Abstract
Thirty-one strains of Klebsiella pneumoniae (including 10 duplicates) from 21 septicemic pediatric patients (age, <2 months) were studied during a 4-month period (June to October 1996) in which the fatality rate was 62% (13 of 21). These isolates identified by the API 20E system yielded the same biotype. Pulsed-field gel electrophoresis experiments revealed the same clone in 31 strains. The isolates were multidrug-resistant but were still susceptible to ciprofloxacin, imipenem, and cefoxitin. A 135-kb plasmid was harbored in all of the isolates. No transconjugants were obtained that were resistant to ampicillin, cefotaxime, tetracycline, or gentamicin. Isoelectric focusing for β-lactamases was performed on all strains, and three bands with pIs of 5.4, 7.6, and 8.2 were obtained. Of these, the pI 8.2 β-lactamase had an extended-spectrum β-lactamase phenotype. PCR amplification of both TEM- and SHV-type genes was obtained. The sequence analysis of the SHV PCR product indicated a mutation corresponding to the SHV-5 β-lactamase.
Klebsiella pneumoniae is an important hospital-acquired pathogen with the potential of causing severe morbidity and mortality in pediatric patients. Several outbreaks of infection caused by K. pneumoniae isolates that are simultaneously resistant to broad-spectrum cephalosporins and aminoglycosides have been widely reported (8, 9, 18, 21). Some of these multidrug-resistant isolates produce extended-spectrum β-lactamases (ESBLs) that are able to hydrolyze expanded-spectrum cephalosporins (e.g., ceftriaxone, cefotaxime, and ceftazidime), aztreonam, and related oxyimino-β-lactams. Most of these enzymes are TEM- or SHV-type β-lactamases in which the substitution of one or more amino acids has altered the configuration of the active site (7, 14, 19). Most of the plasmids determining ESBLs are large (≥80 kb) and encode multiple resistances (10). Little is known about the ESBL-producing Enterobacteriaceae in Mexico (23). The present work is a characterization of 31 ESBL-producing K. pneumoniae isolates from a nosocomial outbreak occurring in a hospital in Cuernavaca, Mexico.
MATERIALS AND METHODS
Patients and bacterial strains.
The General Hospital in the state of Morelos is a secondary-care facility with 100 beds. The neonatal intensive care unit has 8 beds. An outbreak was suspected in the neonatal intensive care unit due to an increased number of isolates of K. pneumoniae from blood cultures during a 4-month period. A retrospective epidemiological investigation included 74 children less than 2 months old hospitalized from June to October 1996.
Twenty-one clinical isolates were from blood; another 10 duplicate strains were from sites other than blood (urine and catheter tip) corresponding to 10 patients. In total there were 31 clinical isolates identified as K. pneumoniae by the API 20E system (BioMerieux, Merck).
Susceptibility testing.
The antimicrobial susceptibility was initially determined with the MicroScan (Dade) system, using the Combo 14 panel. Subsequently, MICs of several β-lactams were determined by agar dilution on Mueller-Hinton agar following the recommendations of the National Committee for Clinical Laboratory Standards (17). MICs were determined alone or in combination with clavulanic acid at 2 μg/ml.
Antimicrobials.
Standard powders of the following were provided by the companies indicated: cefotaxime and cefpirome, Hoechst-Marion-Roussel, Romainville, France; ceftazidime, GlaxoWellcome, Mexico City, Mexico; aztreonam and cefepime, Bristol-Myers Squibb, Mexico City, Mexico; clavulanic acid, SmithKline Beecham Pharmaceuticals, Mexico City, Mexico, rifampin, tetracycline, and gentamicin were obtained from Sigma, St. Louis, Mo.
Plasmid isolation and conjugation experiments.
DNA was extracted from clinical isolates according to the method described by Kieser (11). DNA was visualized after vertical electrophoresis in 0.7% agarose gels stained with ethidium bromide. Matings were performed as described by Miller (15), using the Escherichia coli J53-2 (F− pro met Rifr) strain. In all cases, transconjugants were selected on Luria agar supplemented with rifampin (100 μg/ml) in combination with cefotaxime (1 μg/ml), ampicillin (50 μg/ml), tetracycline (15 μg/ml), gentamicin (16 μg/ml).
TEM- or SHV-specific PCR and DNA sequencing.
