Abstract
Ankylosing spondylitis (AS) is an autoimmune disease with unknown aetiology. To unravel the mechanisms mediating AS pathogenesis, we profiled peripheral blood mononuclear cells (PBMCs) from AS patients and healthy subjects using 10X single‐cell RNA sequencing. The frequencies of immune cell subsets were evaluated by flow cytometry. NK cells were purified from PBMCs using isolation kit and were examined for gene expression by RT‐qPCR. Plasma levels of cytolytic molecules were examined by enzyme‐linked immunosorbent assay. Compared to healthy controls, AS patients showed a significant decrease in total NK cells as well as CD56dim NK subset, whereas CD56bright NK cells were increased. Additionally, impaired expression of cytotoxic genes in NK cells of AS patients was observed by bioinformatics algorithm and verified by RT‐qPCR and flow cytometry. Consistent with changes in transcriptomics, we found decreased plasma levels of granzymes, but not granulysin, in AS patients. Furthermore, Pearson correlation analysis revealed a negative correlation between plasma GZMB levels and disease activity (r = −0.5275, p = 0.0358). No correlation was observed between plasma cytolytic molecules and biochemical indexes (ESR and CRP). Our findings uncover altered NK cell subsets and cytotoxic profiles in peripheral circulation of AS patients at single‐cell resolution.
Keywords: ankylosing spondylitis, granzyme B, natural killer cells, peripheral blood mononuclear cells, single‐cell RNA sequencing
1. INTRODUCTION
Ankylosing spondylitis (AS) is a branch of spondyloarthritis (SpA), characterized by long‐term rheumatic inflammation of unknown origin. The current diagnostic system tends to classify SpA into two categories: peripheral SpA (mainly affecting the extremities, associated with psoriasis, inflammatory bowel disease or preceding infection) and axial SpA (mainly affecting the spine, such as AS). 1 Chronic inflammatory back pain is a characteristic complaint of most patients with AS, presenting as a dull or vague ache that often worsens in the morning or evening. 2 Over time, osteophyte formation and ligament ossification induced by prolonged inflammatory irritation contribute to irreversible structural damage. 3 In some severe cases, complete fusion of the spine causes kyphosis, limited thoracic mobility and complications such as cardiopulmonary and digestive dysfunction. For now, there is no cure for AS, but reasonable physical exercise and specific medication, such as JAK inhibitor, can effectively alleviate pain and prevent the condition from worsening. 4 , 5
Although there is no satisfactory explanation for the disease mechanism, many hypotheses have been proposed from clinical practice or animal experiments. HLA‐B27 is present in up to 70–80% of patients and its association with AS is still considered to be important. 6 Unlike other HLA‐B alleles, due to the specificity of the molecular structure, heavy chain of HLA‐B27 readily forms dimers and oligomers that not only bind to immune‐related receptors (e.g. KIR3DL2) on the cell surface but also trigger unfolded protein responses, resulting in inflammatory responses. 7 Imbalance of immune cell populations is also thought to be a hallmark of AS. 8 Previous studies have shown that the proportion of CD4 lineage T cell subsets, such as Th17 and Th22, that secrete inflammatory cytokines is elevated in the peripheral circulation of AS patients. 9 , 10 In another line of research, some scholars linked AS to the changes in cytotoxic cell profiles. A large‐scale genotyping of immune‐related loci involving 10,619 AS cases and 15,145 controls identified four CD8+ lymphocyte‐associated SNPs, including EOMES, IL7R, RUNX3 and ZMIZ1. 11 By analyzing epigenetic, RNA sequencing and protein expression data, Li et al. 12 showed that AS‐associated loci were enriched in immune cell types (e.g., monocytes and T cells) and that proteins encoded by genes downregulated in AS patients were enriched in CD8+ T cells and natural killer (NK) cells. Gracey et al. 13 found a changed cytotoxic cell profile in AS patients whose joint fluid had a significantly increased number of CD8+ T cells with activated phenotype. In addition to AS, studies of other diseases in SpA family (such as psoriatic arthritis and SpA with Crohn’s disease) consistently support a pathogenic role for CD8+ T cells, suggesting that cytotoxicity‐related factors may play a neglected role in the development of SpA. 14 , 15 Despite the updated knowledge of AS in recent years, research on the role of NK cells in the pathogenesis and pathophysiology of AS has been less satisfactory.
