Abstract
Mutations that prevent the expression of the hepatitis B e antigen frequently emerge in the immunoreactive phase of infection. The predominant mutation, the precore G→A-1896 mutation, is restricted by the variability at position 1858 and is rare in strains with cytosine at nucleotide 1858. The C-1858 variant is characteristic of genotype A. It also occurs in genotypes C and F, but not in B, D, or E, explaining the geographical variation in the prevalence of precore mutants. C-1858 strains have been frequently observed in southeast Asia, but have not been phylogenetically characterized. By sequencing eight complete hepatitis B virus genomes, C-1858 variants of east Asian origin were found to constitute a phylogenetic entity within genotype C that probably diverged several hundred years ago. Further study of the distribution of this variant is warranted.
Hepatitis B virus (HBV) is an important cause of chronic hepatitis, liver cirrhosis, and hepatocellular cancer, and more than 350 million people worldwide (mainly in the developing countries) carry the virus. HBV has been classified into six genotypes (A to F) (24, 27), which have been associated with different geographical areas: A with Europe and sub-Saharan Africa, B and C with east Asia, D with the Mediterranean and Middle East regions, E with western Africa, and F with the Americas (16, 26). Recently, an additional genotype, G, has been observed in France and the United States (30).
The precore region of HBV contains a signal sequence that directs the precore protein to the endoplasmic reticulum, resulting in the secretion of the final protein, hepatitis B e antigen (HBeAg), to the blood (29). The function of this protein is not fully understood, but it is considered to modulate the immune response of the host during the early phase of infection (23). Later, in the immunoactive or “viral clearance” phase, HBeAg is instead a major target antigen (10), and the frequent emergence in the precore region of mutations that prevent synthesis of HBeAg is considered to be due to immunological escape. These mutations, the predominant one of which creates a premature stop codon through a G→A mutation at nucleotide (nt) 1896 (7), confuse the interpretation of HBeAg serology and may possibly influence the clinical course of infection or the response to interferon (6, 20, 32).
The prevalence of precore mutations shows considerable geographical variation. It has been demonstrated that the low prevalence in areas where genotype A predominates, such as northern Europe, is due to the presence in this genotype of cytosine at nt 1858 (C-1858) (15). Because nt 1858 and 1896 form a base pair in a pregenomic RNA loop (14), the G→A mutation does not evolve in strains with C-1858 instead of thymine (T-1858) (15, 17, 21). T-1858 predominates in all non-A genotypes, but C-1858 has been found in a proportion of genotype C and genotype F strains (2, 16).
The C-1858 in genotype C occurs in two variants: one with C-1856 and CCC for proline, and another with T-1856 and TCC for serine, at precore codon 15 (1, 8, 16). It has been speculated that the TCC variant represents a mutation that influences HBeAg expression by effects on the signal sequence, but this has not been verified (4, 8). Although it is now well established that nt 1858 is critical for the emergence of HBeAg-minus precore mutations, the clinical importance of nt 1858 variants is insufficiently studied. However, published data indicate an impact on HBeAg seroconversion (21), liver damage (20), interferon response (33), and core promoter mutations (9). It is not known if any of the nt 1858 variants confers any advantage for the virus; in vitro data indicate that there is no difference in the rate of replication (15). Furthermore, it is unclear if C-1858 variants in genotype C strains are due to sporadic T→C mutations, or if they represent one or more distinct phylogenetic groups within this genotype. The latter issue was the topic of the present study.
MATERIALS AND METHODS
Serum samples.
Eight HBeAg-positive HBV carriers with C-1858 variants of genotype C, all of southeast Asian origin, were identified previously (16, 19). Serum samples from these eight carriers, designated ea1 to ea8 (GenBank accession no. AF223954-61), were analyzed in the present study. Three carriers (ea2, ea3, and ea6, all from Vietnam) had TCC for serine, and 5 carriers (ea1, ea4, and ea5 from Vietnam; ea7 from Malaysia; and ea8 from Thailand) had CCC for proline at precore codon 15 (nt 1856 to 1858). The carriers were unrelated, except for ea3 and ea6, who were siblings. One genotype C genome with T-1858 originating from Thailand (975484) was sequenced and included in the phylogenetic analysis, as were 30 sequences from databases presented in Fig. 1.
