Abstract
Objectives: N6-methyladenosine (m6A) is hypothesized to play a role in the regulation of pathogenesis of myocardial infarction (MI). This study was designed to compare m6A-tagged transcript profiles to identify mRNA-specific changes on pathophysiological variations after MI.
Methods: N6-methyladenosine methylated RNA immunoprecipitation sequencing (MeRIP-seq) and RNA sequencing (RNA-seq) were interacted to select m6A-modified mRNAs with samples collected from sham operated and MI rat models. m6A methylation regulated mRNAs were interacted with apoptosis/angiogenesis related genes in GeneCards. Afterwards, MeRIP-quantitative real-time PCR (MeRIP-qRT-PCR) was performed to measure m6A methylation level of hub mRNAs. m6A methylation variation was tested under different oxygen concentration or hypoxic duration in H9c2 cells and HUVECs. In addition, Western blot and qRT-PCR were employed to detect expression of hub mRNAs and relevant protein level. Flow cytometry and Tunel assay were conducted to assess apoptotic level. CCK-8, EdU, and tube formation assay were performed to measure cell proliferation and tube formation ability.
Results: Upregulation of Mettl3 was firstly observed in vivo and in vitro, followed by upregulation of m6A methylation level. A total of 567 significantly changed m6A methylation peaks were identified, including 276 upregulated and 291 downregulated peaks. A total of 576 mRNAs were upregulated and 78 were downregulated. According to combined analysis of MeRIP-seq and RNA-seq, we identified 26 significantly hypermethylated and downregulated mRNAs. Based on qRT-PCR and interactive analysis, Hadh, Kcnn1, and Tet1 were preliminarily identified as hub mRNAs associated with apoptosis/angiogenesis. MeRIP-qRT-PCR assay confirmed the results from MeRIP-seq. With the inhibition of Mettl3 in H9c2 cells and HUVECs, downregulated m6A methylation level of total RNA and upregulated expression of hub mRNAs were observed. Increased m6A level was verified in the gradient context in terms of prolonged hypoxic duration and decreased oxygen concentration. Under simulated hypoxia, roles of Kcnn1 and Tet1 in angiogenesis and Hadh, Tet1, and Kcnn1 in apoptosis were further confirmed with our validation experiments.
Conclusion: Roles of m6A-modified mRNA transcripts in the context of MI were preliminarily verified. In the context of m6A methylation, three hub mRNAs were validated to impact the process of apoptosis/angiogenesis. Our study provided theoretical basis and innovative targets for treatment of MI and paved the way for future investigations aiming at exploring upstream epigenetic mechanisms of pathogenesis after MI.
Keywords: myocardial infarction, m6A methylation, mRNA, angiogenesis, apoptosis
1 Introduction
Myocardial infarction (MI), due to the reduction or interruption of the blood supply of the coronary artery, always results in ischemia of the corresponding myocardium leading to myocardial necrosis (Saleh and Ambrose, 2018). It is characterized by an elevated ST-segment in the electrocardiogram, and is one of the most common causes of death worldwide (Lu et al., 2015). At present, the reperfusion therapy to restore the blood circulation of the heart has become a common method for the treatment of MI, however the subsequent reperfusion injury would impair endothelial function and aggravate myocardial cell death (Thygesen et al., 2007; Puymirat et al., 2019). Upon these concerns, exploration of strategies on compensating myocardial cell regeneration and death after MI becomes the most warranted task in this research field. In addition, understanding the microscopic regulations in MI might be essential to reveal the pathophysiological mechanisms behind MI and might shed light on uncovering novel therapies for the treatment of MI.
Robust studies have explored the mechanisms of myocardial cell regeneration and death after MI. Integrin-linked kinase (ILK) has been reported to be an important factor regulating apoptosis and angiogenesis. In hypoxic condition, upregulation of ILK increased phosphorylation of protein kinase B and mammalian target of rapamycin, resulting in enhanced mesenchymal stem cells (MSCs) survival and vascular endothelial growth factors expression level. In addition, transplantation of MSCs rich in ILK could further improve angiogenesis at 3 weeks (Zeng et al., 2017). Exosomes derived from TIMP2-modified MSCs significantly increased the expression of antiapoptotic bcl-2, followed by decreased proapoptotic Bax and pro-caspase9 level, and finally attenuated apoptosis in MI injury via Akt/Sfrp2 pathway in vivo (Ni et al., 2019). However, the upstream regulation of these pathways has not been well documented. Recent studies on epigenetic regulation have revealed the relationships between epigenetic modifications and cardiovascular diseases. Epigenetics, including the reversible modification of DNA and protein, were proven to independently regulate gene expression of DNA and protein. It was not until recently that RNA modification was believed to be the third layer of epigenetics, regulating RNA processing and metabolism. There has been uncovered with more than 100 modifications in RNAs, including 5-methylcytosine (m5C), N6-methyladenosine (m6A), and N1-methyladenosine (m1A). Among which, m6A methylation was demonstrated to be the common and abundant internal modification of eukaryotic messenger RNA (mRNA) (Meyer et al., 2012). It is a dynamic reversible process regulated by methyltransferases (writers), demethylases (erasers), and binding proteins (readers). ALKBH5 was responsible for reducing m6A methylation and ALKBH5 (one of the demethylases) knockout mice exhibited decreased cardiac regenerative ability and cardiac function after neonatal apex resection (Han et al., 2021). The expression of METTL3 was increased in cardiac fibrotic tissue with chronic myocardial infarction. It promoted proliferation of cardiac fibroblasts, fibroblast-to-myofibroblast transition, and collagens accumulation, while silence of METTL3 (one of the methyltransferases) alleviated cardiac fibrosis in MI mice (Li et al., 2021). These findings emphasized the importance of m6A methylation in individual mRNAs and provided new insights into therapeutic strategies. Nonetheless, there is limited knowledge of the whole picture of m6A modification on mRNAs after MI and how performance of m6A methylated mRNAs on the downstream functional phenotypes.
Upon the above concerns, we aimed to systematically compare the m6A-tagged transcript profiles of heart tissue from rat MI models with those from sham operated (SO) rats to identify gene-specific changes in mRNA methylation and contribute to the development of MI. Specifically, we first observed the change of methyltransferases in vivo and vitro followed by the verification of m6A methylation in the gradient context of prolonged hypoxic duration and decreased oxygen concentration. We then identified m6A methylation regulated mRNAs between the SO group and the MI group by combined analysis of N6-methyladenosine methylated RNA immunoprecipitation sequencing (MeRIP-seq) and RNA sequencing (RNA-seq), then top 10 hypermethylated and downregulated mRNAs were selected and validated in rats through quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). Afterward, we interacted the hub mRNAs with genes related to apoptosis and angiogenesis in open-source datasets. We finally validated the role of methyltransferase in the regulation of m6A methylation and selected hub mRNAs in angiogenesis and apoptosis. With the exploration of potential roles for the m6A modified mRNA transcripts in the physiological and pathological mechanisms underlying MI, a theoretical basis and innovative targets could be identified for the treatment of MI.
2 Materials and Methods
2.1 Models
2.1.1 Animal MI Model
Male Sprague-Dawley (SD) rats aged 10 weeks and weighing 250–300 g were collected from Beijing Vital River Laboratory Animal Technology Co. Ltd. The current study was carried out in accordance with the guidelines of the Chinese Council on Animal Protection and approved by the Institutional Animal Care and Use Committee of Nanjing Medical University (reference number of 10091). SD rats were randomly divided into the sham operated (SO) group and the myocardial infarction (MI) group. Ligation of left anterior descending coronary artery was conducted in the MI group and myocardial ischemia was confirmed with elevated ST-segment in electrocardiogram. However, string went through the corresponding myocardial region without ligation in the SO group. Finally, heart samples were collected from the infarction area in the MI group and the corresponding region in the SO group. Heart samples were refrigerated at −80°C for further detection.
2.1.2 Cell Hypoxic Model
H9c2 cells were cultured in Dulbecco’s modified eagle medium (Gibico, Waltham, CA) supplemented with 10% fetal bovine serum (Gibico, Waltham, CA) and 1% of penicillin/streptomycin at 37°C in an incubator with 95% humid air and 5% CO2 (Wang M. et al., 2020). Human umbilical vein endothelial cells (HUVECs) were maintained in endothelial cell growth medium-2 bullet kit (Lonza, Basel, BS, CH) at 37°C in an incubator with 95% humid air and 5% CO2 (Chen et al., 2020). To simulate myocardial ischemia, H9c2 cells and HUVECs were managed with hypoxia. Specifically, they were placed in a hypoxic incubator containing 94% N2, 5% CO2, and 1% O2 for 24 h. Meanwhile, the control group was maintained in a normal atmosphere of 95% air and 5% CO2 at 37°C (Zhu et al., 2021). Afterward, H9c2 cells and HUVECs were cultured in consistent 1% O2, 2% O2, and 5% O2 hypoxic condition for 24 h to explore the impact of different concentrations of oxygen on m6A methylation level. These cells were also cultured with 1% oxygen concentration for 12, 24, and 48 h to explore the impact of different hypoxic duration on m6A methylation level. H9c2 cells and HUVECs were then treated with 25 μM Mettl3 inhibitor (STM2457) for 24 h to explore the role of Mettl3 on m6A methylation level (Yankova et al., 2021).
