USPs and NF-κB signalling.
In response to TNF, TNFR conjugates with TRADD, recruiting the kinase RIPK1 and ubiquitin E3 ligase TRAF2/5. TRAF2 associates with cIAP1 and 2 to modify multiple components in TNFR1 complex with Lys63 polyubiquitin (left side of the figure). Stimulation of Toll-like receptors (TLR) or interleukin-1 receptor (IL-1R) induces the arrangement of the Myddosome complex consisting of MyD88, IRAK4, IRAK1. Subsequently, IRAK1 is phosphorylated by IRAK4 and subsequently recruits the ubiquitin E3 ligase TRAF6 (middle side of the figure). Upon the stimulation of IL-17, IL-17R engages Act1 to mediate the recruitment of TRAF6. In all these cases, ubiquitination serves to recruit TAB2/TAB3/TAK1 and then NEMO/IKK kinase complexes, activating NF-κB signalling (right side of the figure). USPs including CYLD, USP7, USP15, USP16, USP19 and USP25 counteract the NF-κB signalling. CYLD removes Lys63-linked polyubiquitin chains from several substrates such as TNFR, TRADD, RIP1, TRAF2/5, cIAP1/2, MyD88 and TRAF6, negatively regulating NF-κB activation. USP15 deubiquitinates Lys48-linked polyubiquitination to maintain TAB2 stability and enhance NF-κB signalling induced by TNF and IL-1β. Similarly, upon TNF and IL-1β stimulation, USP19 removes Lys63- and Lys27-linked polyubiquitin chains from TAK1 and negatively regulates the activation of NF-kB. USP16 deubiquitinates Lys33-linked polyubiquitination from IKKβ and activates NF-kB. In IL-17 induced signalling, USP25 deubiquitinates Lys63-linked ubiquitination of TRAF6 mediated by Act1 and negatively regulates NF-κB signalling. Instead of regulating signals in the upstream of NF-κB, USP7 interacts with NF-κB subunits and deubiquitinates Lys48-linked ubiquitin chains, inhibiting p65 degradation and promoting NF-κB signalilng.
In addition to CYLD, other USPs also regulate the NF-κB signalling pathway. USP15 can remove Lys48-linked ubiquitination chains and subsequently inhibit the degradation of TAB2 and TAB3 to hamper autophagy cargo receptor 1-mediated selective autophagic degradation [38]. In accordance with the results, USP15 overexpression in 293T cells enhances the interleukin (IL)-1β- and the TNF-α-mediated phosphorylation of two NF-κB activation hallmarks, the IKK complex and IκBα, while USP15 knockdown suppresses NF-κB activation and downstream gene transcription [38]. USP19, an endoplasmic reticulum–anchored DUB, is implicated in regulating endoplasmic reticulum-associated protein degradation and damage repair and is mediated by IL-1β and TNF-α at the TAK1 level, similar to USP15. USP19 negatively regulates TAK1-TAB1-dependent, rather than IKK-β- and p65-mediated, NF-κB activation with specific deconjugation of Lys63- and Lys27-linked polyubiquitin chains from TAK1 [39]. USP16 expression levels are significantly raised in macrophages of IBD patients and higher in inflammatory sections [40]. Mass spectrometry reveals that USP16 specifically binds to IKK-α and IKK-β. Under lipopolysaccharide or TNF-α stimulation, competing with NEMO, USP16 specifically deubiquitinates the Lys33-linked polyubiquitination of IKKs and promotes IKK-β-mediated p105 phosphorylation without direct IκBα or p65 phosphorylation, leading to an autoimmune response and IBD colon cancer progression [40]. USP16 levels may be enhanced by p65 overexpression on the binding sites of USP16-NF-κB2/3/4 on USP16 mRNA [41]. In addition, mRNA expression of inflammatory cytokines such as TNF, IL-1β, -12α, -12β and -23α, is elevated in the colons of USP16-deficient mice [40]. The dysregulation of nucleotide-binding oligomerization domain-containing protein 2 (NOD2), a significant CD pathogenesis genetic risk factor and a bacterial cell wall component sensor, in the colon leads to a chronic relapsing inflammatory disorder and microbial infection control failure, thereby initiating systemic immune responses and aberrant inflammation [42]. USP8 suppresses NOD2 activation stimulated by Staphylococcus aureus in the NF-κB pathway, thus reducing the expression of pro-inflammatory factor IL-8 [43]. USP25 is also a significant modulator in regulating NF-κB signalling. Upon IL-17 stimulation, USP25 deubiquitinates the Lys63-linked TRAF5 and TRAF6 ubiquitination mediated by Act1. Overexpression of USP25 inhibits the IL-17-mediated phosphorylation and degradation of IκBα, thereby negatively regulating NF-κB signalling [44]. USP7 knockdown inhibits p65 recruitment to the IL-6 and TNF-α promoters, suggesting USP7 regulates NF-κB at the proximal point near the promoter in place of the point upstream of Toll-like receptor (TLR)- or TNFR-mediated signalling pathways. USP7 inhibits p65 ubiquitination and Lys48-linked ubiquitin chain-mediated p65 degradation and can directly interact with NF-κB, including the p50, p52, c-Rel and RelB subunits [45]. These researches collectively represent the essential functions of USPs in affecting NF-κB signalling pathway component activation.
USPs and the TGF-β signalling pathway
The immunosuppressive cytokine TGF-β signalling pathway has been studied with respect to potent regulatory and inflammatory activity [46]. The TGF-β ligand binds to specific TGF-β type I or II receptors on the surfaces of immune and epithelial cells to initiate TGF-β signalling. Type I receptors are recruited to type II receptors and activated via phosphorylation modification. Activated type I receptors then phosphorylate and activate the intracellular transduction signalling pathway via SMAD-dependent and -independent noncanonical pathways [47]. In the canonical SMAD-dependent pathway, TGF-β type I receptors phosphorylate SMAD2 and SMAD3, subsequently assembling SMAD4, which transfers into the nucleus to control target gene expression associated with the TGF-β signalling pathway [48]. In SMAD-independent pathways, TGF-β receptors activate other signalling modules, for instance, mitogen-activated protein kinases (MAPKs), phosphatidylinositol 3-kinase-Akt and p21 activated kinase 2 [49].