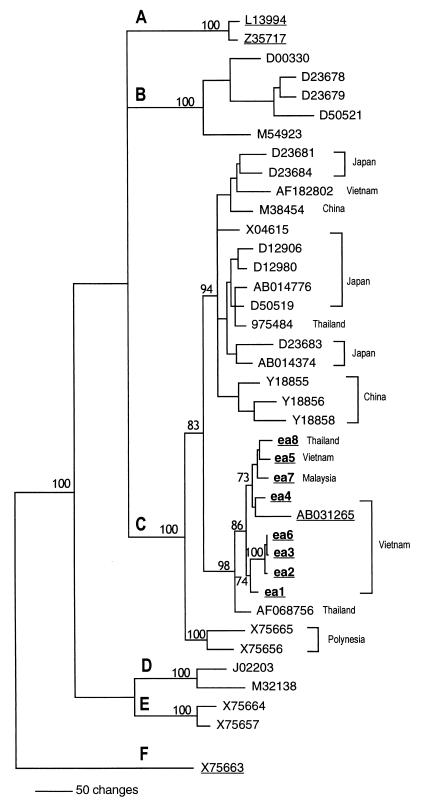
FIG. 1.
Phylogenetic tree from distance matrix and neighbor-joining analysis of the complete HBV genome using the PAUP software. The analysis included the eight C-1858 strains (ea1 to ea8) and one T-1858 strain (975484), as well as 30 sequences obtained from the GenBank/EMBL/DDBJ databases (represented by their accession numbers). Bootstrap values are shown. Sequences with C-1858 are underlined.
Sequencing and phylogenetic analysis.
Eight overlapping segments, covering the entire HBV genome, were amplified by PCR and directly sequenced with the aid of an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems) as described previously (11). The sequences were phylogenetically compared to database sequences representing all genotypes by using PAUP software, version 4.0b3a (Sinauer Associates, Inc., Sunderland, Mass.). Tree construction was done by distance matrix and neighbor-joining analyses, as well as by maximum-likelihood analysis. Recombination was investigated by using SimPlot (22) (distributed by the author at http://www.med.jhu.edu/deptmed/sray/download/).
Nucleotide sequence accession number.
The GenBank accession numbers of the sequences reported in this paper are AF223954-61 and AF330110.
RESULTS
As shown in Fig. 1, all of the analyzed sequences with C-1858 clustered on one branch of genotype C together with a previously reported strain (AB031265) that also was found to carry C-1858. The nucleotide difference between the sequences with C-1858, including AB031265, ranged between 0.02 and 1.84%. The C-1858 strains differed from genotype C strains with T-1858 by 1.68 to 5.14%. The three sequences with TCC at precore codon 15 (T-1856) formed a subcluster on the C-1858 branch, with nucleotide differences between 0.02 and 0.6%.
In the HBsAg region, all sequences ea1 to ea8 had L-110 and R-160, typical for serotype ar. One sequence (ea2) showed R-122 characteristic for serotype ayr; the remaining sequences had K-122, typical for serotype adr. In the pre-S1 region, two strains, ea3 and ea6, showed identical 18-nt deletions between nt 2847 to 2864 involving the start codon.
The previously reported sequence with C-1858 (AB031265) was from Vietnam, like six of the ea strains. The T-1858 strain (AF068756) that was most similar to the C-1858 strains (54 nt differences, 1.68%) originated from Thailand. The time that has elapsed since the divergence of the C-1858 variants can be estimated on the basis of the observed 54-nt difference between the C-1858 strains and this T-1858 strain. Assuming a rate of 5 × 10−5 mutations per site per year as described by Orito et al. (28), the C-1858 and T-1858 variants in genotype C would have diverged about 150 years ago, but since the mutation rate in the population may be even lower (11, 25), the divergence may be of older date.