Understanding gene expression differences between phenotypes is crucial for transcriptomics studies. As a novel method for reliable assessment of transcript abundance, single‐cell RNA sequencing (scRNA‐seq) focuses on gene expression in individual cells, thus providing higher resolution and better understanding of the status of different cell populations. Here, we isolated peripheral blood mononuclear cells (PBMCs) from AS patients and healthy controls and examined a subset of participants (n = 3 per group) using 10X scRNA‐seq technology to explore the heterogeneity in immune cell populations and cytotoxic profiles between the two groups. Pilot data generated from scRNA‐seq were followed up with flow cytometry, RT‐qPCR and enzyme‐linked immunosorbent assay (ELISA) in our larger cohort.
2. MATERIAL AND METHODS
2.1. Study subjects
In this study, two cohorts totaling 29 AS patients and 29 healthy controls were enrolled. Detailed characteristics of the participants were summarized in Table S1. All patients with AS fulfilled the modified New York criteria, and active disease was defined by Bath Ankylosing Spondylitis Disease Activity Index (BASDAI) ≥4. 16 This study was approved by the Ethics Committees of Hangzhou Xiaoshan Hospital of Traditional Chinese Medicine. All participants were informed of the details of this experiment and signed consent forms.
2.2. Isolation of PBMCs from fresh peripheral blood
We isolated PBMCs using Lymphoprep density gradient medium (#07801, Stem Cell Technologies) according to the user guide. Then, enriched PBMCs were washed twice with PBS and stained with 0.4% Trypan blue solution (#PB180423, Procell Life Science & Technology) to assess cell viability.
2.3. cDNA library construction and 10X single‐cell sequencing
Briefly, single cells, reverse transcription reagents, Gel Beads containing barcoded oligonucleotides and oil were combined on a microfluidic chip to form Gel Bead‐in‐Emulsions. Subsequently, libraries (comprising standard Illumina paired‐end constructs, Read 1, and Read 2) were constructed using Chromium Single Cell 3′ gene expression kit (version 3.1). With the help of Gene Denovo Technology, we performed 10X single‐cell sequencing on Illumina NovaSeq 6000 at a sequencing depth of ~50,000 reads per cell.
2.4. Data processing pipelines
After sequencing, the original BCL files were converted to FASTQ files using Cell Ranger (version 3.1.0). Upon completion of gene expression quantification, we transferred the output data to Seurat R package (version 3.2.3) for subsequent analysis. 17
To further exclude unwanted cells, we set strict quality control criteria: the number of genes identified in a single cell was between 500 and 4000; the number of UMIs in a single cell <20,000 and the percentage of mitochondrial gene expression in a single cell <10%. Principal component analysis was performed to reduce the number of gene dimensions, and UMAP (uniform manifold approximation and projection) algorithm was run to visualize cells in a two‐dimensional space. Thereafter, marker genes of each cluster were identified using “FindAllMarkers” function embedded in Seurat R package. To become a cluster‐specific differentially expressed gene (DEG), there were three conditions to be met. First, |log2 fold‐change| ≥0.25. Second, the gene was expressed in more than 25% of cells in the target cluster. Third, adjusted p‐value ≤0.05. Cell‐type annotation was automatically inferred by “SingleR” package 18 and then manually checked according to known cell‐linage‐specific genes.
2.5. Function enrichment analysis of marker genes
Gene Ontology (GO) and KEGG are internationally standardized databases that identify enriched biological functions and pathways by comparison with genome‐wide background. We used clusterProfiler R package 19 in RStudio (version 1.2.1335) and ClueGO plugin 20 in Cytoscape software (version 3.8.2) for GO and KEGG analysis, respectively. Results with adjusted p‐value <0.05 were retained according to the hypergeometric test algorithm.