2.2 MeRIP-Seq and Bioinformatic Analysis
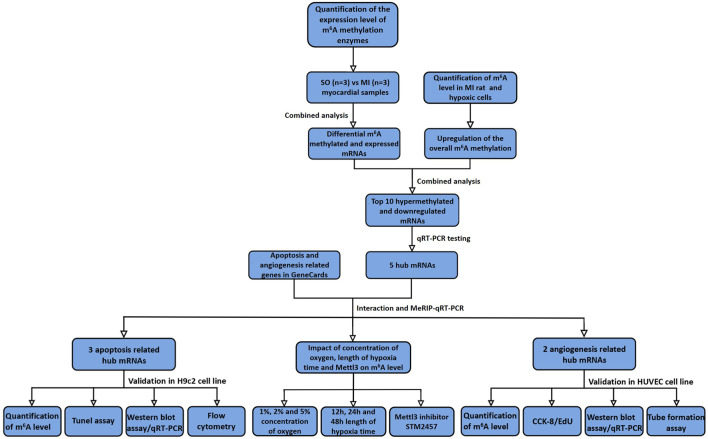
The detailed procedure of MeRIP-Seq analysis and validation of newly discovered hub mRNAs on apoptosis and angiogenesis are demonstrated in Figure 1.
FIGURE 1.
Flowchart of study procedure. SO, sham operated; MI, myocardial infarction; qRT-PCR, quantitative reverse transcription polymerase chain reaction; MeRIP-qRT-PCR, MeRIP-quantitative real-time PCR; CCK-8, cell counting kit-8; EdU, 5-ethynyl-20-deoxyuridine.
2.2.1 Methylated RNA Immunoprecipitation Sequencing
Total RNA was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s procedure. The RNA amount and purity of each sample was quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE). The RNA integrity was assessed by Bioanalyzer 2100 (Agilent, Santa Clara, CA) with RIN number >7.0 and confirmed by electrophoresis with denaturing agarose gel. Afterward, Epicentre Ribo-Zero Gold Kit (Illumina, San Diego, CA) was used to deplete ribosomal RNA (rRNA) from total RNA. The ribosomal-depleted RNA was fragmented into small pieces using Magnesium RNA Fragmentation Module (New England Biolabs, Ipswich, MA). Then the cleaved RNA fragments were incubated with m6A-specific antibody (Synaptic Systems, Walldorf, BW, DE) in immunoprecipitation (IP) buffer (50 mM Tris-HCl, 750 mM NaCl, and 0.5% Igepal CA-630). Subsequently, the IP RNA was reverse transcribed to create cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, Waltham, CA). Eluted fragments containing m6A and untreated input control fragments were converted to construct the strand-specific cDNA library by dUTP method (Dominissini et al., 2013). The average insert size of the final cDNA library was 300 ± 50 bp. We finally performed 2 × 150 bp paired-end sequencing (PE150) with an illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) (Meng et al., 2014; Li M. et al., 2019).
2.2.2 RNA-Seq
Total RNA was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA). For RNA-seq analysis, rRNA was depleted, according to the protocol of the Epicentre Ribo-Zero Gold Kit (Illumina, San Diego, CA). Subsequently, the ribosomal-depleted RNA was fragmented into small pieces using Magnesium RNA Fragmentation Module (New England Biolabs, Ipswich, MA). The fragments were converted to construct the strand-specific cDNA library with dUTP method and were sequenced by illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) (Wang Q. et al., 2020).
2.2.3 Bioinformatic Analysis
Fastp software was used to remove the reads containing adaptor contamination, low quality bases, and undetermined bases with default parameters (Chen et al., 2018). Then sequence quality of IP and input samples were verified with fastp. HISAT2 were used to map reads to the reference genomeRattus norvegicus (Version 101) (Kim et al., 2015). Mapped reads of IP and input libraries were then provided with “exomePeak” package (Meng et al., 2014). m6A peaks from the corresponding libraries were visualized with IGV software (Broad Institute, Cambridge, MA) (Thorvaldsdóttir et al., 2013). MEME and HOMER software were used for de novo and known motif findings followed by localization of the motif with respect to peak summit (Bailey et al., 2009). The information of m6A peaks was obtained by intersection with gene architecture using “ChIPseeker” package (Yu et al., 2015). Then StringTie was used to obtain expression for all mRNAs from input libraries by calculating Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) (Pertea et al., 2015). The differentially expressed mRNAs were selected with |log2FC|>1 and p value < 0.05 by “edgeR” package (Robinson et al., 2010).
2.3 Validation
Expression of top 10 hypermethylated and downregulated mRNAs was first validated with qRT-PCR testing. Subsequently, the m6A RNA methylation level was measured in histological and cellular level. qRT-PCR and Western blot were performed to verify the expression of genes and corresponding proteins. MeRIP-quantitative real-time PCR was performed to measure m6A methylation level of hub genes. Flow cytometry and Tunel assay were carried out to assess the apoptosis level. Cell counting kit-8 (CCK-8), 5-ethynyl-20-deoxyuridine (EdU), and tube formation assay were performed to test cell proliferation and tube formation ability.
2.3.1 Quantification of Total m6A Methylation Level
m6A RNA methylation level was detected using the EpiQuik™ m6A RNA Methylation Quantification kit (Epigentek, New York, NY) according to the manufacturer’s protocol (Liu et al., 2020). Briefly, a negative control and a standard curve consisting of six different concentrations (ranged from 0.02 to 1 ng of m6A) were prepared. There was 200 ng of total RNA used for each reaction. After RNA binding to the 96-well plates, diluted capture anti-m6A antibodies were added, then 100 µl of developer solution to each well, and incubated at room temperature for 10 min without light. There was 100 µl of stop solution added afterward to each well to stop enzyme reaction. The optical density at 450 nm was measured using a microplate reader (Thermo Fisher Scientific, Waltham, CA). Percentage of m6A within the total RNA was calculated (Dang et al., 2020; Liu et al., 2020).
2.3.2 Screening Strategy for Angiogenesis and Apoptosis Related Genes
We first obtained m6A methylation regulated hub genes by qRT-PCR, then, the hub genes were interacted with genes related to angiogenesis and apoptosis in the GeneCards. Results were visualized by Venn diagram (Liang et al., 2019).
2.3.3 MeRIP-Quantitative Real-Time PCR
RNA sample from myocardial tissue and H9c2 cells was fragmented (300 nt) after incubation with fragmentation buffer at 94°C for 4 min. A total of 5% of fragmented RNA was saved as input control. The procedure of m6A-IP sample preparation was similar to that of MeRIP-seq (Deng et al., 2021). Finally, both input control and m6A-IP samples were subjected to qRT-PCR with gene-specific primers listed in Supplementary Table S1.
2.3.4 qRT-PCR
Total RNA was extracted from cells and myocardial tissues and complementary DNA (cDNA) was synthesized using total RNA with the PrimeScript™ RT reagent kit with gDNA Eraser (TaKaRa, Kusatsu, Japan) according to the manufacturer’s instructions. qRT-PCR was performed with the Pro-17 Steponeplus system (Applied Biosystems, Carlsbad, CA) (Saeidi et al., 2018). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control to normalize the expression of genes. There are 25 pairs of primer listed in Supplementary Table S2. Relative expression of differentially expressed genes were analyzed with the 2−△△CT method (Jozefczuk and Adjaye, 2011).
2.3.5 Western Blot Analysis
H9c2 cells and HUVECs were lysed using RIPA lysis buffer (Beyotime, Shanghai, China). Protein concentration was determined using the BCA Protein Assay kit (New Cell and Molecular Biotech, Suzhou, China). Specifically, the corresponding protein was separated with 7.5 and 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Afterward, the membranes were blocked with 5% skim milk for 2 h in room temperature (Qiu et al., 2020). They were incubated with primary antibodies against GAPDH (Proteintech, Wuhan, China); Tet1 (Signalway Antibody, College Park, MD); Hadh (Signalway Antibody, College Park, MD); Kcnn1 (Signalway Antibody, College Park, MD); Caspase3 (Proteintech, Wuhan, China); and Cleavd-caspase3 (Cell Signaling Technology, Danvers, MA); bcl-2 (Abcam, Cambridge, MA); Bax (Abcam, Cambridge, MA); HIF-1α (Abcam, Cambridge, MA); and Mettl3 (Proteintech, Wuhan, China) overnight at 4°C. Then, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Abcam, Cambridge, MA) for 1 h. The bands were analyzed with chemiluminescence Western blotting detection system (Tanon, Shanghai, China) (Gu et al., 2018; Ren et al., 2018; Li X. et al., 2020).
2.3.6 Flow Cytometry
Annexin V-Fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining kit (Vazyme, Nanjing, China) were used to identify apoptotic H9c2 cells. Flow cytometry analysis was performed through Cytoflex (Beckman Coulter, Brea, CA) and data was analyzed using FlowJo software (Tree Star, SanCarlos, CA). The second quadrant (FITC+/PI+) showed the late apoptotic cells and the fourth quadrant (FITC+/PI-) presented the early apoptotic cells (Luo et al., 2019; Zhang et al., 2019; Wei J. et al., 2020; Li Y. et al., 2020).