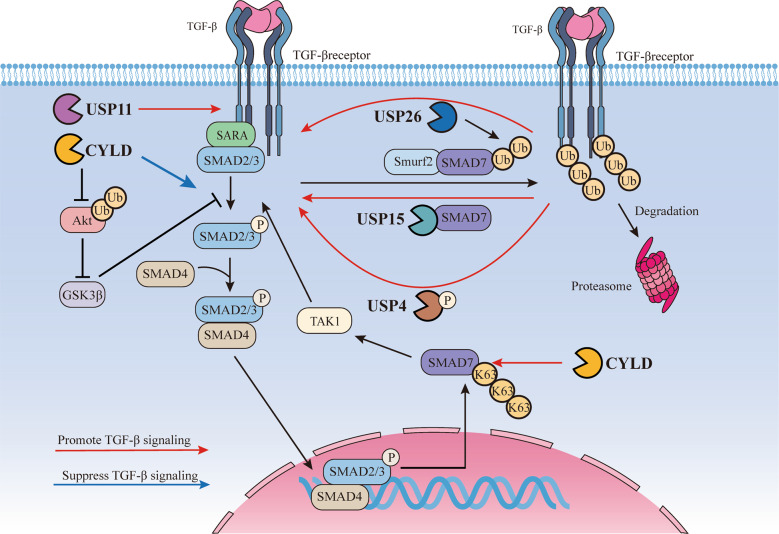
The negatively regulated inhibition of receptor-activated SMADs by ubiquitin ligases is counteracted by USPs through deubiquitination (Fig. 3). USP15, a TGF-β signalling pathway regulating factor in mammalian cells [50], operates both nucleus- and cytoplasm-localised downstream SMAD activation, which targets ubiquitinated receptor-activated SMADs [50]. Similar to TGF-β type I receptor associated with USP15, rather than binding to the receptor directly, USP15 binds to the receptor by SMAD7, a scaffold that also recruits SMAD-specific E3 ubiquitin protein ligase 2 (SMURF2). USP15 can remove the polyubiquitin chains of the SMURF2-ubiquitinated receptor [51]. The TGF-β signalling pathway exhibits enhanced expression of both SMAD7 and USP16; the latter, involved in negative regulation of the TGF-β signalling pathway [52], can deubiquitinate as well as stabilise the former, resulting in SMAD7-SMURF2 interaction stabilisation. USP4, with strong TGF-β signalling-inducing capability, is relocated from the nucleus to the cytoplasm and membrane by AKT-mediated phosphorylation and directly binds to, and reverses the ubiquitination of, the activated TGF-β type I receptor [19]. USP11 is a protein which participates in TGF-β signalling by regulating multiple responses, including regulatory T (Treg) and T helper (Th)17 cell induction, which is implicated in colon cancer [53]. In stable and peripherally induced Treg cells, USP11 expression is enhanced and increases TGF-β signal sensitivity in receptor-stimulated CD4+ T cells. However, increasing exogenous TGF-β or IL-2 levels do not affect USP11 expression in CD4+ T cells [53]. In addition, USP11 has been shown to affect TGF-β type II receptor stability via deubiquitination [54]. CYLD is a DUB which controls the TGF-β signalling pathway and decreases SMAD3 protein stability in an Akt-glycogen synthase kinase3β-hsc70-interacting protein-dependent manner, thus inhibiting TGF-β signalling [55]. CYLD-deficient T cells exhibit an elevated TGF-β responsiveness capacity and promote AP-1, TAK1 and p38 activation. Furthermore, CYLD deubiquitinates SMAD7 Lys63-linked polyubiquitination chains, regulating activation of the SMAD7-TAK1-TAB2/3 complex and transcription factor activator protein 1 [56].
Fig. 3. USPs and TGF-β signalling.
TGF-β ligands bind to the TGF-β receptors and activate the SMADs proteins. USPs including USP4, USP11, USP15, USP26 and CYLD are associated with TGF-β signalling. Upon AKT-mediated phosphorylation, phosphorylated USP4 recruits to the activated TGF-β type I receptor directly and reverses receptor ubiquitination, leading to TGF-β signalling. USP11 deubiquitinates and stabilizes TGF-β type II receptor to promote TGF-β signalling. Reversed the polyubiquitination by SMAD specific E3 ubiquitin protein ligase 2 (Smurf2) and SMAD7, USP15 is recruited to the TGF-β type I receptor with scaffold protein SMAD7 as well and suppresses polyubiquitination and degradation of the receptor, positively regulating TGF-β signalling. USP26 is recognized as a negative regulator that deubiquitinates SMAD7 and stabilizes the interaction of SMAD7 and Smurf2, negatively regulating TGF-β signalling. CYLD decreases SMAD3 protein stability via an Akt-glycogen synthase kinase3β-hsc70-interacting protein-dependent manner, suppressing the activation of TGF-β signalling. CYLD also deubiquitinate Lys63-linked polyubiquitin chains of SMAD7 to regulate the activation of SMAD7-TAK1-TAB2/3 complex and transcription factor activator protein 1.
USPs and other signalling pathways
In addition to the NF-κB and TGF-β pathways, the USP deubiquitinating role is essential to other signalling pathways. USP11 promotes the MAPK signalling pathway in a PPP1CA-dependent manner [57]. PPP1CA, one of the three catalytic subunits of protein phosphatase 1, is implicated in tumorigenesis via the MAPK signalling pathway [58]. A previous research demonstrated that elevated USP11 in colorectal cancer cells could protect PPP1CA from proteasome-mediated degradation, thus activating the MAPK pathway and promoting colon cancer cell growth and metastasis [57]. Moreover, per the critical protein ubiquitination function in TLR signalling, several USPs regulate TLR-mediated signalling via crosstalk with other pathways. USP25 inhibits TLR4-dependent activation of NF-κB and MAPKs to regulate innate immune responses by deubiquitinating adaptor protein TRAF3 via the coiled-coil domains, along residues 644 to 1055 [59]. USP15 interacts with Hrd1, an E3 ligase, in microphages to regulate IkBα degradation and pro-inflammatory cytokine production in the TLR4-mediated signalling pathway [60]. In small intestine tumour tissues or USP25-deficient colons, the level of Wnt-related gene products is significantly reduced compared to those in USP25-sufficient colons subjected to Wnt3a stimulation, demonstrating that USP25 participates in Wnt signalling pathway regulation during colonic tumourigenesis [61]. Finally, USP28 controls cellular activation levels of the Notch pathway, which maintains intestinal homoeostasis and controls intestinal stem cell development and differentiation [62]. Notch signalling regulator alterations can lead to aberrant Notch activation and subsequent severe complications, including intestinal inflammation due to secretory cell deficiency [63]. USP28 recognises and deubiquitylates Notch receptor 1, regulating the gene expression of Notch signalling pathway downstream molecules, such as Hes1, Hes5 and delta-like canonical notch ligand 1 [64]. Insufficient USP28 levels may deactivate excessive intestinal hyperplasia and impaired intestinal differentiation [65].