2.6. Flow cytometry
Peripheral blood mononuclear cells were isolated from fresh blood and resuspended at 10 × 106/ml in flow cytometry staining buffer (#420201, BioLegend). Then, cells were incubated with fluorescent conjugated antibodies, including APC‐Cy7‐CD3 (#557757, BD Pharmingen) and BV421‐CD56 (#562751, BD Pharmingen), on ice for 20 min in the dark. To detect the expression of intracellular cytotoxic molecules, cells were fixed and permeabilized using Fixation and Permeabilization Buffer Set (#88‐8824, eBioscience), and then stained with GZMB antibody (#515406, BioLegend) or IgG1 κ (#400136, BioLegend) as an isotype control. After washing twice by centrifugation at 350 g for 5 min, cells were resuspended in 0.5 ml of staining buffer and harvested using CytoFlex S (Beckman Coulter). Data analysis was performed using FlowJo software (version 10).
2.7. Isolation of NK cells and RNA extraction
Natural killer cells were harvested by using human NK cell isolation kit (#17955, Stem Cell Technologies). To assess the efficiency of NK cell sorting, enriched cells were analyzed by flow cytometry. Total RNA was extracted from NK cells using RNA‐quick purification kit (#RN001, Yi Shan Biotechnology), followed by RNA quality assay (Nanodrop, Thermo Scientific) and first‐strand cDNA synthesis (#K1622, Thermo Scientific). Amplification of cDNA was performed on ABI‐7500 system (Applied Biosystems). Primer sequences for RT‐qPCR were shown in Table S2. The results of gene expression were calculated using the method.
2.8. Detection of cytolytic molecules in plasma
Plasma levels of cytotoxic granules, including GZMA, GZMB and granulysin, were quantified using ELISA kits (#EK1162, #EK1114 and # EK1280, BOSTER Biological Technology). Each plasma sample was diluted with equal volume of dilution buffer prior to testing, and duplicate analyses were performed to ensure the reliability of the assay.
2.9. Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 7.0). Comparisons between AS patients and healthy controls were made using unpaired t‐test, and results were shown as mean ± SEM. Receiver operating characteristic (ROC) curve was adopted to estimate the predictive ability of a specific gene. Area under the ROC curve (AUC) >0.8 implies that the gene is able to distinguish between patients and healthy controls and may be a valuable diagnostic biomarker for AS. Correlations between plasma levels of cytotoxic particles and BASDAI were calculated using Pearson correlation analysis. All tests with p‐value <0.05 were considered significant.
3. RESULTS
3.1. scRNA‐seq identified major immune cell populations in peripheral blood
We harvested PBMCs from three AS patients and three healthy subjects by gradient density centrifugation and profiled them based on 10X Genomics platform (Figure 1A). A total of 53,823 cells were recovered according to the quality control criteria mentioned in “Methods” section, of which 24,810 cells were from the AS group and 29,013 cells from the control (Table S3). The number of genes and UMIs detected per cell and the percentage of mitochondrial genes were all within the normal range (Figure S1). Unbiased clustering of PBMCs yielded 20 clusters, covering 10 different cell types that were identified based on classical surface markers of immune cells (Figure 1B, C, Table 1).