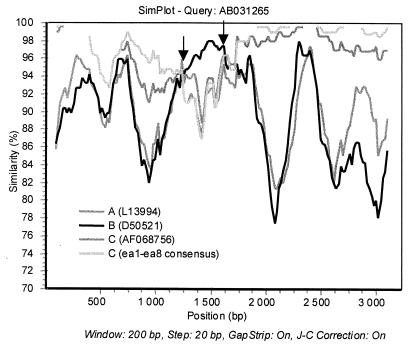
None of the sequences ea1 to ea8 showed signs of recombination. However, the sequence AB031265 carried a recombinant part, nt 1260 to 1639, originating from genotype B, as shown by analysis by SimPlot (Fig. 2). This recombination was further supported by separate phylogenetic tree construction of the segments nt 1640 to 1259 (placing AB031265 in genotype C with a 100% bootstrap value) and nt 1260 to 1639 (placing AB031265 in genotype B with a 85% bootstrap value, not shown).
FIG. 2.
SimPlot analysis comparing the C-1858 genotype C sequence AB031265 with sequences representing genotypes A, B, and C (including a consensus sequence of ea1 to ea8). The graph reveals recombination in AB031265 with a segment between nt 1260 and 1639 (indicated by arrows) originating from genotype B.
DISCUSSION
The findings of this study indicate that C-1858 strains from southeast Asia, including those with CCC at precore codon 15 (coding for proline), which is the variant also seen in genotype A, and those with TCC at codon 15 (coding for serine), have a common phylogenetic origin and represent a subgroup within genotype C. The data suggest that mutations at nt 1858 are not emerging frequently, but that there may be only one significant C-1858 branch, which diverged from the major genotype C branch at least 150 years ago. The TCC variants seem to have evolved from this CCC branch at a later stage through a C→T mutation at nt 1856.
Both CCC and TCC variants have been observed by others in HBV carriers from Hong Kong (1, 4, 8, 21). We have previously found CCC and TCC variants in 12% of southeast Asian HBV carriers (16). In the studies from Hong Kong, the CCC and TCC variants were observed at frequencies as high as 15 and 25%, respectively, of all HBV carriers (21). The high frequencies and the diverse geographic origin indicate that infection with the CCC and TCC variants may be widely spread throughout southeast Asia. Considering that the prevalence of HBV infection in this region reaches 10% or more, the number of carriers with C-1858 in genotype C may be higher than in genotype A, which is prevalent in western Europe and sub-Saharan Africa, despite the fact that C-1858 is present in almost 100% of genotype A sequences.
Recombination was not observed in any of the sequences reported here (ea1 to ea8). However, we found that a previously reported C-1858 sequence showed signs of recombination with genotype B in the segment nt 1260 to 1639. Recombination has been reported recently by others as well as by us (3, 5, 12) and may be more common than previously recognized.
Although C-1858 has a radical impact on the emergence of mutations that prevent the expression of HBeAg (i.e., the G→A-1896 mutation), it is unclear if or how this influences the course of infection. Although loss of HBeAg in general is not dependent on “HBeAg-minus” mutations, but rather reflects viral clearance, the presence of C-1858 might delay HBeAg seroconversion, and this has in fact been observed (21). A continued but undetectable expression and secretion of HBeAg in patients with C-1858 precore wild-type infection might also influence the immune pathogenesis. Moreover, Chen et al. recently described that, during HBe seroconversion, C-1858 strains more frequently developed core promoter mutations (9), which have been associated with more severe liver damage (13, 18, 31).
It is unclear if variation at nt 1858 itself has any significant effect on propagation of virus. In vitro, the replicative capacities appear to be equal in T-1858 and C-1858 strains (15). In the presence of an efficient host immune response, the potential of developing precore mutations (which may prolong viral replication) might give T-1858 strains an advantage. However, the absence of T-1858 from genotype A indicates that this conceivable advantage is of little importance for propagation and spread of the virus in the population. This may be explained as follows. (i) The emergence of precore mutations in most cases is paralleled by a radically reduced infectiousness (and thus chance of further spread). (ii) An intact precore region is required for HBeAg synthesis and induction of tolerance and hence for a long-lasting, highly infectious stage. The finding of the present study that C-1858 strains in genotype C do not represent sporadic mutations underlines the genetic stability of nt 1858 variants, supporting the idea that, despite its impact on precore mutation events in individual carriers, the variability at nt 1858 is of little importance for the evolution of HBV.