2.3.7 Tunel Analysis
TUNEL BrightGreen Apoptosis Detection kit (Vazyme, Nanjing, China) was used for Tunel staining based on the manufacturer’s instructions. Briefly, the H9c2 was fixed in 4% PFA at 4°C for 25 min. Then the cells were incubated with Proteinase K (20 μg/ml) for 5 min. After incubation with 1×equilibration buffer for 30 min, the cells were treated with BrightGreen Labeling Mix and Recombinant TdT Enzyme for 1 h at 37°C. The nucleus was stained with DAPI (2 μg/ml) away from light for 10 min (Shi et al., 2021). The Tunel staining images were photographed with a fluorescence microscope (Bio-tek, Winusky, VT) (Zhang et al., 2016; Li T. et al., 2019; Luo et al., 2019; Sharma et al., 2021).
2.3.8 Cell Counting Kit-8
Cell suspension with a concentration of 5×103 each well was digested with trypsin and inoculated into a 96-well plate with 100 μL per well (Yu et al., 2021). After conventional culture for 24, 48, and 72 h, 10 μl CCK-8 solution (MedChemExpress, Shanghai, China) was added to each well and incubated for another 2 h. The absorbance value at 450 nm was finally measured by enzyme labeling instrument (Chen et al., 2019; Chen and Ling, 2019; Li X. et al., 2020; Zhou et al., 2020; Sun et al., 2021).
2.3.9 EdU Assay
EdU incorporation assay kit (Ribobio, Guangzhou, China) was used for the measurement of cell proliferation (Liu et al., 2018). The cell suspension with a concentration of 8×103 each well was digested with trypsin and inoculated into a 96-well plate with 100 μl per well, followed by the addition of 50 μM EdU diluent. After 2 h, cells were fixed in 4% paraformaldehyde, cultivated with 100 μl of 0.5% Triton X-100 and mixed with 100 μL of 1×Apollo® 567 fluorescent staining solution (Xu et al., 2019). The cell nucleus was finally subjected to DAPI staining in a dark environment. Images were finally obtained from Cytation1 (Bio-tek, Winusky, VT).
2.3.10 Tube Formation Assay
Matrigel (BD Biosciences, Franklin Lake, NJ) was used for tube formation assay to assess the tube-forming ability of HUVECs (Cheng et al., 2017). Briefly, it was dissolved at 4°C overnight, and each well of the 96-well plate was then coated with 50 μl of Matrigel. The plate was then left to polymerize at 37°C for 1 h incubation. The HUVECs (2×104 cells/100 μl) were then seeded into each well. After 6 h of incubation at 37°C, tube formation was photographed at ×50 magnification. Tube formations were calculated with the number of branches using ImageJ software (National Institutes of Health, Bethesda, MD) (Li et al., 2017; Hanlon et al., 2019).
2.4 Statistical Analysis
Data were presented as mean with standard deviation. Results of qRT-PCT, MeRIP-qRT-PCR, total m6A methylation level, tube formation assay, and flow cytometry were analyzed with Student’s t-test. CCK-8 assay data across 24, 48, and 72 h was analyzed with two-way analysis of variance (ANOVA). Statistical significance was considered with p value less than 0.05. All analyses were performed using SPSS 22.0 (International Business Machines Corporation, Armonk, NY) and GraphPad Prism 9 (GraphPad, San Diego, CA).
3 Results
3.1 Overview of Methylated RNA Immunoprecipitation Sequencing
In the MeRIP-seq library, on average 86,616,150 and 90,985,883 valid reads were obtained in two groups of myocardial samples, while 81,704,807 and 90,279,353 valid reads were obtained in the RNA-seq library (Supplementary Table S3). In myocardial IP samples, the average mapping ratios of valid reads in the SO and the MI groups were 90.79 and 91.50%, respectively. The average mapping ratios of valid reads were 92.53 and 91.78% in the input samples (Supplementary Table S4). The valid data that mapped to the reference genome can be defined as the alignment to exon, intron, and intergenic according to the regional information. The average ratios of IP and input samples to exons were 67.91 and 55.20% in the SO group while 68.34 and 52.29% in the MI group, respectively (Supplementary Figure S1).
3.2 Mettl3 Induced Upregulation of m6A Methylation Level in Myocardial Tissue, H9c2 Cells, and HUVECs
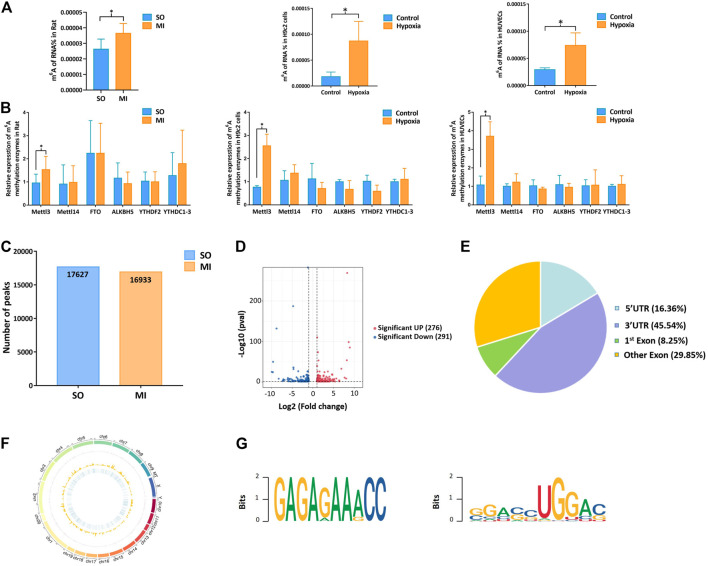
Upregulation of Mettl3 was first observed in vivo (rat MI model) and in vitro (hypoxic H9c2 cells and HUVECs), followed by upregulation of m6A methylation level (Figures 2A,B). Through the analysis of MeRIP-seq, we identified 17,627 distinct m6A peaks in 9889 mRNAs in the SO samples and 16,933 distinct m6A peaks in 9764 mRNAs in the MI samples (Figure 2C). There were 567 significantly variated peaks identified, in which 276 peaks were upregulated and 291 were downregulated (Figure 2D). The top 20 differentially m6A methylated peaks are shown in Table 1. We analyzed the distribution patterns of differentially m6A methylated peaks. A total of 45.54% of the m6A methylated peaks harbored in the 3′UTR and 16.36% enriched in the 5′UTR (Figure 2E). All differentially m6A methylated peaks within mRNAs were mapped to chromosomes (Figure 2F). The top five chromosomes harboring the most m6A peaks were listed as follows: chr1 (646), chr10 (436), chr11 (130), chr12 (176), and chr13 (170). Finally, we conducted motif prediction for samples and demonstrated the predicted motif in Figure 2G.
FIGURE 2.
Mettl3 induced upregulation of m6A methylation level in myocardial tissue, H9c2 cells, and HUVECs. (A) m6A methylation level of MI rat and hypoxic cells; (B) expression of m6A methylation enzymes in myocardial tissue samples and cells by qRT-PCR; (C) number of peaks in the SO and MI group; (D) volcano plots for differentially methylated m6A peaks; (E) distribution of differentially methylated m6A peaks in the SO and MI group; (F) distribution patterns of differentially m6A methylated peaks on chromosomes; (G) sequence motif for the m6A peak regions. UTR, untranslated region; Exon, expressed region; SO, sham operated; MI, myocardial infarction. *p < 0.05.
TABLE 1.
Top 20 differentially methylated mRNAs
| mRNA | Chromosome | Peak region | Peak start | Peak end | p-value | Log2FC | Up/Down |
|---|---|---|---|---|---|---|---|
| Bach1 | chr11 | 3′ UTR | 27,397,948 | 27,398,514 | 6.30E-86 | 8.96 | Up |
| Rrn3 | chr10 | 3′ UTR | 3,213,487 | 3,213,996 | 9.99E-270 | 8.32 | Up |
| Senp5 | chr11 | Exon | 72,064,886 | 72,067,150 | 2.45E-09 | 8.06 | Up |
| LOC100910130 | X | Exon | 156,395,518 | 156,397,956 | 1.00E-11 | 7.68 | Up |
| Hnrnph3 | chr20 | Exon | 27,176,410 | 27,177,224 | 1.26E-33 | 6.91 | Up |
| Gspt1 | chr10 | Exon | 4,412,071 | 4,426,636 | 8.50E-03 | 6.39 | Up |
| Bdkrb1 | chr6 | 3′ UTR | 129,439,859 | 129,440,455 | 1.20E-04 | 6.38 | Up |
| Scart1 | chr1 | Exon | 212,701,635 | 212,704,224 | 7.20E-02 | 5.70 | Up |
| Natd1 | chr10 | 5′ UTR | 45,065,415 | 45,071,643 | 2.90E-02 | 5.39 | Up |
| Gata6 | chr18 | Exon | 2,417,213 | 2,417,483 | 6.46E-08 | 5.18 | Up |
| Tap2 | chr20 | 5′ UTR | 3,995,544 | 3,996,581 | 7.94E-26 | −9.94 | Down |
| Nfat5 | chr19 | 5′ UTR | 38,533,016 | 38,533,316 | 3.98E-25 | −9.69 | Down |
| Sp3 | chr3 | Exon | 59,646,075 | 59,646,253 | 1.25E-50 | −9.63 | Down |
| Cd248 | chr1 | Exon | 220,353,416 | 220,354,637 | 1.26E-50 | −8.79 | Down |
| AABR07048992 | chr5 | 5′ UTR | 99,032,868 | 99,033,107 | 1.00E-132 | −8.25 | Down |
| Nfs1 | chr3 | 5′ UTR | 151,687,398 | 151,688,149 | 6.76E-09 | −7.34 | Down |
| Rpusd2 | chr3 | Exon | 110,836,072 | 110,836,252 | 2.18E-04 | −7.17 | Down |
| Ranbp2 | chr20 | Exon | 28,047,206 | 28,047,481 | 9.77E-06 | −6.81 | Down |
| Capns1 | chr1 | Exon | 91,063,753 | 91,063,928 | 6.30E-11 | −6.04 | Down |
| Raver2 | chr5 | Exon | 119,938,132 | 119,938,341 | 1.50E-02 | −5.95 | Down |
FC, fold change.