FIGURE 1.
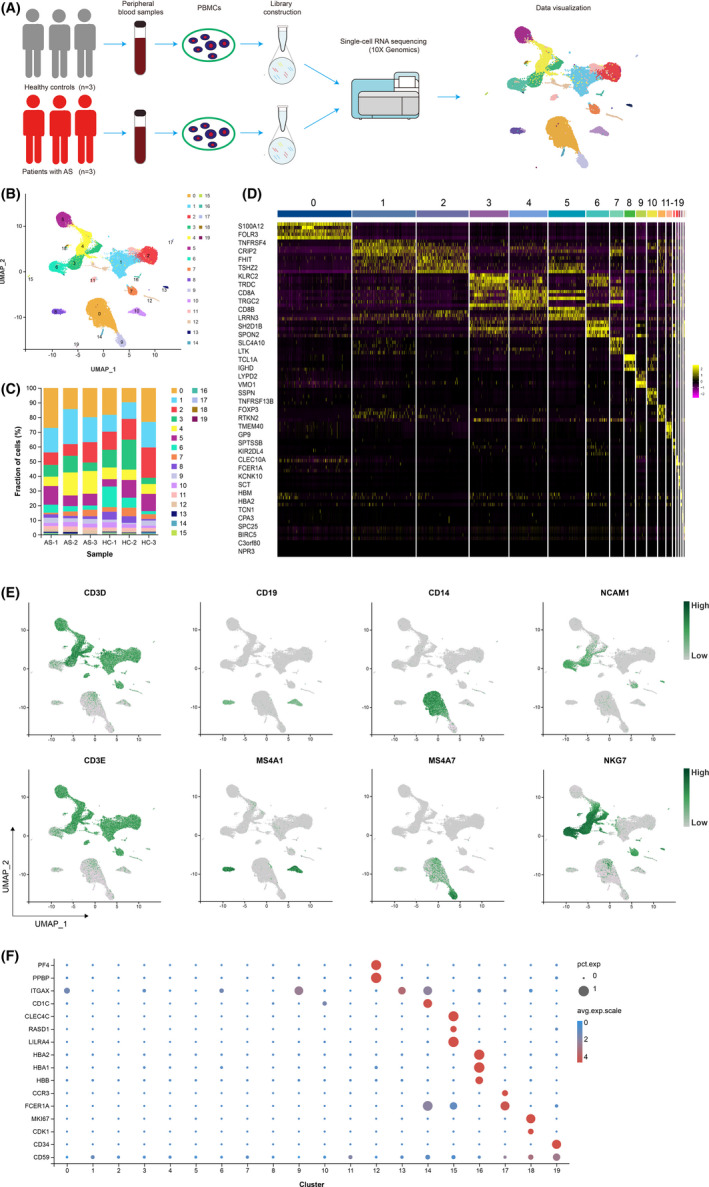
Single‐cell transcriptome profiling of PBMCs from AS patients (n = 3) and controls (n = 3). (A) Schematic diagram of the experimental workflow. (B) Two‐dimensional UMAP visualization of PBMCs resulted in 20 clusters. (C) The fractions of cell clusters in each sample. (D) Top 2 differentially expressed genes that were upregulated in each cluster were visualized in Heatmap. (E) Expression of marker genes for T cells, B cells, monocytes and NK cells (left to right). (F) Bubble plot shows the expression levels of key linage defining genes among all clusters. The size of bubble indicates the percentage of cells expressing a specific gene, and the colour of bubble indicates the average level of gene expression
TABLE 1.
Cell type and number of each cluster
| Cluster | Cell type | Classical markers | Number of cells |
|---|---|---|---|
| 0 | Classical monocytes | CD14 | 9798 |
| 1 | T cells | CD3D, CD3E | 8797 |
| 2 | T cells | CD3D, CD3E | 7216 |
| 3 | T cells | CD3D, CD3E | 5500 |
| 4 | T cells | CD3D, CD3E | 5343 |
| 5 | T cells | CD3D, CD3E | 5176 |
| 6 | Natural killer cells | NCAM1, NKG7 | 3208 |
| 7 | T cells | CD3D, CD3E | 1885 |
| 8 | B cells | CD19, MS4A1 | 1436 |
| 9 | Non‐classical monocytes | FCGR3A, MS4A7 | 1329 |
| 10 | B cells | CD19, MS4A1 | 1317 |
| 11 | T cells | CD3D, CD3E | 1022 |
| 12 | Megakaryocytes | PF4, PPBP | 806 |
| 13 | Natural killer cells | NCAM1, NKG7 | 327 |
| 14 | Conventional dendritic cells | ITGAX | 207 |
| 15 | Plasmacytoid dendritic cells | CLEC4C, RASD1, LILRA4 | 156 |
| 16 | Erythrocytes | HBA1, HBA2, HBB | 103 |
| 17 | Granulocytes | CCR3, FCER1A | 90 |
| 18 | MKI67+ proliferating cells | MKI67, CDK1 | 88 |
| 19 | Haemopoietic stem cells | CD34, CD59 | 19 |
Specifically, T cells comprised seven clusters highly expressing CD3D and CD3E; B cells were characterized by CD19 and MS4A1 expression; classic and non‐classical monocytes were marked by the presence of CD14 and MS4A7, respectively; NK cells showed expression of NCAM1 (CD56) and lack of CD3 molecules (Figure 1D, E). Specific genes in cluster 12 include platelet‐associated factor PF4 and PPBP, both proteins belonging to the CXC family and released from the alpha granules of activated platelets, indicating that this cluster consists of megakaryocytes (Figure 1F). Commonly, dendritic cells are classified as “conventional dendritic cells (cDCs)” vs. “plasmacytoid dendritic cells (pDCs)”. 21 Here, we used CD11C (ITGAX) to distinguish cDCs (cluster 14) and selected three markers (CLEC4C, RASD1 and LILRA4) to define pDCs (cluster 15) (Figure 1F). In addition, four small clusters were annotated as erythrocytes (cluster 16), granulocytes (cluster 17), MKI67+ proliferating cells (cluster 18) and haematopoietic stem cells (cluster 19) according to their respective markers (Figure 1F, Table 1). Apart from the widely known identity markers mentioned above, we obtained substantial novel cluster‐specific genes that will be informative for future single‐cell studies (Table S4).