USPs are also involved in regulating the Hippo pathway, a conserved kinase cascade that has been implicated in colon development, intestinal regeneration and homoeostasis [66]. Yes-associated protein (YAP)1 and the transcriptional co-activator with PDZ-binding motif, both downstream effectors of the Hippo pathway, are the key transcriptional co-activators that regulate target gene expression and tissue growth. YAP is elevated in IBD and is mainly found in intestinal epithelial cell (IEC) nuclei in the villi and crypts of the small intestine and colon [67, 68]. USP9X strongly interacts with large tumour suppressor kinase 1, an important YAP negative regulator, consequently stimulating the Hippo pathway [69]. In addition, USP47 interacts with YAP, promotes YAP transcriptional activity and protein stability, and is the interactor for β-Trcp, an E3 ubiquitin ligase, leading to NLR family pyrin domain-containing (NLRP)3 proteasomal degradation via Lys27-linked polyubiquitination at Lys380. YAP binds to, and promotes activation of, NLRP3, which competitively inhibits β-TrCP1-mediated polyubiquitination [70], demonstrating YAP’s function in immune disorders and inflammatory diseases. This suggests that the USP47-YAP signalling axis in the Hippo pathway may mediate NLRP3 activation and regulate immune and inflammatory responses.
USPs and IBD pathogenesis
Although the causes of IBD pathogenesis remains elusive, many studies have revealed that genetic susceptibility, intestinal epithelial barrier, mucosal immune response, and gut microbiota are associated with an altered homoeostasis, leading to the aggravation of gastrointestinal inflammation. In this chapter, we summarise the role of USPs in the pathogenesis of IBD (Table 1).
Table 1.
USPs and pathogenesis of IBD.
| USPs | Pathogenesis of IBD | Expression | Mechanism/major finding | Patients/Model | References |
|---|---|---|---|---|---|
| USP7 | Imbalance of intestinal immunity | Deficiency | Decreased ability of resolving inflammation | Adoptive-transfer-induced colitis | [102] |
| USP8 | Imbalance of intestinal immunity | Deficiency | Disturbed T cell homoeostasis, impaired T cell regulatory function, predominance of CD8+ γδ T cells | T cell-specific USP8-deficient colitis in mice | [103] |
| USP9X | Defect of intestinal barrier | Decrease | Decreased FBW7, tissue damage | DSS-induced colitis | [91] |
| USP9X | Defect of intestinal barrier | Inactive | Impaired intestinal regeneration | Colitis-associated intestinal cancer | [91] |
| Usp22 | Defect of intestinal barrier | Deficiency | Moderate and severe epithelial damage; increases local and systemic inflammation | DSS-induced colitis | [90] |
| Usp22 | Imbalance of intestinal immunity | Deficiency | Increases IL-6 levels; increases local immune cell infiltration and systemic inflammation in CD45-positive immune regulatory cells | DSS-induced colitis; inflammation-associated CRC murine model | [90] |
| USP25 | Defect of intestinal barrier | Deficiency | Increased Paneth cells and IECs, epithelial damage | DSS-induced colitis | [61] |
| USP25 | Imbalance of intestinal immunity | Deficiency | Higher expression of IRF-dependent genes and genes related to inflammatory cytokines and chemokines | DSS-induced colitis | [61] |
| USP25 | Imbalance of intestinal immunity | Deficiency | increased phosphorylated p38 and p65, decreased TRAF3 and elevated IL-6 and TNF-α | Colitis infected by Citrobacter rodentium | [61] |
| CYLD | Defect of intestinal barrier | Deficiency | Histologic damage, greater leucocyte infiltration, histologic damage, and increased intestinal epithelial dysplasia | AOM and DSS-induced colitis-associated cancer model | [34] |
| CYLD | Defect of intestinal barrier | Deficiency | Enhanced bacterial dissemination, greater submucosal oedema and broader mucosal impairment and ulceration | Citrobacter rodentium induced colitis | [88] |
| CYLD | Imbalance of intestinal immunity | Deficiency | Higher concentration of IL-18 | Colitis infected by Citrobacter rodentium | [88] |
| CYLD | Imbalance of intestinal immunity | High/low gene expression | Low/high production of IL-18 | Patients with UC | [88] |
| CYLD | Defect of intestinal barrier | Inactive | Decreased intestinal epithelial cell death | Colitis in mice by FADD deficiency in IECs | [89] |
| CYLD | Imbalance of intestinal immunity | Deficiency | Increased cytokines including IL-10 | Colitis with transferred T cell | [33] |
| sCYLD | Imbalance of intestinal immunity | Increase | Enhanced SMAD7 translocation, impaired suppressive function of Treg cells | sCYLD/SMAD7 mice | [98] |
| CYLD | Disturbance of gut microbiota | Decrease | Increased invasion and intracellular replication of AIEC bacteria. | Transfected T84 IECs | [77] |
| CYLD | Disturbance of gut microbiota | Increase | Decreased number of AIEC bacteria | Transfected T84 IECs | [77] |
| USP1 | Genetic susceptibility | – | SNP (rs1748195) in USP1 gene is associated with CD risk | Patients with CD | [83] |
| USP3 | Genetic susceptibility | – | Polymorphisms in USP3 genes are associated with both CD and UC | Patients with IBD | [77] |
| USP4, | Genetic susceptibility | – | Polymorphisms in USP4 genes are associated with both CD and UC | Patients with IBD | [74, 77, 82] |
| USP3, USP5, USP15, USP19, USP39 | Genetic susceptibility | – | Polymorphisms in USP5, 15, 18, 39 genes are associated with UC | Patients with UC | [82] |
| USP25 | Genetic susceptibility | – | Two SNPs, rs7278277 and rs2242830, are associated with CD and IBD, respectively | Patients with IBD | [79] |
| USP40 | Genetic susceptibility | – | Polymorphisms in USP4 genes are associated with both CD and UC | Patients with IBD | [74, 77, 82] |
| USP44 | Genetic susceptibility | – | USP44 methylation relevant to neoplasia associated IBD | Patients with neoplasia associated IBD | [81] |
| CYLD | Genetic susceptibility | – | The most important gene of ubiquitin proteasome system that associated with CD | Patients with CD | [76–78] |
USPs and genetic susceptibility
Since 2001, numerous studies on the genetic component of IBD susceptibility have identified over 240 IBD-related loci [71]. Genetic susceptibility data has helped shape the emerging understanding of IBD as a system-level dysfunction in both mucosal immune and commensal ecosystems and thus, identify therapeutic intervention targets [3]. Studies have shown that ubiquitin- and deubiquitin-related genes have strong associations with IBD, for instance, a coding variant *333F in Clorf106 increases IBD risk [72]; a single-exon finger E3 ubiquitin-protein ligase, RNF186 is associated with UC susceptibility [72], and the protein-truncating R179X variant in RNF186 exerts a protective effect against UC [73]. Furthermore, polymorphisms in the human DUB gene A20/TNFAIP3 locus are associated with IBD and negatively regulate inflammatory cytokines [74]; associations of CARD9 rs10870077 SNP to CD an UC are also observed [75]. Except these gene associations, USP gene variants that have been identified as related to IBD susceptibility include variants of CYLD [76, 77]. In a multicentre cohort study, CYLD was demonstrated to be the most significant CD-related ubiquitin-proteasome system gene. The single nucleotide CYLD polymorphism rs12324931 demonstrates the strongest association with CD. Genetic stratification research also indicates that CYLD likely plays a vital role in CD patients [78].