To amplify TEM-related genes from clinical isolates, the oligonucleotide primers OT1 and OT2 described by Arlet and Philippon (1) were used for PCR. For SHV-specific PCR, primers SE5 (5′-GGT CGG AAT TCA GGA GGT TGA CTA TGC GTT ATA TTC GCC TG-3′) and SB3 (5′-GGT GCG GAT CCT TAT TAG CGT TGC CAG TGC TC-3′), including the signal peptide and the complete SHV gene, were designed and synthesized. The amplification conditions were as follows: 94°C for 5 min; then 35 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min; and, finally, one cycle at 72°C for 15 min. The product of this amplification was used to determine the nucleotide sequence with the fluorescence-based Taq FS Dye terminator cycle sequencing kit using the same primers. Sequence analysis was performed with Genetics Computer Group software and BLASTx searching (of the EMBL, SwissProt, and PIR databases) (25).
Omp and SDS-PAGE.
Preparation of outer membrane protein (Omp) from clinical isolates and separation of the proteins by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (acrylamide-bis, 30:3) and 8 M urea were performed according to the method of Piddock et al. (20).
PFGE typing.
For pulsed-field gel electrophoresis (PFGE) typing, whole-cell DNA was obtained according to the method of Miranda et al. (16). DNA was digested with XbaI restrictase (Gibco, BRL, Gaithersburg, Md.) and separated in 1% agarose gel (Pulsed Field-Certified; Pronadisa, Madrid, Spain), with a Gene-Path System (Bio-Rad, Hercules, Calif.). Strain number 917 isolated from a patient in the same hospital 2 months before the outbreak, was included as a control for comparison. The gel was stained with ethidium bromide and visualized with the Gel-Doc system (Bio-Rad Laboratories) according to the criteria of Tenover et al. (24).
IEF and bioassay.
Isoelectric focusing (IEF) was conducted according to the method described by Matthew et al. (13) using a Phast system minigel with a pH range of 3 to 10 (Pharmacia). Extracts from TEM-1-, SHV-1-, and SHV-5-producing strains were used as standards for pIs of 5.4, 7.6, and 8.2, respectively. To determine the ESBLs coded in the strains, a bioassay was performed as described by Silva et al. (22).
RESULTS
Bacterial strains.
During the study period, 29 clinical isolates were recovered from 54 blood cultures (53.7%) obtained from 74 children. Twenty-one isolates were identified as K. pneumoniae, 3 isolates were Escherichia coli, 3 were Enterobacter sp., 1 was Pseudomonas aeruginosa, and 1 was Staphylococcus aureus. In addition, 10 K. pneumoniae strains were obtained from sites other than blood (urine and catheter tip), corresponding to 10 patients with blood culture positive for K. pneumoniae. All 31 of these isolates had the same biotype: 5215773. Thirteen of 21 patients died of sepsis (lethality rate, 62%).
Susceptibility.
Antimicrobial susceptibilities using the MicroScan (Dade) system with the Combo 14 panel revealed that all strains in the study were susceptible to cefoxitin, ciprofloxacin, and imipenem. Subsequently, MICs of several β-lactam antibiotics and clavulanic acid as an inhibitor were determined by agar dilution. MICs for the clinical isolates are presented in Table 1. The clinical isolates were resistant to all the cephalosporins and the monobactam tested. The β-lactam inhibitor clavulanic acid (2 μg/ml) reduced the MICs of these antibiotics 10- to 12-fold, indicating that an ESBL was produced by the clinical isolates.
TABLE 1.
Antimicrobial susceptibilities of K. pneumoniae against different β-lactams and inhibitors
| Drug(s)b | MIC (μg/ml)a of:
|
|
|---|---|---|
| K. pneumoniae | E. coli ATCC | |
| Cefotaxime | 256 | 0.125 |
| Cefotaxime-clavulanic acid | 0.06 | 0.06 |
| Ceftazidime | >512 | 0.125 |
| Ceftazidime-clavulanic acid | 1 | 0.06 |
| Aztreonam | >512 | 0.125 |
| Aztreonam-clavulanic acid | 0.125 | 0.06 |
| Cefpirome | 128 | 0.125 |
| Cefpirome-clavulanic acid | 0.06 | 0.06 |
| Cefepime | 64 | 0.06 |
| Cefepime-clavulanic acid | 0.06 | 0.06 |
| Imipenem | 1 | 0.125 |
Mode values from three independent determinations.
When it is indicated, a constant concentration (2 μg/ml) of clavulanic acid was used.