3.2. NK cells were depleted in AS patients
To understand which cell population distinguish AS patients from healthy controls, we first attempted to compare the percentage of each cell type between the two groups. Due to the limited number of samples examined by scRNA‐seq, we failed to identify the cell types that were uniquely enriched in AS (Table S5). Previous evidence, however, suggests that patients with AS do have multiple immune perturbations or dysregulation, such as marked expansion of certain T cell subset 22 and monocytes 23 in the peripheral circulation.
In the current study, we mainly focused on NK cells and probed their alterations in AS condition, considering that the role of NK cells in the pathogenesis of AS is poorly understood but well studied in some other autoimmune diseases, such as systemic lupus erythematosus (SLE). 24 For this purpose, we carried out flow cytometry and observed a significant reduction of CD3−CD56+ NK cells in PBMCs, with the mean percentage of NK cells in patients (4.96%) being much lower than in controls (10.53%) (p < 0.001) (Figure 2).
FIGURE 2.
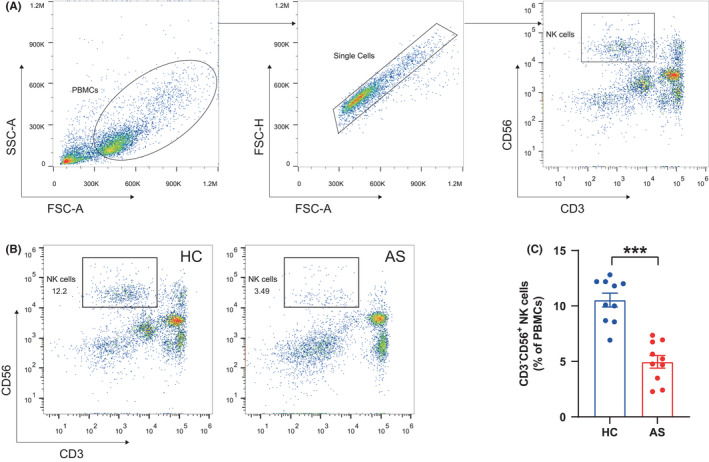
Reduction of total NK cells in peripheral blood of AS patients. (A) Gating strategy of CD3−CD56+ NK cells. (B) Representative flow cytometry plots showing CD3−CD56+ NK cells. (C) Proportions of CD3−CD56+ NK cells in PBMCs of healthy controls (HCs, n = 10) and AS patients (n = 10). Horizontal lines and error bars show the mean ± SEM. ***p < 0.001
3.3. CD56dim NK cell subset was reduced in AS patients
Natural killer cells, also known as large granular lymphocytes, are cytotoxic lymphocytes that have the ability to recognize and kill harmful cells without the involvement of MHC molecules and antibodies and are therefore critical to the innate immune system. In the initial clustering atlas, clusters 6 and 13 were authenticated as NK cells (3535 in total), of which 1182 cells were from AS patients and 2353 cells were from controls (Figure 1B and Table S5). To analyse the NK cell population at a finer scale, we extracted all cells from clusters 6 and 13 and performed a secondary clustering analysis using the Seurat R package. As a result, two subsets of NK cells (named NK0 and NK1) were generated, exhibiting heterogeneous transcriptome properties with distinctive marker genes (Figure 3A, B and Table S6). In terms of the NK0 subset, the preferential expression of cytotoxic genes (such as GZMB and NKG7) as well as FCGR3A (an important receptor for initiating antibody‐dependent cellular cytotoxicity) implied a strong killing capacity, much like that previously reported for CD56dim NK cells 25 (Figure 3C, D). NK1 subset was not dominant in cell numbers; however, this subset was enriched in genes such as GPR183, IL7R, SELL and TCF7, which are important for lymphocyte activation, migration and functional regulation, indicating its identity as CD56bright NK cells 26 (Figure 3C, E).