Meta-analysis has detected an IBD-relevant single nucleotide polymorphism near USP25, which is located on chromosome 21q11 [79] and, according to genome-wide association studies (GWASs), displays genome-wide significant associations for IBD patients of various ethnicities, including African American, Caucasian, and Korean [74, 79, 80]. An ImmunoChip analysis demonstrated USP25 is related to CD in Korean populations [80]. In a methylation-specific curve biopsy analysis, USP44, encoded by a gene on chromosome 12, was identified as a possible DNA methylation signature contributing to the early detection of colorectal cancer associated with IBD. However, the correlation between USP44 methylation and IBD-associated colorectal cancer was only detected in IBD patients with associated neoplasia, and not in patients with sporadic colorectal cancer [81]. GWASs by Jostins et al. and Cleynan et al. have revealed a USP4 association with both UC and CD and a USP40 association with CD only [74, 82]; the latter study also identified five USPs: USP19, USP15, USP39, USP3 and USP5, associated with UC only [82]. However, a later study by Cleynan et al. reached a slightly different conclusion that USP3 and USP40 are associated with both UC and CD, but USP4 is only related to UC [77]. A trans-ethnic association study has revealed that USP1 single nucleotide polymorphisms are associated with CD in both European and non-European populations [83].
USPs and the intestinal epithelial barrier
The intestinal epithelial barrier is composed of multiple physical, cellular, and chemical components, primarily contributed by intact IEC coherence—robust intercellular junctions including tight junctions, adherens junctions, and desmosome—and by mucus secreted by goblets [84, 85]. Acting as the guardian of the mucosal surface, the intestinal epithelial barrier not only regulates the bidirectional flow of water, macromolecules, and ions between the lumen and the host, but also protects intestinal cells from antigens and microbes in the lumen [85]. Intestinal barrier dysregulation will contribute to IBD pathology. Both CD and UC have common features, including epithelial breaks, reduced tight junction expression, and glandular atrophy [86]. Impairment of the intestinal barrier function generates increased permeability, resulting in an aberrant interaction between luminal pathogens and the intestinal mucosal immune system [87]. In genetically susceptible individuals, impaired gut barrier dysregulates immune responses and perpetuates chronic autoinflammation, contributing to IBD [84].
Studies have revealed that CYLD-deficient mice are susceptible to colitis. Upon azoxymethane and DSS administration, CYLD-deficient mice demonstrate increased colonic epithelium dysplastic changes, greater leucocyte infiltration, and histologic damage [34]. In CYLD-deficient mice with Citrobacter rodentium-induced severe inflammation, intestinal barrier disruption is demonstrated by enhanced bacterial dissemination, greater submucosal oedema, broader mucosal impairment and ulceration [88]. Endoscopic and histological analyses of mice with an IEC-specific deficiency of FADD, an adaptor protein involved in apoptosis initiation, have revealed severe colitis with epithelial erosion, mucosal thickening and transmural inflammation that may be due to CYLD catalytic activity. CYLD inhibition protects epithelial cells from death and prevents commensal bacterial infection and intestinal epithelial barrier disruptions that trigger colitis development [89]. The abnormal response of mice CYLD-deficient T cells spontaneously develops colitis and displays an inflammatory phenotype characterised by mucosa thickening, inflammatory cell infiltration, and goblet cell depletion, and also exhibits substantial weight loss and high pro-inflammatory cytokine levels in the colon [33]. In an azoxymethane and DSS-induced colitis-associated cancer model, CYLD-deficient mice are susceptible to colonic inflammation with histological damage, greater leucocyte infiltration, increased dysplastic changes and more mucosal ulcers in the intestinal epithelium [34].
Other USPs also function in the intestinal epithelial barrier in IBD cases. Kosinsky et al. demonstrated that USP22 maintains the intestinal epithelial integrity in DSS-induced colitis. In this mouse model, the USP22-deficient group presented more severe epithelial damage [90]. USP9X-deficient mice with DSS-induced colitis exhibit decreased body weight, indicating impaired gut regeneration. In a colitis-associated intestinal cancer model, USP9X inactivation impairs intestinal regeneration. In DSS-induced colitis, the USP9x and FBW7 protein levels are obviously decreased in the peak phase of colitis and return to normal levels during later phases. Notably, decreased FBW7 protein levels are a consequence of reduced USP9x expression. In accordance with these findings, USP9x suppresses c-MYC via FBW7 transcriptional activity, controlling intestinal tissue homoeostasis during tissue maintenance, damage, and repair [91]. USP25, expressed ubiquitously in various intestinal cell types, plays an essential role in intestinal inflammation [92, 93]. When subjected to DSS-induced colitis, the intestines of USP25-deficient mice exhibit increased quantities of Paneth and goblet cells in the villi, and greater damage to the remaining epithelium and intestinal epithelium than those of USP25-sufficient mice [61]. TLRs overexpress in goblet cells, Paneth cells and IECs [94]. Therefore, increased secretory cell levels and elevated TLR signalling in secretory cells and IECs may contribute to the protection against DSS-induced colitis in USP25-deficient intestines [61].