Typing by genomic DNA PFGE analysis.
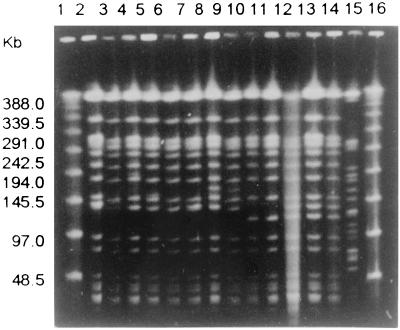
The macrorestriction genomic DNA analysis was carried out with the 31 clinical isolates of K. pneumoniae (including 10 duplicates). The XbaI restriction patterns showed one single clone (type A) with three subtypes (1 to 3), indicating a clonal origin of the outbreak. Figure 1 shows type A (slots 2 to 7), type A1 (slots 8 and 9), type A2 (slots 10 and 11), type A3 (slots 12 to 14), and a nonrelated strain with type B (slot 15).
FIG. 1.
Representative agarose gel from PFGE of XbaI-digested genomic DNAs of ESBL-producing K. pneumoniae isolates. Lanes 1 and 16, molecular size marker of lambda ladder (Bio-Rad); lane 2, 907; lane 3, 912; lane 4, 913; lane 5, 918; lane 6, 919; lane 7, 920; lane 8, 906; lane 9, 921; lane 10, 909; lane 11, 910; lane 12, 905; lane 13, 922; lane 14, 923; and lane 15, 917 (nonrelated strain).
Plasmid pattern and resistance transfer.
All 31 clinical isolates harbored one plasmid of 135 kb, and most of them (29 of 31) harbored a second plasmid of 105 kb. Twenty-four of 31 isolates harbored a third plasmid of 80 kb. The cefotaxime, ampicillin, tetracycline, and gentamicin resistance transfer experiments were carried out with the 31 clinical isolates. There were no transconjugants grown on Luria agar in combination with four selected drugs.
IEF of β-lactamases and detection of cefotaximase activity.
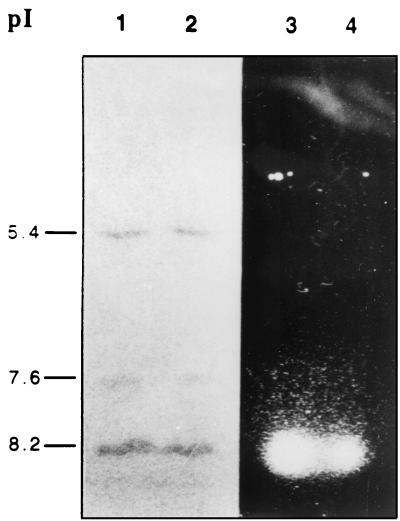
pI values of β-lactamases identified by analytical IEF in extracts from the 31 clinical isolates were identical in all cases (Fig. 2 [slots 1 and 2 show only two representative strains]). The strains were found to express three enzymes with pIs of 5.4, 7.6, and 8.2. The 8.2 pI β-lactamase was subsequently shown to possess cefotaximase activity in the bioassay (Fig. 2, slots 3 and 4).
FIG. 2.
IEF of β-lactamases on polyacrylamide gel with nitrocefin and detection of ESBL. Lanes 1 and 3, extract from clinical isolate of K. pneumoniae R910; lanes 2 and 4, extract from clinical isolate of K. pneumoniae 912. Lanes 1 and 2 were developed with nitrocefin; lanes 3 and 4 show the results of the bioassay.
Identification of an ESBL-encoding gene by specific PCR and DNA sequencing.
The results of PFGE, plasmid profile, mating experiments, and β-lactamase pattern analysis strongly suggested correspondence to the same clonal strain. Clinical isolate 912 was selected as being representative of the group. The total genomic DNA preparation from clinical isolate 912 was used in specific PCR with SE5 and SB3 primers (see Materials and Methods). The PCR product of the expected size of 0.9 kb (representing the complete gene of blaSHV, including the signal peptide region) was completely sequenced, and the amino acid sequence was deduced. A doublet of mutations responsible for the Gly238Ser and Glu240Lys amino acid substitutions was detected when this sequence compared with the sequence of the SHV-1 β-lactamase (2), which corresponds to the blaSHV-5 gene (4).
Determination of the TEM-encoding β-lactamase.