FIGURE 3.
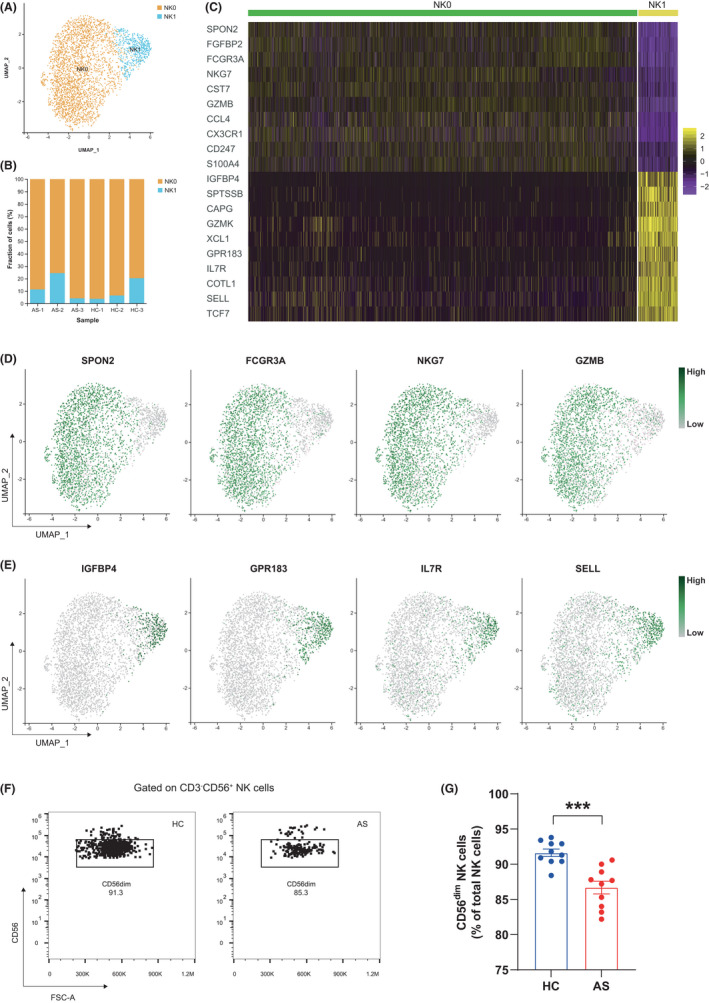
CD56dim NK cell subset was diminished in AS patients. (A) Two‐dimensional UMAP visualization of NK cells resulted in two subsets. (B) The fractions of NK cell subsets in each sample. (C) Top 10 differentially expressed genes that were upregulated in each NK cell subset were visualized in Heatmap. (D) UMAP plots of the specific marker genes in NK0 subset (CD56dim). (E) UMAP plots of the specific marker genes in NK1 subset (CD56bright). (F) Representative flow cytometry plots showing CD56dim NK cells. (G) Proportions of CD56dim NK cells in total NK cells of HCs (n = 10) and AS patients (n = 10). Horizontal lines and error bars show the mean ± SEM. ***p < 0.001
Reviewing previous studies, we discovered that patients with certain immune diseases, such as SLE and multiple sclerosis, have dysfunctional NK cells. 27 To see whether this condition exists in AS, we then compared the differences in NK cell composition between patients with AS and healthy controls. From Table S7 and Figure 3B, it seems that the NK compartment within AS patients was skewed towards CD56bright (NK1) phenotype (except for the AS‐3 case). To follow‐up on this hypothesis, we quantified the circulating NK subsets in our larger cohort. Based on the flow cytometry results, we noted that the CD56dim (NK0) subset was diminished in AS patients, with an average of 86.69% of total NK cells compared to 91.62% in healthy controls (p < 0.001) (Figure 3F, G).