3.3 Hub mRNAs Associated with Angiogenesis and Apoptosis
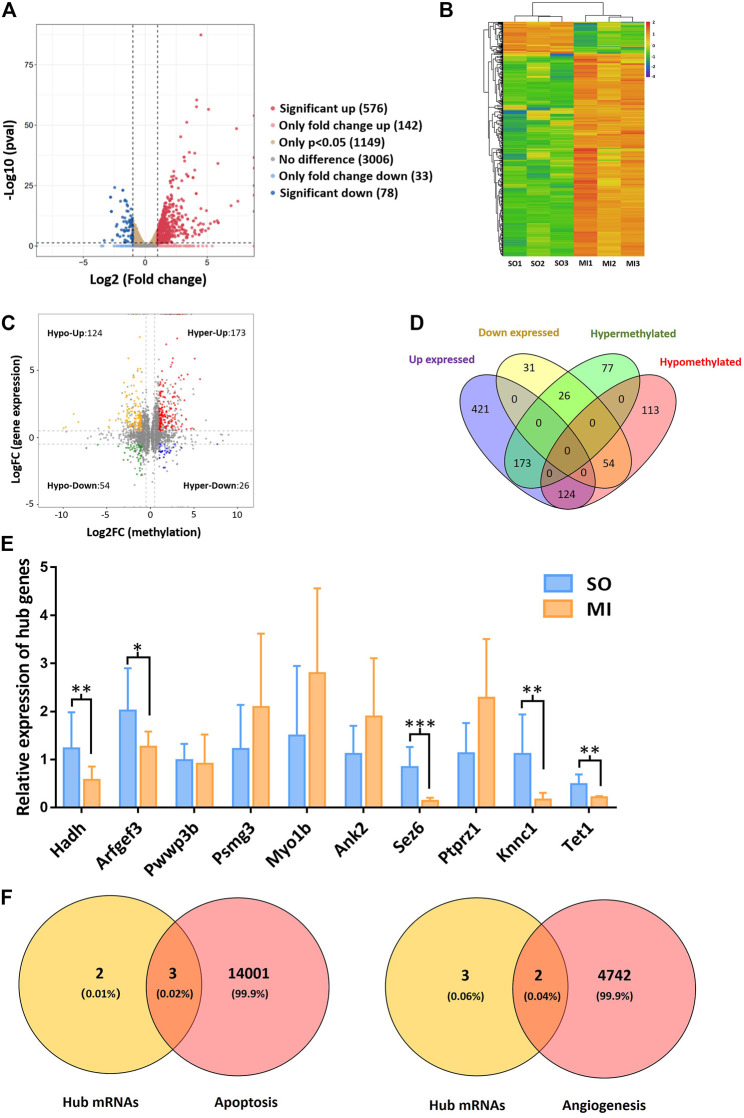
Based on the results of RNA-seq, 576 mRNAs were significantly upregulated, and 78 mRNAs were downregulated. The top 20 differentially expressed mRNAs are listed in Table 2. Volcano plots and heatmap plots of differentially expressed mRNAs are shown in Figures 3A,B. Combined analysis of m6A methylation and mRNAs expression levels was performed according to the following thresholds: |log2FC|>1, p < 0.05 for m6A methylation and |log2FC|>0.5, p < 0.05 for mRNAs expression. As a result, we obtained 377 mRNAs where their m6A peaks and mRNA expression both changed significantly. The relationships between m6A methylation and mRNAs expression are shown in the four-quadrant graph and Venn diagram (Figures 3C,D). Accordingly, there were 124 significantly hypomethylated and upregulated mRNAs, 26 significantly hypermethylated and downregulated mRNAs, 173 significantly hypermethylated and upregulated, and 54 significantly hypomethylated and downregulated mRNAs. The top 10 hypermethylated and downregulated mRNAs are list in Table 3. Meanwhile, we provided the detailed information of the top 10 hypermethylated and upregulated mRNAs, the top 10 hypomethylated and downregulated mRNAs, and the top 10 hypomethylated and upregulated mRNAs in Supplementary Tables S5–S7. Expression of hub mRNAs, including Hadh, Arfgef3, Sez6, Psmg3, Kcnn1, Tet1, Myo1b, Ptprz1, Ank2, and Pwwp3b were compared between the SO and the MI groups by qRT-PCR testing. Among them, the expression of Hadh, Arfgef3, Sez6, Kcnn1, and Tet1 were significantly downregulated in the MI group (Figure 3E). After interaction of the above three hub mRNAs with apoptosis and angiogenesis-related genes in GeneCards, Hadh, Tet1, and Kcnn1 were finally identified to be significantly associated with apoptosis, and Tet1 and Kcnn1 were significantly correlated to angiogenesis (Figure 3F).
TABLE 2.
Top 20 differentially expressed mRNAs.
| mRNA | Chromosome | Gene start | Gene end | p-value | Log2FC | Up/Down |
|---|---|---|---|---|---|---|
| Gm47305 | chr6 | 91,680,317 | 91,680,386 | 8.33E-67 | 12.06 | Up |
| Ccn4 | chr7 | 107,695,215 | 107,723,772 | 1.10E-23 | 8.30 | Up |
| Mmp12 | chr8 | 5,606,592 | 5,616,493 | 1.27E-15 | 8.28 | Up |
| Col11a1 | chr2 | 216,863,428 | 217,056,523 | 1.12E-57 | 7.95 | Up |
| Grem1 | chr3 | 105,203,309 | 105,214,989 | 1.22E-16 | 7.65 | Up |
| Comp | chr16 | 20,798,437 | 20,807,070 | 1.57E-68 | 7.59 | Up |
| Card14 | chr10 | 108,440,950 | 108,468,310 | 3.02E-19 | 7.48 | Up |
| Angptl7 | chr5 | 165,312,130 | 165,316,652 | 2.84E-49 | 7.37 | Up |
| Fam180a | chr4 | 62,844,971 | 62,860,446 | 4.73E-33 | 7.36 | Up |
| Ccn5 | chr3 | 160,207,913 | 160,219,331 | 1.24E-134 | 7.18 | Up |
| Ptprh | chr1 | 72,810,545 | 72,859,161 | 1.54E-05 | −6.83 | Down |
| Fam111a | chr1 | 229,003,961 | 229,019,527 | 3.80E-07 | −4.96 | Down |
| Gp6 | chr1 | 73,040,901 | 73,064,641 | 2.80E-03 | −4.22 | Down |
| 7SK | chr17 | 90,799,609 | 90,799,914 | 2.00E-06 | −3.90 | Down |
| AABR07069186 | chr8 | 10,985,553 | 10,988,077 | 4.10E-04 | −3.47 | Down |
| Gzmc | chr15 | 35,392,162 | 35,394,792 | 3.81E-04 | −3.41 | Down |
| Sec14l5 | chr10 | 10,591,251 | 10,629,735 | 2.36E-13 | −3.34 | Down |
| Pdzk1 | chr2 | 198,965,685 | 198,999,323 | 5.50E-03 | −3.31 | Down |
| Klk12 | chr1 | 99,706,780 | 99,711,004 | 3.10E-02 | −3.25 | Down |
| Alas2 | X | 23,167,696 | 23,187,341 | 2.90E-04 | −3.18 | Down |
FC, fold change.
FIGURE 3.
Hub mRNAs associated with angiogenesis and apoptosis. (A) Volcano plots for differentially expressed mRNAs; (B) heatmap plots for differentially expressed mRNAs; (C) four-quadrant graph for m6A methylation regulated mRNAs; (D) Venn diagram showing the relationships between m6A methylation and mRNAs expression; (E) validation of the top 10 hypermethylated and downregulated mRNAs in MI rats; (F) Venn diagram showing the relationship between hub mRNAs and apoptosis/angiogenesis. Hypo-Up, hypomethylated and upregulated; hyper-Up, hypermethylated and upregulated; Hypo-Down, hypomethylated and downregulated; hyper-Down, hypermethylated and downregulated. SO, sham operated; MI, myocardial infarction. *p < 0.05, **p < 0.01, ***p < 0.001.
TABLE 3.