USPs and intestinal immunity
The intestinal immune system consists of innate and adaptive immune responses. Innate immune cells, equipped with germline-encoded receptors which recognise conserved pathogen-associated molecular patterns, include macrophages, granulocytes and dendritic cells. Adaptive immune cells include adaptive B and T lymphocytes, which contain antigen-specific receptors with tailored responses to multitudinous antigens, and mucosal-associated invariant T cells, which demonstrate a limited diversity of antigen receptors [2, 95]. The immune system maintains an intestinal homoeostasis balance between tolerance toward commensal microflora and aggressive responses to invading pathogens, which relies on mucosal immune cells, commensal bacteria, and IECs. Under physiological conditions, macrophages and Treg cells secrete TGF-β and IL-10 to induce immunotolerance [96]. However, under pathological conditions, complex networks of elevated pro-inflammatory cytokines, such as IL-1, -4, -6, -12, and -23, enhancement of Th1, Th2 and Th17 cells, and activation of other immune cells, cooperatively trigger IBD [96].
Studies have shown that CYLD levels in IBD patient mucosae are lower than those in normal samples [77, 97]. CYLD deubiquitinates the NLRP6 Lys63-linked polyubiquitin chains and inhibits the NLRP6-apoptosis-associated speck-like protein comprising a CARD inflammasome complex, which prevents active IL-18 concentration in colonic epithelial cells [88]. Following Citrobacter rodentium infection, CYLD-deficient mice exhibit a substantially higher IL-18 concentration, indicating that severe mice colitis depends on elevated IL-18 production. In accordance with these findings, in UC patients, CYLD mRNA expression is inversely correlated with IL-18 protein levels [88]. Moreover, CYLD catalytic activity inhibition may prevent chronic inflammation and maintain physiological immune homoeostasis [89]. CYLD-deficient IBD mouse models exhibit IBD-like phenotypes with inflammatory CD4+ T cell infiltration in inflamed areas lacking colonic patches, crypt damage and muscularis layer thickening. In transferred T cell models, the loss of CYLD induces autoimmunity and colitis. The in vitro hyper-response to T cell receptor stimulation in CYLD-deficient T cells results in hyperproliferation and markedly greater aberrant production of cytokines, including IFN-γ, indicating CYLD’s pivotal role in impeding activation of peripheral T cells. CYLD-deficient mice also express elevated levels of T cell-derived cytokines, for example, IL-4, -10 and -12, in the colon [33]. Increased CYLD splicing and SMAD7 expression are discovered in the intestinal T cells of CD patients. CYLD splicing impairs Treg and Th17 cell differentiation, increases Th1 cell polarisation, enhances SMAD7 translocation to the nucleus with cleavage of Lys63-linked polyubiquitin chains, and inhibits TGF-Β signalling. Co-overexpression of CYLD splicing and SMAD7 in T cells impairs Treg cell suppressive functions, resulting in abnormal T cell homoeostasis and triggering fatal colitis. However, these effects have not been examined in CYLD-knockout SMAD7 mice [98].
In IBD, the NLRP3 inflammasome is upregulated in the colonic mucosa [99]. The NLRP3 inflammasome serves as a central node in intestinal microbiota and inflammatory response interactions, and deubiquitination is an essential step in NLRP3 inflammasome activation, which leads to IBD pathogenesis [100]. USP7 and USP47 act as NLRP3 positive regulators, interfering with Lys63 polyubiquitin modification and regulating NLRP3 activation in microphages at a post-transcriptional level [101]. USP7 inhibition by its specific inhibitor P22077 prevents NLRP3-containing oligomer formation and decreases the release of ATP-dependent IL-1β, IL-18 and caspase-1 [101]. Highly regulated and active in Treg cells, USP7 reduces Foxp3 polyubiquitination and conversely enhances Foxp3 protein expression. In an adoptive-transfer-induced colitis model, Treg cell USP7 loss abrogates the capacity to resolve inflammation, suggesting the role of USP7 in adaptive immunity [102]. Mice with colitis developed by T cell-specific USP8 deficiency exhibit disturbed T cell homoeostasis, impaired regulatory T cell function, and predominance of CD8+ γδ T cells, revealing the critical role of T cell development and homoeostasis [103]. USP22-deficient mice in a DSS-induced colitis model display increased local immune cell infiltration and higher serum IL-6 levels. Inflammation-associated colorectal cancer murine models exhibit increased local immune cell infiltration and systemic inflammation in CD45-positive immune regulatory cells [90]. In DSS-induced colitis models, USP25-deficient mice exhibit increased leucocytes, CD8+ T cells, and CD11b+F4/80+ macrophages, and decreased infiltration of CD4+IL-17A+ T cells and CD11b+Gr-1+ neutrophil in the colonic lamina propria. USP25 loss and DSS-induced colitis resistance in nonhematopoietic cells are prevalent in immune cell homoeostasis of the colon lamina propria. This study further demonstrates that USP25 deficiency positively alters cytokine expression and protects mice against DSS-induced colitis. In addition, Citrobacter rodentium-infected USP25-deficient mice present raised concentration of phosphorylated p38 and p65 and of IL-6 and TNF-α, and decreased concentration of TRAF3 in colon tissues and sera [61].
USPs and gut microbiota
The focus in the past decade on the multiplicity and complexity of gut microbiota has provided growing insights into the pathogenic and commensal microorganisms that establish and disrupt intestinal homoeostasis [104, 105]. The indigenous intestinal microbiota is considered be a major trigger of IBD [106]. Up to five major bacteria phyla, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and Verrucomicrobia, contain thousands of anaerobic species that colonize the intestine with a gradient driven by bile acid, oxygen and antimicrobial peptides from the proximal to the distal [107]. A systematic prospective study has revealed that IBD patients are more likely to have intramucosal bacteria in the colon and terminal ileum than control subjects [108]. Multiple studies have revealed global changes in the gut microbiota composition of IBD patients, including decreased levels of members of the Firmicutes phylum, specifically Faecailbacterium prausnitzii in CD patients, and increased levels of members of the Proteobacteria phylum, including Escherichia coli in IBD patients, suggesting an essential role of dysbiosis in disease state [109]. Recently, studies of glycosylation in the intestinal interactions of gut microbiota suggest the pivotal role of post-translational modification in IBD [110]. As one such complex post-translational modification, deubiquitination by USPs may also function in dysbiosis-mediated IBD. CD-associated adherent-invasive E. coli LF82 bacteria modulate CYLD protein levels, which subsequently decreases CYLD expression in transferred T84 cells, leading to increased adherent-invasive E. coli invasion and intracellular replication. Conversely, CYLD overexpression in T84 cells reduces adherent-invasive E. coli bacteria levels 24 h after infection [77]. CYLD can also dissociate Lys63-linked polyubiquitin chains from NLRP6 and hampers its activation [88]. Colitogenic microbiota with altered bacterial populations are present in the intestine of NLRP6-deficient mice, replacing intestinal microbiota in immunocompetent mice [111]. This emphasizes the role of CYLD in gut microbiota regulation and intestinal homoeostasis via NLRP6 deubiquitination.