ACKNOWLEDGMENTS
Anki Gusdal is acknowledged for her skillful technical assistance.
This project was supported by grants from the Swedish Medical Association.
REFERENCES
- 1.Akarca U S, Greene S, Lok A S. Detection of precore hepatitis B virus mutants in asymptomatic HBsAg-positive family members. Hepatology. 1994;19:1366–1370. [PubMed] [Google Scholar]
- 2.Arauz-Ruiz P, Norder H, Visona K A, Magnius L O. Genotype F prevails in HBV infected patients of hispanic origin in Central America and may carry the precore stop mutant. J Med Virol. 1997;51:305–312. [PubMed] [Google Scholar]
- 3.Bollyky P L, Rambaut A, Harvey P H, Holmes E C. Recombination between sequences of hepatitis B virus from different genotypes. J Mol Evol. 1996;42:97–102. doi: 10.1007/BF02198834. [DOI] [PubMed] [Google Scholar]
- 4.Boner W, Schlicht H J, Hanrieder K, Holmes E C, Carman W F. Further characterization of 2 types of precore variant hepatitis B virus isolates from Hong Kong. J Infect Dis. 1995;171:1461–1467. doi: 10.1093/infdis/171.6.1461. [DOI] [PubMed] [Google Scholar]
- 5.Bowyer S M, Sim J G. Relationships within and between genotypes of hepatitis B virus at points across the genome: footprints of recombination in certain isolates. J Gen Virol. 2000;81:379–392. doi: 10.1099/0022-1317-81-2-379. [DOI] [PubMed] [Google Scholar]
- 6.Brunetto M R, Stemler M, Bonino F, Oliveri F, Rizetto M, Verme G, Will H. A new hepatitis B virus strain in patients with severe anti-HBe positive chronic hepatitis B. J Hepatol. 1990;10:258–261. doi: 10.1016/0168-8278(90)90062-v. [DOI] [PubMed] [Google Scholar]
- 7.Carman W, Jacyna M R, Hadziyannis S, Karayiannis P, McGarvey M J, Makris A, Thomas H C. Mutation preventing formation of HBeAg in patients with chronic hepatitis B infection. Lancet. 1989;ii:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 8.Carman W F, Ferrao M, Lok A S, Ma O C, Lai C L, Thomas H C. Precore sequence variation in Chinese isolates of hepatitis B virus. J Infect Dis. 1992;165:127–133. doi: 10.1093/infdis/165.1.127. [DOI] [PubMed] [Google Scholar]
- 9.Chan H L, Hussain M, Lok A S. Different hepatitis B virus genotypes are associated with different mutations in the core promoter and precore regions during hepatitis B e antigen seroconversion. Hepatology. 1999;29:976–984. doi: 10.1002/hep.510290352. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari C, Penna A, Bertoletti A, Fiaccadori F. Cell mediated immune response to hepatitis B virus nucleocapsid antigen. Arch Virol Suppl. 1993;8:91–101. doi: 10.1007/978-3-7091-9312-9_10. [DOI] [PubMed] [Google Scholar]
- 11.Hannoun C, Horal P, Lindh M. Long-term mutation rates in the hepatitis B virus genome. J Gen Virol. 2000;81:75–83. doi: 10.1099/0022-1317-81-1-75. [DOI] [PubMed] [Google Scholar]
- 12.Hannoun C, Norder H, Lindh M. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J Gen Virol. 2000;81:2267–2272. doi: 10.1099/0022-1317-81-9-2267. [DOI] [PubMed] [Google Scholar]
- 13.Hou J, Lau G K, Cheng J, Cheng C C, Luo K, Carman W F. T1762/A1764 variants of the basal core promoter of hepatitis B virus: serological and clinical correlations in Chinese patients. Liver. 1999;19:411–417. doi: 10.1111/j.1478-3231.1999.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 14.Knaus T, Nassal M. The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 1993;21:3967–3975. doi: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J-S, Tong S-P, Wen Y-M, Vitvitski L, Zhang Q, Trépo C. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol. 1993;67:5402–5410. doi: 10.1128/jvi.67.9.5402-5410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindh M, Andersson A S, Gusdal A. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus — large-scale analysis using a new genotyping method. J Infect Dis. 1997;175:1285–1293. doi: 10.1086/516458. [DOI] [PubMed] [Google Scholar]