Top 10 hypermethylated and downregulated mRNAs.
| mRNA | Chromosome | m6A regulation | Regulation | FPKM of MI input | FPKM of SO input |
|---|---|---|---|---|---|
| Hadh | chr2 | Up | Down | 87.12 | 249.88 |
| Arfgef3 | chr1 | Up | Down | 0.40 | 1.10 |
| Pwwp3b | X | Up | Down | 0.53 | 1.11 |
| Sez6 | chr10 | Up | Down | 0.35 | 0.71 |
| Psmg3 | chr12 | Up | Down | 1.28 | 2.62 |
| Kcnn1 | chr16 | Up | Down | 0.85 | 1.70 |
| Tet1 | X | Up | Down | 0.29 | 0.58 |
| Myo1b | chr9 | Up | Down | 12.53 | 21.41 |
| Ptprz1 | chr4 | Up | Down | 0.25 | 0.41 |
| Ank2 | chr2 | Up | Down | 11.74 | 18.99 |
FPKM, fragments per kilobase of exon model per million mapped fragments; MI, myocardial infarction; SO, sham operated.
3.4 m6A Methylation Influenced by Hypoxia in a Time- and Dose-Dependent Pattern
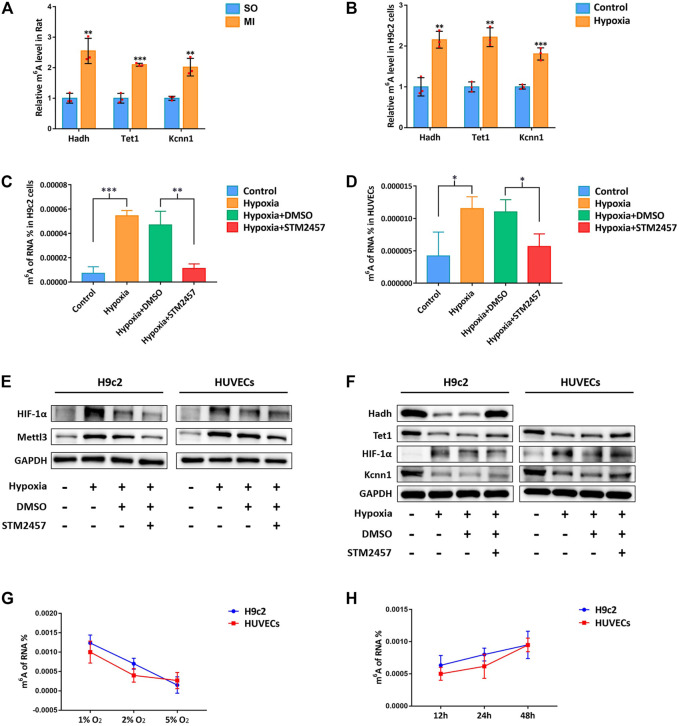
Upon our previous observation, m6A methylation level was upregulated by Mettl3 in vivo and in vitro (Figures 2A,B). We afterward detected upregulated m6A methylation level of hub mRNAs with MeRIP-qRT-PCR, which further confirmed our results from transcriptome-wide MeRIP-seq analysis (Figures 4A,B). With the inhibition of Mettl3 in H9c2 cells and HUVECs, downregulated m6A methylation level of total RNA and upregulated expression of hub mRNAs were detected (Figures 4C–F). After treating with different oxygen concentration (1% O2, 2% O2, and 5% O2) for 24 h, m6A methylation level in H9c2 cells and HUVECs progressively decreased with the increase of oxygen concentration (Figure 4G). Afterward, these cells were treated with different hypoxic duration (12, 24, and 48 h) at 1% of oxygen. m6A methylation level in H9c2 cells and HUVECs gradually increased with prolonged hypoxic duration (Figure 4H). Variation of m6A methylation in the simulated hypoxic context demonstrated a time- and dose-dependent pattern.
FIGURE 4.
m6A methylation influenced by hypoxia in a time- and dose-dependent pattern. (A) MeRIP-qRT-PCR validation of hub mRNAs in MI rat; (B) MeRIP-qRT-PCR validation of hub mRNAs in H9c2 cells; (C) m6A methylation levels of H9c2 cells with Mettl3 inhibitor; (D) m6A methylation levels of HUVECs with Mettl3 inhibitor; (E) Mettl3 protein level of H9c2 cells and HUVECs with Mettl3 inhibitor. HIF-1α was used as positive control. GAPDH was used as loading control; (F) protein level of hub mRNAs in H9c2 cells and HUVECs with Mettl3 inhibitor; (G) m6A methylation levels of H9c2 cells and HUVECs with different concentrations of oxygen; (H) m6A methylation levels of H9c2 cells and HUVECs with different hypoxic duration. SO, sham operated; MI, myocardial infarction; DMSO, dimethyl sulfoxide. *p < 0.05, **p < 0.01, ***p < 0.001. *STM2457 is the inhibitor of Mettl3.
3.5 m6A Methylation Regulated Hub mRNAs Play a Role on Apoptosis in vitro
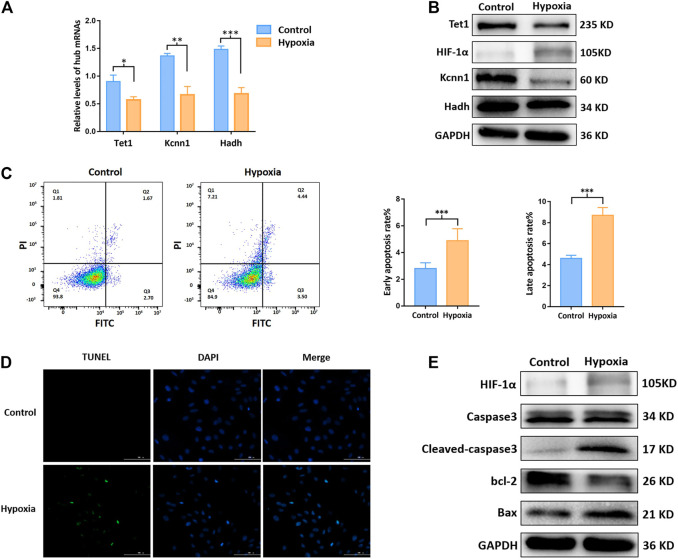
As presented in Figures 5A,B, Hadh, Tet1, and Kcnn1 were significantly downregulated after hypoxia. The apoptosis rate of cardiomyocytes exposed to hypoxia increased significantly (Figures 5C,D). As compared to the control group, the expressions of cleaved-caspase3 and Bax increased significantly, while the expressions of bcl-2 decreased in the hypoxia group based on Western blot (Figure 5E).
FIGURE 5.
m6A methylation regulated hub mRNAs play a role on apoptosis in vitro. (A) Expression of apoptosis related mRNAs by qRT-PCR; (B) protein level of Tet1, Kcnn1, and Hadh by Western blot; (C) apoptosis rate by flow cytometry; (D) apoptotic status by Tunel staining; (E) protein level (Caspase3, cleaved-caspase3, bcl-2, Bax, and HIF-1α) by Western blot. *p < 0.05, **p < 0.01, ***p < 0.001.
3.6 m6A Methylation Regulated Hub mRNAs Play a Role on Angiogenesis in vitro
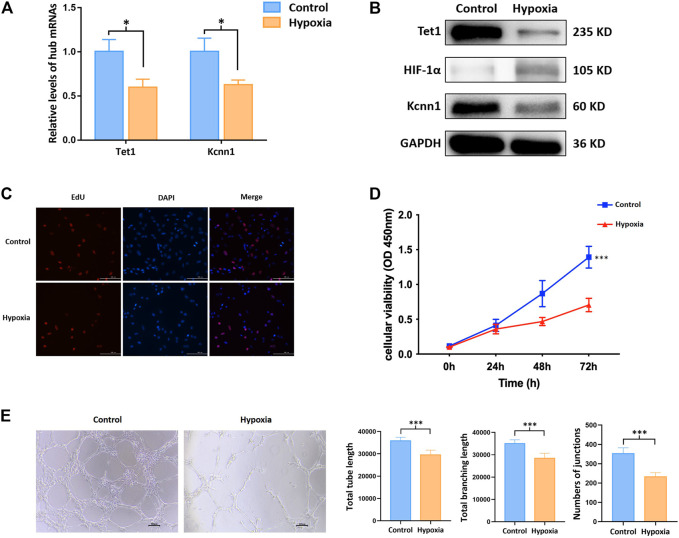
Kcnn1 and Tet1 determined by qRT-PCR and Western blot were significantly decreased in the hypoxia group (Figures 6A,B). The effects of hypoxia on cell proliferation and angiogenesis were revealed by CCK-8, EdU, and tube formation assay. Cell viability and EdU-positive cells were significantly decreased due to hypoxia (Figures 6C,D). All indicators of tube formation ability, including the total branching points, total tube length, and total loops, were markedly decreased in the hypoxia group (Figure 6E).
FIGURE 6.
m6A methylation regulated hub mRNAs play a role on angiogenesis in vitro. (A) Expression of angiogenesis related mRNAs by qRT-PCR; (B) protein level of Tet1, Kcnn1, and HIF-1α by Western blot; (C) cell proliferation by EdU assay; (D) cell viability by CCK-8 assay; (E) tube formation. *p < 0.05; ***p < 0.001.