Conclusion
Here, we have summarised the regulatory roles of USPs in several signalling pathways and the relationship between USPs and IBD pathogenesis. Certain USPs are key IBD regulators in pro-inflammatory signalling pathways and play important roles in regulating protein activation during post-translational modification. The majority of recent IBD studies has chiefly focused on the proliferation of cellular substances, including pro-inflammatory cytokine production, epithelial-mesenchymal transformation, and aberrant fibrotic growth. However, the role of protein degradation has remained unaddressed, especially the DUB inverse process that counteracts regulatory protein proteolysis. After modification of the ubiquitination process by E1/E2/E3, ubiquitinated proteins either undergo degradation or deubiquitination by DUBs. As the largest DUB subfamily, the role of USPs should not be overlooked. Two aspects of the regulatory role of USPs are associated with the proteolytic process: impeding the normal hydrolyzation affecting the cellular metabolic cycle or blocking the natural apoptosis of the regulatory protein. We have summarised findings regarding the pivotal role of USPs in susceptibility genetics, intestinal epithelial barrier function, immunity, and gut microbiota associated with IBD. However, our current knowledge is far from adequate. The exact signals that trigger chronic inflammation are yet to be explored. Elucidating the USP control of protein modification in the regulation of protein activation and degradation provides a novel approach in identifying effective inflammatory response targets and the treatment of inflammation. Thus, targeting USPs to clarify the underlying molecular mechanism may be an effective strategy to treat IBD, and more extensive research on USPs is needed to explore their specific mechanisms.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (#81870374, #82070538), Guangzhou Science and Technology Department (#202002030041), and Guangdong Science and Technology Department (#2017A030306021).
Author contributions
SZ, MC and RC jointly designed the theme and content structure. RC and XP wrote the manuscript. SZ, LL and ZZ conducted the editing and critical revision. XP and LL drew the figure. All the co-authors have read and approved the final manuscript for publication.
Data availability
All data included in this review are available upon request by contact with the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Dr. Angelo Peschiaroli.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rirong Chen, Xiaobai Pang.
References
- 1.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–78.. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 2.Chang JT. Pathophysiology of inflammatory bowel diseases. N. Engl J Med. 2020;383:2652–64.. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 3.Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–39.. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu H, Sun SC. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457–83. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–92.. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 6.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 7.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–97.. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–208.. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 10.Pozhidaeva A, Bezsonova I. USP7: Structure, substrate specificity, and inhibition. DNA Repair. 2019;76:30–9. doi: 10.1016/j.dnarep.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–64.. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 12.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–86.. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Reverter D. Molecular mechanisms of DUBs regulation in signaling and disease. Int J Mol Sci. 2021;22:986.. doi: 10.3390/ijms22030986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai J, Culley MK, Zhao Y, Zhao J. The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein Cell. 2018;9:754–69.. doi: 10.1007/s13238-017-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, Guo J, North BJ, Wang B, Cui CP, Li H, et al. Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochimica et Biophysica Acta Rev Cancer. 2019;1872:188312. doi: 10.1016/j.bbcan.2019.188312. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46:662–73. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sussman RT, Stanek TJ, Esteso P, Gearhart JD, Knudsen KE, McMahon SB. The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2) J Biol Chem. 2013;288:24234–46.. doi: 10.1074/jbc.M113.469783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basar MA, Beck DB, Werner A. Deubiquitylases in developmental ubiquitin signaling and congenital diseases. Cell Death Differ. 2021;28:538–56.. doi: 10.1038/s41418-020-00697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, et al. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat Cell Biol. 2012;14:717–26.. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- 20.Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34:307–25. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–54.. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 22.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–56.. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr., Mackenzie F, Newman EM, et al. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–70. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 24.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 26.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel DK, Eden K, Ringel VM, Allen IC. Emerging roles for noncanonical NF-κB signaling in the modulation of inflammatory bowel disease pathobiology. Inflamm Bowel Dis. 2016;22:2265–79. doi: 10.1097/MIB.0000000000000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atreya I, Atreya R, Neurath MF. NF-kappaB in inflammatory bowel disease. J Intern Med. 2008;263:591–6. doi: 10.1111/j.1365-2796.2008.01953.x. [DOI] [PubMed] [Google Scholar]
- 29.Sato Y, Goto E, Shibata Y, Kubota Y, Yamagata A, Goto-Ito S, et al. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat Struct Mol Biol. 2015;22:222–9. doi: 10.1038/nsmb.2970. [DOI] [PubMed] [Google Scholar]
- 30.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–6. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 32.Draber P, Kupka S, Reichert M, Draberova H, Lafont E, de Miguel D, et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 2015;13:2258–72. doi: 10.1016/j.celrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204:1475–85. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JM, et al. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Investig. 2006;116:3042–9. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–83.. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 37.Lee BC, Miyata M, Lim JH, Li JD. Deubiquitinase CYLD acts as a negative regulator for bacterium NTHi-induced inflammation by suppressing K63-linked ubiquitination of MyD88. Proc Natl Acad Sci USA. 2016;113:E165–71. doi: 10.1073/pnas.1518615113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Q, Cheng C, Wei Y, Yang J, Zhou W, Song Q, et al. USP15 potentiates NF-κB activation by differentially stabilizing TAB2 and TAB3. FEBS J. 2020;287:3165–83.. doi: 10.1111/febs.15202. [DOI] [PubMed] [Google Scholar]