The same genomic DNA preparation of isolate 912 which produced the β-lactamase with a pI of 5.4 was used in PCR with primers OT1 and OT2 (1), which are specific for the blaTEM family of genes. A PCR product of the expected size of 503 bp was obtained (data not shown).
Omp pattern.
In order to determine an alteration in the permeability of the cell in the clinical isolate of K. pneumoniae 912 with the multidrug-resistant phenotype, the Omp pattern was analyzed. The Omp pattern was similar to that of a susceptible strain of K. pneumoniae ATCC 13883 (data not shown).
DISCUSSION
The high level of mortality arising from intrahospital infections with multidrug-resistant strains of K. pneumoniae, as seen in this study, is reason for alarm. It appears that the lack of control of contamination sources and hygiene has caused the dissemination of the microorganism among patients. However, there was no direct evidence of the source of contamination or transmission of the multidrug-resistant strains, although it was probably due to transitory hand contact through hospital personnel (6). Hence, it is important to differentiate individual strains and characterize the acquisition of resistance mechanisms in this type of microorganism. The PFGE technique has proven to be a trustworthy method to determine the clonal origin of strains (9, 18). Using this tool and the biotype, we demonstrated that the causal agent of this intrahospital outbreak was a single clone of K. pneumoniae. After the outbreak, the policy of the hospital was to change its antimicrobial therapy scheme and to increase attention to handwashing in order to combat these infections.
Because plasmids are not a stable part of the bacterial genome, they are not necessarily good indicators of the dissemination of a strain. The differences in the number of plasmids in the clinical isolates could have occurred because of spontaneous changes in the population, so that the 135-kb plasmid was stabilized due to the selective pressure of the antibiotics. This nonconjugative plasmid may be responsible for the production of SHV-5 ESBL and the cefotaxime resistance in these strains. Such an outbreak of ESBL-producing organisms, in which clonally related isolates are characterized by a different plasmid profile, has already been described (21).
Reports of broad-spectrum β-lactamase types TEM and SHV, mediated by plasmids, have proliferated with the increased identification of K. pneumoniae strains and with the use of broad-spectrum cephalosporins during recent years. One of these was due to the SHV-5 β-lactamase (3, 5, 26). Three different β-lactamases were identified during this study outbreak: a β-lactamase with a pI of 5.4 that corresponds to the TEM-type family by evidence obtained with oligo-specific PCR experiments; a second enzyme with a pI of 8.2 which corresponded to SHV-5; and a β-lactamase with a pI of 7.6, produced by all the isolates, which could be a chromosomally SHV-1-encoded K. pneumoniae enzyme, as previously reported by Leung et al. (12). Prior studies with clinical isolates from other hospitals in Mexico demonstrated the transfer of two β-lactamases, TEM- and SHV-derived enzymes that were encoded by conjugative high-molecular-weight plasmids (23). In the present work, all the clinical isolates of K. pneumoniae possessed a nonconjugative 135-kb plasmid that might encode both TEM and SHV-5 β-lactamases.
In conclusion, this is one of the first reports from Mexico in which the causative organism of an intrahospital outbreak of infection with multidrug-resistant K. pneumoniae associated with a high mortality rate has been characterized at the molecular level. Knowing the resistance mechanisms in this type of clinical isolate will allow the proposal of improved therapeutic measures and the rational use of antibiotics, which are indispensable tools in fighting infectious diseases produced by bacteria. More-prudent use of antibiotics and control of the spread of these resistant organisms are necessary.
ACKNOWLEDGMENTS
We are indebted to P. Bradford and C. Conde for their meticulous review of the manuscript. We thank T. Rojas for her technical assistance.
This study was supported in part by federal resources from the Consejo Nacional de Ciencia y Tecnología (CONACYT) (grants 1892N-P and 30938M) and the Instituto Nacional de Salud Pública.