4 Discussion
DNA methylation and histone modification have been extensively investigated in genetic and cellular biology (Panneerdoss et al., 2018). However, roles of reversible RNA methylation in the cardiovascular field have been under development in recent years (Mongelli et al., 2020). In the hypoxia/reoxygenation-treated cardiomyocytes, low-expression of METTL3 and over-expression of ALKBH5 were observed to inhibit autophagy and apoptosis (Song et al., 2019). In addition, over-expression of ALKBH5 was verified to reduce infarct size, restore cardiac function, and facilitate cardiomyocyte proliferation after MI (Han et al., 2021). These promising results preliminarily revealed the roles of m6A methylation on mRNAs in ischemic heart diseases. However, information regarding systematic and comprehensive analysis is limited. In the current study, we observed the overall m6A methylation level was significantly upregulated with the upregulation of Mettl3 in MI rat and hypoxic cells. We demonstrated the landscape changes of m6A methylation on mRNA regulated apoptosis and angiogenesis in MI. Meanwhile, we explored the impact of oxygen concentration and hypoxic duration on m6A methylation. Upon which, several hypermethylated and downregulated mRNAs were successfully identified after MI by combined analysis of MeRIP-seq and RNA-seq. It further provided potential targets for treatment and scientific purposes. However, how and what performance of these m6A methylated mRNAs may have on MI need to be further elaborated.
Followed by the newly proposed mechanisms of upstream methylation on mRNA in this research field, the final destination is to compensate or improve the downstream functional impairment launched by MI. According to the literature review, apoptosis plays a role in the process of tissue damage after MI, which appears to have pathological and therapeutic implications (Krijnen et al., 2002). Therefore, the upcoming question in front of us was whether the m6A methylated mRNAs would play a role in the regulation of cardiomyocyte apoptosis. In this regard, we interacted our newly identified m6A methylated mRNAs and apoptosis related genes in the GeneCards and three of them were correlated to apoptosis (e.g., Tet1, Hadh, and Kcnn1). Over-expression of Tet1 increased cell apoptosis and inhibited cell growth in osteosarcoma cells (Teng et al., 2019). Over-expression of Hadh linked to cell apoptosis in acute myeloid leukemia (Wei L. et al., 2020). However, the relationship between Kcnn1 and apoptosis has not been validated according to previous studies. We afterward validated the m6A methylation regulated mRNAs on apoptosis in hypoxic H9c2 cells and found the m6A methylation level was upregulated in hypoxic H9c2 cells. Our results of MeRIP-qRT-PCR demonstrated that m6A methylation of three hub mRNAs was significantly upregulated, while their expression levels were downregulated in MI rat and hypoxic cells. In addition, inhibition of Mettl3 downregulated the overall m6A level and upregulated the expression of Hadh, Tet1, and Kcnn1 in hypoxic H9c2 cells. Apoptosis was accompanied with variations of hub proteins, for instance, decrease of bcl-2 and increase of Bax, Caspase3, and cleaved-caspase3. Taken together, these three hub mRNAs regulated by m6A methylation were preliminarily verified to impact the process of cardiomyocyte apoptosis after MI.
As the exacerbation of myocardial injury, it is likely leading to heart failure and sudden death (Zouggari et al., 2013). Early revascularization in the infarcted region is essential for promoting the survival of myocardial cells, reducing infarct size, and improving the prognosis of patients with MI (Keeley et al., 2003; Guo et al., 2017). Upon this perspective, angiogenesis has been reported to be an integral and indispensable part of the myocardial healing process following ischemic events (Cochain et al., 2013). The inhibition of angiogenesis accelerated heart failure in a murine model and various treatments were proposed to ameliorate infarction size, left ventricular remodeling, and cardiac function after MI via promoting angiogenesis in animal models of MI (Shiojima et al., 2005; Zeng et al., 2010; Shindo et al., 2016; Gou et al., 2020). In the past several decades, attentions have been shifted to investigate the mechanical associations between angiogenesis and epigenetics. mRNAs are responsible for protein translation and can directly regulate the synthesis of proteins such as vascular endothelial growth factor in the context of MI. However, the upstream regulation of mRNAs has not been well clarified. In the current study, two m6A methylation regulated mRNAs (e.g., Kcnn1 and Tet1) were detected by interaction analysis of five hub mRNAs and angiogenesis-related genes. Kcnn1 is one of the members in the calcium activated potassium channel subfamily. It was previously reported to be associated with the development of atrial fibrillation while its role in angiogenesis in the context of MI has not been reported (Park et al., 2014; Rahm et al., 2021). On the other hand, Tet1, as one of translocation enzymes mediating 5-methylcytosine (5mC) hydroxylation, was reported to participate in the facilitation of DNA demethylation (Gomes et al., 2020). In addition, inhibition of Tet1 expression may contribute to tumor growth and angiogenesis in vivo (Si et al., 2019). Upon this condition, we explored the impact of Kcnn1 and Tet1 on angiogenesis in hypoxia-induced HUVECs to further verify their roles after MI. Not surprisingly, in the context of hypermethylated and downregulated Kcnn1 and Tet1, CCK-8, EdU, and tube formation assay potentially indicated that hypoxia inhibits endothelial cell proliferation and angiogenesis. Meanwhile, we found the overall level of m6A methylation of total RNA was upregulated in hypoxic endothelial cells. Inhibition of Mettl3 downregulated the overall m6A level and upregulated the expression of Tet1 and Kcnn1 in hypoxic HUVECs. These results suggested that m6A methylated Kcnn1 and Tet1 may play an essential role in angiogenesis. Further studies to be conducted in knockdown and over-expression models would be helpful to validate the role of m6A methylation on mRNAs in the process of angiogenesis.
Similar to other MeRIP-seq and RNA-seq studies, the reproducibility of MeRIP-seq was considered poor. Fortunately, heart tissues of our animal model were consistently collected from similar regions in a standard environment, which guaranteed the homogeneity at the histological level to some extent. In addition, we performed a rescue experiment to verify the reproducibility of our sequencing results. However, it needs to be further verified with well-designed in vitro or in vivo studies. Finally, our results supplemented information of m6A methylated mRNAs in MI, which was conducive to the broadening of research ideas in this direction and provided reference for further studies.
5 Conclusion
The overview of upstream epigenetic changes after MI were demonstrated with emphasizing the essential role of m6A methylated mRNAs after MI. Five m6A methylation regulated mRNAs were newly identified by interaction analysis of MeRIP-seq and RNA-seq. In the context of m6A methylation, three of hub mRNAs were validated to impact the process of cardiomyocyte apoptosis and angiogenesis. Our study paved the way for future investigations aiming at exploring the upstream epigenetic mechanisms in the pathogenesis of MI.
Acknowledgments
The authors acknowledge the Jiangsu province hospital core facility center for providing experimental instruments, research assistants here for their valuable help, the medical staff from the Department of Rehabilitation Medicine in the First Affiliated Hospital of Nanjing Medical University.
Data Availability Statement
The data presented in the study are deposited in the GEO repository, accession number GSE189593.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Nanjing Medical University.