- 39.Lei CQ, Wu X, Zhong X, Jiang L, Zhong B, Shu HB. USP19 inhibits TNF-α- and IL-1β-triggered NF-κB activation by deubiquitinating TAK1. J Immunol. 2019;203:259–68. doi: 10.4049/jimmunol.1900083. [DOI] [PubMed] [Google Scholar]
- 40.Yu JS, Huang T, Zhang Y, Mao XT, Huang LJ, Li YN, et al. Substrate-specific recognition of IKKs mediated by USP16 facilitates autoimmune inflammation. Sci. Adv. 2021;7:eabc4009.. doi: 10.1126/sciadv.abc4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S, Wang J, Guo S, Huang D, Lorigados IB, Nie X, et al. Transcriptional activation of USP16 gene expression by NFκB signaling. Mol brain. 2019;12:120. doi: 10.1186/s13041-019-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al Nabhani Z, Dietrich G, Hugot JP, Barreau F. Nod2: The intestinal gate keeper. PLoS Pathog. 2017;13:e1006177. doi: 10.1371/journal.ppat.1006177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warner N, Burberry A, Pliakas M, McDonald C, Núñez G. A genome-wide small interfering RNA (siRNA) screen reveals nuclear factor-κB (NF-κB)-independent regulators of NOD2-induced interleukin-8 (IL-8) secretion. J Biol Chem. 2014;289:28213–24. doi: 10.1074/jbc.M114.574756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong B, Liu X, Wang X, Chang SH, Liu X, Wang A, et al. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat Immunol. 2012;13:1110–7. doi: 10.1038/ni.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colleran A, Collins PE, O’Carroll C, Ahmed A, Mao X, McManus B, et al. Deubiquitination of NF-κB by ubiquitin-specific protease-7 promotes transcription. Proc Natl Acad Sci USA. 2013;110:618–23.. doi: 10.1073/pnas.1208446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez LG, Kempski J, McGee HM, Pelzcar P, Agalioti T, Giannou A, et al. TGF-β signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat Commun. 2020;11:2608. doi: 10.1038/s41467-020-16363-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colak S, Ten Dijke P. Targeting TGF-β signaling in cancer. Trends Cancer. 2017;3:56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu Rev Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–39. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–75. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 51.Aggarwal K, Massagué J. Ubiquitin removal in the TGF-β pathway. Nat Cell Biol. 2012;14:656–7. doi: 10.1038/ncb2534. [DOI] [PubMed] [Google Scholar]
- 52.Kit Leng Lui S, Iyengar PV, Jaynes P, Isa Z, Pang B, Tan TZ, et al. USP26 regulates TGF-β signaling by deubiquitinating and stabilizing SMAD7. EMBO Rep. 2017;18:797–808. doi: 10.15252/embr.201643270. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Istomine R, Alvarez F, Almadani Y, Philip A, Piccirillo CA. The deubiquitinating enzyme ubiquitin-specific peptidase 11 potentiates TGF-β signaling in CD4(+) T cells to facilitate Foxp3(+) regulatory T and T(H)17 cell differentiation. J Immunol. 2019;203:2388–400. doi: 10.4049/jimmunol.1801689. [DOI] [PubMed] [Google Scholar]
- 54.Jacko AM, Nan L, Li S, Tan J, Zhao J, Kass DJ, et al. De-ubiquitinating enzyme, USP11, promotes transforming growth factor β-1 signaling through stabilization of transforming growth factor β receptor II. Cell Death Dis. 2016;7:e2474. doi: 10.1038/cddis.2016.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, et al. CYLD negatively regulates transforming growth factor-β-signalling via deubiquitinating Akt. Nat Commun. 2012;3:771. doi: 10.1038/ncomms1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y, Thornton AM, Kinney MC, Ma CA, Spinner JJ, Fuss IJ, et al. The deubiquitinase CYLD targets Smad7 protein to regulate transforming growth factor β (TGF-β) signaling and the development of regulatory T cells. J Biol Chem. 2011;286:40520–30. doi: 10.1074/jbc.M111.292961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun H, Ou B, Zhao S, Liu X, Song L, Liu X, et al. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236–47.. doi: 10.1016/j.ebiom.2019.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen M, Wan L, Zhang J, Zhang J, Mendez L, Clohessy JG, et al. Deregulated PP1α phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat Commun. 2018;9:159. doi: 10.1038/s41467-017-02272-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong B, Liu X, Wang X, Liu X, Li H, Darnay BG, et al. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci Signal. 2013;6:ra35. doi: 10.1126/scisignal.2003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu Y, Qiu Y, Chen P, Chang H, Guo L, Zhang F, et al. ER-localized Hrd1 ubiquitinates and inactivates Usp15 to promote TLR4-induced inflammation during bacterial infection. Nat Microbiol. 2019;4:2331–46.. doi: 10.1038/s41564-019-0542-2. [DOI] [PubMed] [Google Scholar]
- 61.Wang X-M, Yang C, Zhao Y, Xu Z-G, Yang W, Wang P, et al. The deubiquitinase USP25 supports colonic inflammation and bacterial infection and promotes colorectal cancer. Nat Cancer. 2020;1:811–25. doi: 10.1038/s43018-020-0089-4. [DOI] [PubMed] [Google Scholar]
- 62.Noah TK, Shroyer NF. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu Rev Physiol. 2013;75:263–88.. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- 63.Sancho R, Cremona CA, Behrens A. Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 2015;16:571–81. doi: 10.15252/embr.201540188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diefenbacher ME, Popov N, Blake SM, Schülein-Völk C, Nye E, Spencer-Dene B, et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Investig. 2014;124:3407–18. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diefenbacher ME, Chakraborty A, Blake SM, Mitter R, Popov N, Eilers M, et al. Usp28 counteracts Fbw7 in intestinal homeostasis and cancer. Cancer Res. 2015;75:1181–6. doi: 10.1158/0008-5472.CAN-14-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong AW, Meng Z, Guan KL. The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol. 2016;13:324–37. doi: 10.1038/nrgastro.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou X, Li W, Wang S, Zhang P, Wang Q, Xiao J, et al. YAP aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell Rep. 2019;27:1176–89.e5. doi: 10.1016/j.celrep.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 69.Toloczko A, Guo F, Yuen HF, Wen Q, Wood SA, Ong YS, et al. Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the hippo pathway. Cancer Res. 2017;77:4921–33.. doi: 10.1158/0008-5472.CAN-16-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang D, Zhang Y, Xu X, Wu J, Peng Y, Li J, et al. YAP promotes the activation of NLRP3 inflammasome via blocking K27-linked polyubiquitination of NLRP3. Nat Commun. 2021;12:2674. doi: 10.1038/s41467-021-22987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Furey TS, Sethupathy P, Sheikh SZ. Redefining the IBDs using genome-scale molecular phenotyping. Nat Rev Gastroenterol Hepatol. 2019;16:296–311. doi: 10.1038/s41575-019-0118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rivas MA, Graham D, Sulem P, Stevens C, Desch AN, Goyette P, et al. A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nat Commun. 2016;7:12342. doi: 10.1038/ncomms12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, et al. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–10. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cleynen I, Vazeille E, Artieda M, Verspaget HW, Szczypiorska M, Bringer MA, et al. Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease. Gut. 2014;63:1265–74. doi: 10.1136/gutjnl-2012-303205. [DOI] [PubMed] [Google Scholar]