- 17.Lindh M, Furuta Y, Vahlne A, Norkrans G, Horal P. Emergence of precore TAG mutation during hepatitis B e seroconversion and its dependence on pregenomic base pairing between nucleotides 1858 and 1896. J Infect Dis. 1995;172:1343–1347. doi: 10.1093/infdis/172.5.1343. [DOI] [PubMed] [Google Scholar]
- 18.Lindh M, Gustavson C, Mårdberg K, Norkrans N, Dhillon P, Horal P. Mutation of nucleotide 1762 in the core promoter region during hepatitis B e seroconversion and its relation to liver damage in hepatitis B e antigen carriers. J Med Virol. 1998;55:185–190. doi: 10.1002/(sici)1096-9071(199807)55:3<185::aid-jmv1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 19.Lindh M, Gonzalez J E, Norkrans G, Horal P. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J Virol Methods. 1998;72:163–174. doi: 10.1016/s0166-0934(98)00026-3. [DOI] [PubMed] [Google Scholar]
- 20.Lindh M, Horal P, Dhillon A P, Furuta Y, Norkrans G. Hepatitis B virus carriers without precore mutations in HBeAg-negative stage show more severe liver damage. Hepatology. 1996;24:494–501. doi: 10.1002/hep.510240305. [DOI] [PubMed] [Google Scholar]
- 21.Lok A S, Akarca U, Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pregenome encapsidation signal. Proc Natl Acad Sci USA. 1994;91:4077–4081. doi: 10.1073/pnas.91.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milich D R, Jones J E, Hughes J L, Price J, Raney A K, McLachlan A. Is a function of the secreted HBeAg to induce immunologic tolerance in utero. Proc Natl Acad Sci USA. 1990;87:6599–6603. doi: 10.1073/pnas.87.17.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norder H, Couroucé A M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 25.Norder H, Ebert J W, Fields H A, Mushahwar I K, Magnius L O. Complete sequencing of a gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 26.Norder H, Hammas B, Lee S D, Bile K, Couroué A M, Mushahwar I K, Magnius L O. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;74:1341–1348. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo R J, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 28.Orito E, Mizokami M, Ina Y, Moriyama E N, Kameshima N, Yamamoto M, Gojobori T. Host-independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci USA. 1989;86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlicht H-J, Schaller H. The secretory core protein of human hepatitis B virus is expressed on the cell surface. J Virol. 1989;63:5399–5404. doi: 10.1128/jvi.63.12.5399-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi R F, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Aoyama K, Ohno N, Iwata K, Akahane Y, Baba K, Yoshizawa H, Mishiro S. The precore/core promoter mutant (T1762A1764) of hepatitis B virus clinical significance and an easy method for detection. J Gen Virol. 1995;76:3159–3164. doi: 10.1099/0022-1317-76-12-3159. [DOI] [PubMed] [Google Scholar]
- 32.Tur-Kaspa R, Klein A, Aharanson S. HBV precore mutants are identical in carriers from various ethnic origins and associated with a range of liver disease severity. Hepatology. 1992;16:1338–1342. doi: 10.1002/hep.1840160606. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Zoulim F, Habersetzer F, Xiong S, Trépo C. Analysis of hepatitis B virus genotypes and pre-core region variability during interferon treatment of HBe antigen negative chronic hepatitis B. J Med Virol. 1996;48:8–16. doi: 10.1002/(SICI)1096-9071(199601)48:1<8::AID-JMV2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]