Author Contributions
X.L., Y.Z., and Y.D. conceived and designed the current study. Y.Z., W.H., Y.C., J.W., X.Z., M.T., and S.W. established MI rat model and collected RNA samples from myocardial tissues. Y.C. and Y.Z. performed data processing, normalization, and bioinformatic analyses. Y.Z. and W.H. completed experimental verification. Y.Z., W.H., and Y.C. drafted the manuscript. Y.Z. and Y.D. contributed to the modification of the manuscript. All authors have read and approved the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Grant number: 81772441, 81902288, 82072546, and 82000529). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.789923/full#supplementary-material
References
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Ling C. H. (2019). Long Noncoding RNA AK027294 Acts as an Oncogene in Non-small Cell Lung Cancer by Up-Regulating STAT3. Eur. Rev. Med. Pharmacol. Sci. 23, 1102–1107. 10.26355/eurrev_201902_17000 [DOI] [PubMed] [Google Scholar]
- Chen J., Chen T., Zhu Y., Li Y., Zhang Y., Wang Y., et al. (2019). circPTN Sponges miR-145-5p/miR-330-5p to Promote Proliferation and Stemness in Glioma. J. Exp. Clin. Cancer Res. 38, 398. 10.1186/s13046-019-1376-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: an Ultra-fast All-In-One FASTQ Preprocessor. Bioinformatics 34, i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. L., Chen Y. X., Zhou J., Li Y., Gong C. Y., Wang X. B. (2020). LncRNA HULC Alleviates HUVEC Inflammation and Improves Angiogenesis after Myocardial Infarction through Down-Regulating miR-29b. Eur. Rev. Med. Pharmacol. Sci. 24, 6288–6298. 10.26355/eurrev_202006_21527 [DOI] [PubMed] [Google Scholar]
- Cheng H.-W., Chen Y.-F., Wong J.-M., Weng C.-W., Chen H.-Y., Yu S.-L., et al. (2017). Cancer Cells Increase Endothelial Cell Tube Formation and Survival by Activating the PI3K/Akt Signalling Pathway. J. Exp. Clin. Cancer Res. 36, 27. 10.1186/s13046-017-0495-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochain C., Channon K. M., Silvestre J.-S. (2013). Angiogenesis in the Infarcted Myocardium. Antioxid. Redox Signaling 18, 1100–1113. 10.1089/ars.2012.4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Xu J., Yang Y., Li C., Zhang Q., Zhou W., et al. (2020). Ling-gui-zhu-gan Decoction Alleviates Hepatic Steatosis through SOCS2 Modification by N6-Methyladenosine. Biomed. Pharmacother. 127, 109976. 10.1016/j.biopha.2020.109976 [DOI] [PubMed] [Google Scholar]
- Deng K., Ning X., Ren X., Yang B., Li J., Cao J., et al. (2021). Transcriptome-wide N6-Methyladenosine Methylation Landscape of Coronary Artery Disease. Epigenomics 13, 793–808. 10.2217/epi-2020-0372 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. (2013). Transcriptome-wide Mapping of N6-Methyladenosine by m6A-Seq Based on Immunocapturing and Massively Parallel Sequencing. Nat. Protoc. 8, 176–189. 10.1038/nprot.2012.148 [DOI] [PubMed] [Google Scholar]
- Gomes A. M., Pinto T. S., Costa Fernandes C. J., Silva R. A., Zambuzzi W. F. (2020). Wortmannin Targeting Phosphatidylinositol 3‐kinase Suppresses Angiogenic Factors in Shear‐stressed Endothelial Cells. J. Cell Physiol 235, 5256–5269. 10.1002/jcp.29412 [DOI] [PubMed] [Google Scholar]
- Gou L., Xue C., Tang X., Fang Z. (2020). Inhibition of Exo-miR-19a-3p Derived from Cardiomyocytes Promotes Angiogenesis and Improves Heart Function in Mice with Myocardial Infarction via Targeting HIF-1α. Aging 12, 23609–23618. 10.18632/aging.103563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Wang J., Wang Y., Xu Y., Zhang Y., Wu W., et al. (2018). MiR-147b Inhibits Cell Viability and Promotes Apoptosis of Rat H9c2 Cardiomyocytes via Down-Regulating KLF13 Expression. Acta Biochim. Biophys. Sin (Shanghai). 50, 288–297. 10.1093/abbs/gmx144 [DOI] [PubMed] [Google Scholar]
- Guo Y., Luo F., Liu Q., Xu D. (2017). Regulatory Non-coding RNAs in Acute Myocardial Infarction. J. Cell. Mol. Med. 21, 1013–1023. 10.1111/jcmm.13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Wang X., Xu Z., Cao Y., Gong R., Yu Y., et al. (2021). ALKBH5 Regulates Cardiomyocyte Proliferation and Heart Regeneration by Demethylating the mRNA of YTHDF1. Theranostics 11, 3000–3016. 10.7150/thno.47354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon M. M., Rakovich T., Cunningham C. C., Ansboro S., Veale D. J., Fearon U., et al. (2019). STAT3 Mediates the Differential Effects of Oncostatin M and TNFα on RA Synovial Fibroblast and Endothelial Cell Function. Front. Immunol. 10, 2056. 10.3389/fimmu.2019.02056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefczuk J., Adjaye J. (2011). Quantitative Real-Time PCR-Based Analysis of Gene Expression. Methods Enzymol. 500, 99–109. 10.1016/b978-0-12-385118-5.00006-2 [DOI] [PubMed] [Google Scholar]
- Keeley E. C., Boura J. A., Grines C. L. (2003). Primary Angioplasty versus Intravenous Thrombolytic Therapy for Acute Myocardial Infarction: a Quantitative Review of 23 Randomised Trials. The Lancet 361, 13–20. 10.1016/s0140-6736(03)12113-7 [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnen P. A., Nijmeijer R., Meijer C. J., Visser C. A., Hack C. E., Niessen H. W. (2002). Apoptosis in Myocardial Ischaemia and Infarction. J. Clin. Pathol. 55, 801–811. 10.1136/jcp.55.11.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Ye J., Zhao G., Hong G., Hu X., Cao K., et al. (2019a). Gas6 Attenuates Lipopolysaccharide-induced TNF-α E-xpression and A-poptosis in H9C2 C-ells through NF-κB and MAPK I-nhibition via the Axl/PI3K/Akt P-athway. Int. J. Mol. Med. 44, 982–994. 10.3892/ijmm.2019.4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Hu P.-S., Zuo Z., Lin J.-F., Li X., Wu Q.-N., et al. (2019b). METTL3 Facilitates Tumor Progression via an m6A-igf2bp2-dependent Mechanism in Colorectal Carcinoma. Mol. Cancer 18, 112. 10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Zhuang Y., Yang W., Xie Y., Shang W., Su S., et al. (2021). Silencing of METTL3 Attenuates Cardiac Fibrosis Induced by Myocardial Infarction via Inhibiting the Activation of Cardiac Fibroblasts. FASEB j. 35, e21162. 10.1096/fj.201903169R [DOI] [PubMed] [Google Scholar]
- Li X., Wang X., Liu Y.-s., Wang X.-d., Zhou J., Zhou H. (2020a). Downregulation of miR-3568 Protects against Ischemia/Reperfusion-Induced Cardiac Dysfunction in Rats and Apoptosis in H9C2 Cardiomyocytes through Targeting TRIM62. Front. Pharmacol. 11, 17. 10.3389/fphar.2020.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhu H., Wei X., Li H., Yu Z., Zhang H., et al. (2017). LPS Induces HUVEC Angiogenesis In Vitro through miR-146a-Mediated TGF-Β1 Inhibition. Am. J. Transl Res. 9, 591–600. [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ren S., Xia J., Wei Y., Xi Y. (2020b). EIF4A3-Induced Circ-BNIP3 Aggravated Hypoxia-Induced Injury of H9c2 Cells by Targeting miR-27a-3p/BNIP3. Mol. Ther. - Nucleic Acids 19, 533–545. 10.1016/j.omtn.2019.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Zhang X., Zou J., Shi Y., Wang Y., Tai J., et al. (2019). Pharmacology Mechanism of Flos Magnoliae and Centipeda Minima for Treating Allergic Rhinitis Based on Pharmacology Network. Drug Dev. Ind. Pharm. 45, 1547–1555. 10.1080/03639045.2019.1635150 [DOI] [PubMed] [Google Scholar]
- Liu T., Yang S., Sui J., Xu S. Y., Cheng Y. P., Shen B., et al. (2020). Dysregulated N6‐methyladenosine Methylation Writer METTL3 Contributes to the Proliferation and Migration of Gastric Cancer. J. Cel Physiol 235, 548–562. 10.1002/jcp.28994 [DOI] [PubMed] [Google Scholar]
- Liu Y., Chen X., Cheng R., Yang F., Yu M., Wang C., et al. (2018). The Jun/miR-22/HuR Regulatory axis Contributes to Tumourigenesis in Colorectal Cancer. Mol. Cancer 17, 11. 10.1186/s12943-017-0751-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu M., Sun R., Zheng Y., Zhang P. (2015). Myocardial Infarction: Symptoms and Treatments. Cell Biochem Biophys 72, 865–867. 10.1007/s12013-015-0553-4 [DOI] [PubMed] [Google Scholar]
- Luo G., Jian Z., Zhu Y., Zhu Y., Chen B., Ma R., et al. (2019). Sirt1 Promotes Autophagy and Inhibits Apoptosis to Protect Cardiomyocytes from Hypoxic Stress. Int. J. Mol. Med. 43, 2033–2043. 10.3892/ijmm.2019.4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Lu Z., Liu H., Zhang L., Zhang S., Chen Y., et al. (2014). A Protocol for RNA Methylation Differential Analysis with MeRIP-Seq Data and exome Peak R/Bioconductor Package. Methods 69, 274–281. 10.1016/j.ymeth.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell 149, 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelli A., Atlante S., Bachetti T., Martelli F., Farsetti A., Gaetano C. (2020). Epigenetic Signaling and RNA Regulation in Cardiovascular Diseases. Ijms 21, 509. 10.3390/ijms21020509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Liu X., Yin Y., Zhang P., Xu Y.-W., Liu Z. (2019). Exosomes Derived from TIMP2-Modified Human Umbilical Cord Mesenchymal Stem Cells Enhance the Repair Effect in Rat Model with Myocardial Infarction Possibly by the Akt/Sfrp2 Pathway. Oxidative Med. Cell Longevity 2019, 1–19. 10.1155/2019/1958941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerdoss S., Eedunuri V. K., Yadav P., Timilsina S., Rajamanickam S., Viswanadhapalli S., et al. (2018). Cross-talk Among Writers, Readers, and Erasers of M 6 A Regulates Cancer Growth and Progression. Sci. Adv. 4, eaar8263. 10.1126/sciadv.aar8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Lee J. S., Ko Y.-G., Lee S. H., Lee B. S., Kang S.-M., et al. (2014). Histological and Biochemical Comparisons between Right Atrium and Left Atrium in Patients with Mitral Valvular Atrial Fibrillation. Korean Circ. J. 44, 233–242. 10.4070/kcj.2014.44.4.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., Salzberg S. L. (2015). StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 33, 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puymirat E., Cayla G., Cottin Y., Elbaz M., Henry P., Gerbaud E., et al. (2019). Twenty-year Trends in Profile, Management and Outcomes of Patients with ST-Segment Elevation Myocardial Infarction According to Use of Reperfusion Therapy: Data from the FAST-MI Program 1995-2015. Am. Heart J. 214, 97–106. 10.1016/j.ahj.2019.05.007 [DOI] [PubMed] [Google Scholar]