- 78.Elding H, Lau W, Swallow DM, Maniatis N. Dissecting the genetics of complex inheritance: linkage disequilibrium mapping provides insight into Crohn disease. Am J Hum Genet. 2011;89:798–805. doi: 10.1016/j.ajhg.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brant SR, Okou DT, Simpson CL, Cutler DJ, Haritunians T, Bradfield JP, et al. Genome-wide association study identifies African-specific susceptibility loci in african americans with inflammatory bowel disease. Gastroenterology. 2017;152:206–17.e2. doi: 10.1053/j.gastro.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang SK, Hong M, Choi H, Zhao W, Jung Y, Haritunians T, et al. Immunochip analysis identification of 6 additional susceptibility loci for Crohn’s disease in Koreans. Inflamm Bowel Dis. 2015;21:1–7. doi: 10.1097/MIB.0000000000000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Azuara D, Aussó S, Rodriguez-Moranta F, Guardiola J, Sanjuan X, Lobaton T, et al. New methylation biomarker panel for early diagnosis of dysplasia or cancer in high-risk inflammatory bowel disease patients. Inflamm Bowel Dis. 2018;24:2555–64.. doi: 10.1093/ibd/izy255. [DOI] [PubMed] [Google Scholar]
- 82.Cleynen I, Jüni P, Bekkering GE, Nüesch E, Mendes CT, Schmied S, et al. Genetic evidence supporting the association of protease and protease inhibitor genes with inflammatory bowel disease: a systematic review. PLoS ONE. 2011;6:e24106. doi: 10.1371/journal.pone.0024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86.. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–53. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 85.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 86.Clayburgh DR, Shen L, Turner JR. A porous defense: the leaky epithelial barrier in intestinal disease. Lab Investig. 2004;84:282–91. doi: 10.1038/labinvest.3700050. [DOI] [PubMed] [Google Scholar]
- 87.Chang J, Leong RW, Wasinger VC, Ip M, Yang M, Phan TG. Impaired intestinal permeability contributes to ongoing bowel symptoms in patients with inflammatory bowel disease and mucosal healing. Gastroenterology. 2017;153:723–31.e1. doi: 10.1053/j.gastro.2017.05.056. [DOI] [PubMed] [Google Scholar]
- 88.Mukherjee S, Kumar R, Tsakem Lenou E, Basrur V, Kontoyiannis DL, Ioakeimidis F, et al. Deubiquitination of NLRP6 inflammasome by Cyld critically regulates intestinal inflammation. Nat Immunol. 2020;21:626–35.. doi: 10.1038/s41590-020-0681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernández-Majada V, Ermolaeva M, et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–4. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 90.Kosinsky RL, Saul D, Ammer-Herrmenau C, Faubion WA, Neesse A, Johnsen SA. USP22 suppresses SPARC expression in acute colitis and inflammation-associated colorectal cancer. Cancers. 2021;13:1817.. doi: 10.3390/cancers13081817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khan OM, Carvalho J, Spencer-Dene B, Mitter R, Frith D, Snijders AP, et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Investig. 2018;128:1326–37.. doi: 10.1172/JCI97325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 2018;175:372–86.e17. doi: 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tabula Muris C, Overall c, Logistical c, Organ c, processing, Library p, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018;562:367–72. [DOI] [PMC free article] [PubMed]
- 94.Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- 95.Branzk N, Diefenbach A. Gut immunity: passing on the baton from innate to adaptive immunity. Curr Biol. 2018;28:R562–r5.. doi: 10.1016/j.cub.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 96.de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 97.Costello CM, Mah N, Häsler R, Rosenstiel P, Waetzig GH, Hahn A, et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays. PLoS Med. 2005;2:e199. doi: 10.1371/journal.pmed.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tang Y, Reissig S, Glasmacher E, Regen T, Wanke F, Nikolaev A, et al. Alternative splice forms of CYLD mediate ubiquitination of SMAD7 to prevent TGFB signaling and promote colitis. Gastroenterology. 2019;156:692–707.e7. doi: 10.1053/j.gastro.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 99.Liu L, Dong Y, Ye M, Jin S, Yang J, Joosse ME, et al. The pathogenic role of NLRP3 inflammasome activation in inflammatory bowel diseases of both mice and humans. J Crohn’s Colitis. 2017;11:737–50. doi: 10.1093/ecco-jcc/jjw219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19:477–89.. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palazón-Riquelme P, Worboys JD, Green J, Valera A, Martín-Sánchez F, Pellegrini C, et al. USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep. 2018;19:e44766.. doi: 10.15252/embr.201744766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Loosdregt J, Fleskens V, Fu J, Brenkman AB, Bekker CP, Pals CE, et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity. 2013;39:259–71.. doi: 10.1016/j.immuni.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dufner A, Kisser A, Niendorf S, Basters A, Reissig S, Schönle A, et al. The ubiquitin-specific protease USP8 is critical for the development and homeostasis of T cells. Nat Immunol. 2015;16:950–60. doi: 10.1038/ni.3230. [DOI] [PubMed] [Google Scholar]
- 104.Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331–45. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 105.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–37. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 106.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–33. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 107.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moussata D, Goetz M, Gloeckner A, Kerner M, Campbell B, Hoffman A, et al. Confocal laser endomicroscopy is a new imaging modality for recognition of intramucosal bacteria in inflammatory bowel disease in vivo. Gut. 2011;60:26–33. doi: 10.1136/gut.2010.213264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573–84. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kudelka MR, Stowell SR, Cummings RD, Neish AS. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat Rev Gastroenterol Hepatol. 2020;17:597–617. doi: 10.1038/s41575-020-0331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gersch M, Gladkova C, Schubert AF, Michel MA, Maslen S, Komander D. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat Struct Mol Biol. 2017;24:920–30. doi: 10.1038/nsmb.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peng H, Yang F, Hu Q, Sun J, Peng C, Zhao Y, et al. The ubiquitin-specific protease USP8 directly deubiquitinates SQSTM1/p62 to suppress its autophagic activity. Autophagy. 2020;16:698–708. doi: 10.1080/15548627.2019.1635381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu X, Lei C, Xia T, Zhong X, Yang Q, Shu HB. Regulation of TRIF-mediated innate immune response by K27-linked polyubiquitination and deubiquitination. Nat Commun. 2019;10:4115. doi: 10.1038/s41467-019-12145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochemical J. 2008;411:249–60. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data included in this review are available upon request by contact with the corresponding author.