REFERENCES
- 1.Arlet G, Philippon A. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB) FEMS Microbiol Lett. 1991;66:19–25. doi: 10.1016/0378-1097(91)90414-6. [DOI] [PubMed] [Google Scholar]
- 2.Barthelemy M, Peduzzi J, Labia R. Complete amino acid sequence of p453-plasmid-mediated PIT-2 β-lactamase (SHV-1) Biochem J. 1988;251:73–79. doi: 10.1042/bj2510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Rosenthal E, Eberlein E, Holley M, Schweighart S. Spread of Klebsiella pneumoniae producing SHV-5 β-lactamase among hospitalized patients. Infection. 1993;21:18–22. doi: 10.1007/BF01739303. [DOI] [PubMed] [Google Scholar]
- 4.Billot-Klein D, Gutmann L, Collatz E. Nucleotide sequence of the SHV-5 β-lactamase gene of a Klebsiella pneumoniae plasmid. Antimicrob Agents Chemother. 1990;34:2439–2441. doi: 10.1128/aac.34.12.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingen E H, Desjardins P, Arlet G, Bourgeois F, Mariani-Kurkdjian P, Lambert-Zechovsky N Y, Denamur E, Philippon A, Elion J. Molecular epidemiology of plasmid spread among extended broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J Clin Microbiol. 1993;31:179–184. doi: 10.1128/jcm.31.2.179-184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun-Buisson C, Legrand P, Philippon A, Montravers F, Ansquer M, Duval J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet. 1987;ii:302–306. doi: 10.1016/s0140-6736(87)90891-9. [DOI] [PubMed] [Google Scholar]
- 7.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French G L, Shannon K P, Simmons N. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and β-lactam–β-lactamase inhibitor combinations by hyperproduction of SHV-5 β-lactamase. J Clin Microbiol. 1996;34:358–363. doi: 10.1128/jcm.34.2.358-363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gniadkowski M, Palucha A, Grzesiowski P, Hryniewicz W. Outbreak of ceftazidime-resistant Klebsiella pneumoniae in a pediatric hospital in Warsaw, Poland: clonal spread of the TEM-47 extended-spectrum β-lactamase (ESBL)-producing strain and transfer of a plasmid carrying the SHV-5-like ESBL-encoding gene. Antimicrob Agents Chemother. 1998;42:3079–3085. doi: 10.1128/aac.42.12.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby G, Mederios A A. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 12.Leung M, Shannon K, French G. Rarity of transferable β-lactamase production by Klebsiella species. Antimicrob Agents Chemother. 1997;39:737–745. doi: 10.1093/jac/39.6.737. [DOI] [PubMed] [Google Scholar]
- 13.Matthew G, Hedges R, Smith J. Types of β-lactamases determined by plasmid in gram-negative bacteria. J Bacteriol. 1979;138:657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medeiros A A. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl. 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 15.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 82–85. [Google Scholar]
- 16.Miranda G, Kelly C, Solórzano F, Leaños B, Coria R, Patterson J E. Use of pulsed-field gel electrophoresis typing to study and outbreak of infection due to Serratia marcescens in a neonatal intensive care unit. J Clin Microbiol. 1996;34:3138–3141. doi: 10.1128/jcm.34.12.3138-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard M/A2. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 18.Pena C, Pujol M, Ardanuy C, Ricart A, Pallares R, Linares J, Ariza J, Gudiol F. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1998;42:53–58. doi: 10.1128/aac.42.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philippon A, Labia R, Jacoby G. Extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piddock L J, Traynor E A, Wise R. A comparison of the mechanisms of decreased susceptibility of aztreonam-resistant and ceftazidime-resistant Enterobacteriaceae. J Antimicrob Chemother. 1990;26:749–762. doi: 10.1093/jac/26.6.749. [DOI] [PubMed] [Google Scholar]
- 21.Rice L B, Eckstein E C, DeVente J, Shlaes D M. Ceftazidime-resistant Klebsiella pneumoniae isolates recovered at the Cleveland Department of Veterans Affairs Medical Center. Clin Infect Dis. 1996;23:118–124. doi: 10.1093/clinids/23.1.118. [DOI] [PubMed] [Google Scholar]
- 22.Silva J, Aguilar C. β-Lactamase bioassay: a simplified method to determine extended-spectrum β-lactamases (ESBL) in Enterobacteria. Arch Med Res. 1997;28:285–287. [PubMed] [Google Scholar]
- 23.Silva J, Aguilar C, Becerra Z, Lopez-Antunano F, Garcia R. Extended-spectrum β-lactamases in clinical isolates of enterobacteria in Mexico. Microb Drug Resist. 1999;5:189–193. doi: 10.1089/mdr.1999.5.189. [DOI] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang X, Shannon K, French G. Mechanism and stability of hyperproduction of the extended-spectrum β-lactamase SHV-5 in Klebsiella pneumoniae. J Antimicrob Chemother. 1997;40:525–532. doi: 10.1093/jac/40.4.525. [DOI] [PubMed] [Google Scholar]