- Qiu L., Zhao Q., Dai L., Zhu A., Xu X., Zhao S., et al. (2020). Long Non-coding RNA DANCR Alleviates Hypoxia-Caused H9c2 Cells Damage through up Regulation of HIF-1α. Artif. Cell Nanomedicine, Biotechnol. 48, 533–541. 10.1080/21691401.2020.1725026 [DOI] [PubMed] [Google Scholar]
- Rahm A. K., Wieder T., Gramlich D., Müller M. E., Wunsch M. N., El Tahry F. A., et al. (2021). Differential Regulation of K Ca 2.1 ( KCNN1 ) K + Channel Expression by Histone Deacetylases in Atrial Fibrillation with Concomitant Heart Failure. Physiol. Rep. 9, e14835. 10.14814/phy2.14835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q., Zhao S., Ren C., Ma Z. (2018). RETRACTED: Astragalus Polysaccharide Alleviates LPS-Induced Inflammation Injury by Regulating miR-127 in H9c2 Cardiomyoblasts. Int. J. Immunopathol Pharmacol. 31, 205873841875918. 10.1177/2058738418759180 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeidi L., Ghaedi H., Sadatamini M., Vahabpour R., Rahimipour A., Shanaki M., et al. (2018). Long Non-coding RNA LY86-AS1 and HCG27_201 Expression in Type 2 Diabetes Mellitus. Mol. Biol. Rep. 45, 2601–2608. 10.1007/s11033-018-4429-8 [DOI] [PubMed] [Google Scholar]
- Saleh M., Ambrose J. A. (2018). Understanding Myocardial Infarction. F1000Res 7, 1378. 10.12688/f1000research.15096.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Iovine C., Agarwal A., Henkel R. (2021). TUNEL Assay-Standardized Method for Testing Sperm DNA Fragmentation. Andrologia 53, e13738. 10.1111/and.13738 [DOI] [PubMed] [Google Scholar]
- Shi W., Sun Y., Wang J., Tang Y., Zhou S., Xu Z., et al. (2021). Trem1 Mediates Neuronal Apoptosis via Interaction with SYK after Spinal Cord Ischemia-Reperfusion Injury. Am. J. Transl Res. 13, 6117–6125. [PMC free article] [PubMed] [Google Scholar]
- Shindo T., Ito K., Ogata T., Hatanaka K., Kurosawa R., Eguchi K., et al. (2016). Low-Intensity Pulsed Ultrasound Enhances Angiogenesis and Ameliorates Left Ventricular Dysfunction in a Mouse Model of Acute Myocardial Infarction. Arterioscler Thromb. Vasc. Biol. 36, 1220–1229. 10.1161/atvbaha.115.306477 [DOI] [PubMed] [Google Scholar]
- Shiojima I., Sato K., Izumiya Y., Schiekofer S., Ito M., Liao R., et al. (2005). Disruption of Coordinated Cardiac Hypertrophy and Angiogenesis Contributes to the Transition to Heart Failure. J. Clin. Invest. 115, 2108–2118. 10.1172/jci24682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y., Liu J., Shen H., Zhang C., Wu Y., Huang Y., et al. (2019). Fisetin Decreases TET1 Activity and CCNY/CDK16 Promoter 5hmC Levels to Inhibit the Proliferation and Invasion of Renal Cancer Stem Cell. J. Cel Mol Med 23, 1095–1105. 10.1111/jcmm.14010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Feng X., Zhang H., Luo Y., Huang J., Lin M., et al. (2019). METTL3 and ALKBH5 Oppositely Regulate m6A Modification of TFEB mRNA, Which Dictates the Fate of Hypoxia/reoxygenation-Treated Cardiomyocytes. Autophagy 15, 1419–1437. 10.1080/15548627.2019.1586246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Chen P., Chen X., Yang W., Chen X., Zhou W., et al. (2021). KIF4A Enhanced Cell Proliferation and Migration via Hippo Signaling and Predicted a Poor Prognosis in Esophageal Squamous Cell Carcinoma. Thorac. Cancer 12, 512–524. 10.1111/1759-7714.13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Ma C., Yu Y., Yi C. (2019). Hydroxyurea Promotes TET1 Expression and Induces Apoptosis in Osteosarcoma Cells. Biosci. Rep. 39, 1. 10.1042/bsr20190456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T., Mesirov J. P. (2013). Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Brief. Bioinform. 14, 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thygesen K., Alpert J. S., White H. D. (2007). Universal Definition of Myocardial Infarction. J. Am. Coll. Cardiol. 50, 2173–2195. 10.1016/j.jacc.2007.09.011 [DOI] [PubMed] [Google Scholar]
- Wang M., Liu J., Zhao Y., He R., Xu X., Guo X., et al. (2020a). Upregulation of METTL14 Mediates the Elevation of PERP mRNA N6 Adenosine Methylation Promoting the Growth and Metastasis of Pancreatic Cancer. Mol. Cancer 19, 130. 10.1186/s12943-020-01249-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu B., Wang Y., Bai B., Yu T., Chu X. m. (2020b). The Biomarkers of Key miRNAs and Target Genes Associated with Acute Myocardial Infarction. PeerJ 8, e9129. 10.7717/peerj.9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Xie Q., Liu X., Wan C., Wu W., Fang K., et al. (2020a). Identification the Prognostic Value of Glutathione Peroxidases Expression Levels in Acute Myeloid Leukemia. Ann. Transl Med. 8, 678. 10.21037/atm-20-3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Zhou Q., Tian H., Su Y., Fu G.-h., Sun T. (2020b). Integrin β3 Promotes Cardiomyocyte Proliferation and Attenuates Hypoxia-Induced Apoptosis via Regulating the PTEN/Akt/mTOR and ERK1/2 Pathways. Int. J. Biol. Sci. 16, 644–654. 10.7150/ijbs.39414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Chen X., Lin K., Zeng K., Liu X., Xu X., et al. (2019). lncRNA SNHG6 Regulates EZH2 Expression by Sponging miR-26a/b and miR-214 in Colorectal Cancer. J. Hematol. Oncol. 12, 3. 10.1186/s13045-018-0690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankova E., Blackaby W., Albertella M., Rak J., De Braekeleer E., Tsagkogeorga G., et al. (2021). Small-molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 593, 597–601. 10.1038/s41586-021-03536-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Lin Y., Ai M.-M., Tan G.-J., Huang J.-L., Zou Z.-R. (2021). Knockdown of Circular RNA hsa_circ_PVT1 Inhibited Laryngeal Cancer Progression via Preventing Wnt4/β-Catenin Signaling Pathway Activation. Front. Cel Dev. Biol. 9, 658115. 10.3389/fcell.2021.658115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., He Q.-Y. (2015). ChIPseeker: an R/Bioconductor Package for ChIP Peak Annotation, Comparison and Visualization. Bioinformatics 31, 2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zeng B., Lin G., Ren X., Zhang Y., Chen H. (2010). Over-expression of HO-1 on Mesenchymal Stem Cells Promotes Angiogenesis and Improves Myocardial Function in Infarcted Myocardium. J. Biomed. Sci. 17, 80. 10.1186/1423-0127-17-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B., Liu L., Wang S., Dai Z. (2017). ILK Regulates MSCs Survival and Angiogenesis Partially through AKT and mTOR Signaling Pathways. Acta Histochem. 119, 400–406. 10.1016/j.acthis.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Zhang D., Wang B., Ma M., Yu K., Zhang Q., Zhang X. (2019). lncRNA HOTAIR Protects Myocardial Infarction Rat by Sponging miR-519d-3p. J. Cardiovasc. Trans. Res. 12, 171–183. 10.1007/s12265-018-9839-4 [DOI] [PubMed] [Google Scholar]
- Zhang D. X., Ma D. Y., Yao Z. Q., Fu C. Y., Shi Y. X., Wang Q. L., et al. (2016). ERK1/2/p53 and NF-Κb Dependent-PUMA Activation Involves in Doxorubicin-Induced Cardiomyocyte Apoptosis. Eur. Rev. Med. Pharmacol. Sci. 20, 2435–2442. [PubMed] [Google Scholar]
- Zhou J., Jiang Y. Y., Chen H., Wu Y. C., Zhang L. (2020). Tanshinone I Attenuates the Malignant Biological Properties of Ovarian Cancer by Inducing Apoptosis and Autophagy via the Inactivation of PI3K/AKT/mTOR Pathway. Cell Prolif 53, e12739. 10.1111/cpr.12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Sun L., Zhao P., Liu Y., Zhang J., Zhang Y., et al. (2021). Macrophage Migration Inhibitory Factor Facilitates the Therapeutic Efficacy of Mesenchymal Stem Cells Derived Exosomes in Acute Myocardial Infarction through Upregulating miR-133a-3p. J. Nanobiotechnol 19, 61. 10.1186/s12951-021-00808-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouggari Y., Ait-Oufella H., Bonnin P., Simon T., Sage A. P., Guérin C., et al. (2013). B Lymphocytes Trigger Monocyte Mobilization and Impair Heart Function after Acute Myocardial Infarction. Nat. Med. 19, 1273–1280. 10.1038/nm.3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the GEO repository, accession number GSE189593.