3.4. Impaired expression of cytotoxic genes in NK cells of AS patients compared to healthy controls
To understand whether the reduction in NK cells was accompanied by altered transcripts, we analysed gene expression levels of NK cells between the two groups. Compared to NK cells from healthy controls, 114 genes were upregulated and 59 genes were downregulated in NK cells of AS patients (Table S8). Notably, genes associated with MHC molecules, which are known to be responsible for antigen presentation in immune responses, showed higher levels in AS patients (Figure 4A). In contrast, genes encoding cytotoxicity‐related molecules or receptors were at low levels, such as granzymes (GZMA, GZMB and GZMM) and killer cell lectin‐like receptors (KLRB1, KLRC1 and KLRC3); some transcription factors related to immunity and inflammatory regulation (e.g. CEBPB, MAF and JUNB) were also downregulated (Table S8). Among these DEGs, we mainly focused on changes in cytotoxic profiles and summarized top five cytotoxic genes in Figure 4B (ranked by adjusted p‐value). Moreover, we isolated NK cells from PBMCs of patients and healthy controls and compared the gene expression levels by RT‐qPCR. The tested up‐ and downregulated DEGs showed changes in the same direction as those observed by scRNA‐seq, with the most pronounced suppression of GZMB expression (Figure 4C and Figure S2A). ROC curves were plotted to assess the ability of these genes to differentiate between AS patients and healthy controls (Figure 4D). Consistent with the RT‐qPCR results, GZMB displayed optimal predictive power with AUC = 0.9333 (p = 0.0048). To verify the reduction of GZMB expression at the protein level, we performed flow cytometry analysis on AS patients and healthy controls (n = 10 in each group) by intracellular staining with GZMB antibody. Significantly, the percentage of GZMB+ NK cells in the total NK cell population was significantly lower in AS patients than in controls (p < 0.01) (Figure 4E, F).
FIGURE 4.
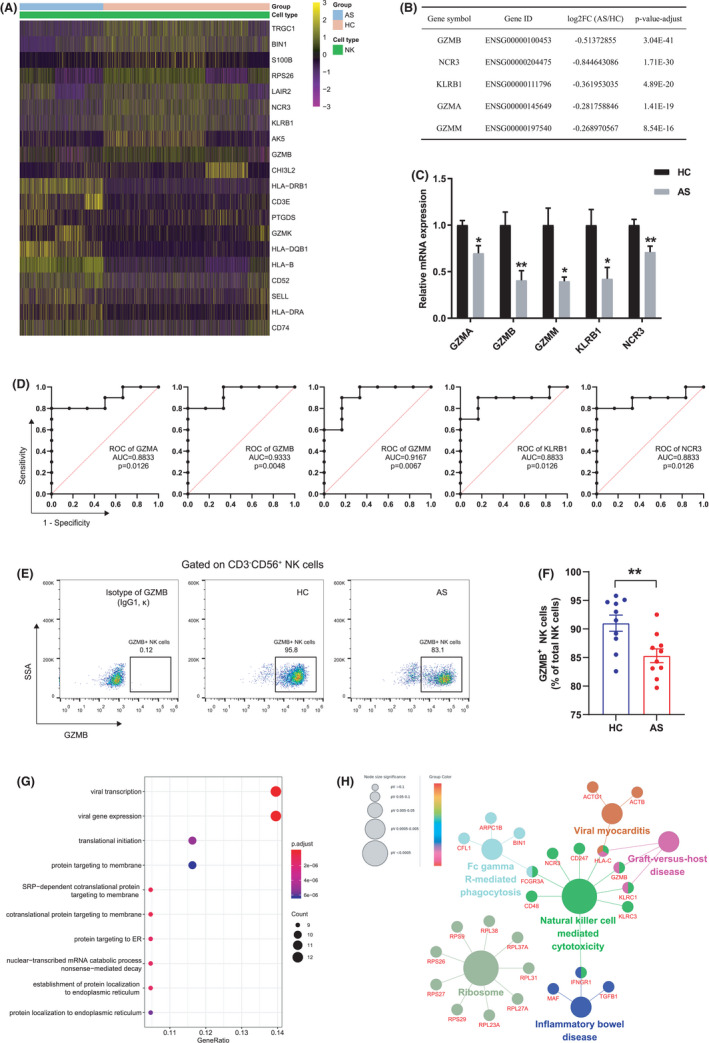
Impaired expression of cytotoxic genes in NK cells of AS patients. (A) Heatmap illustration of the representative up‐ and downregulated genes in NK cells from AS patients. (B) Top 5 downregulated cytotoxicity‐related molecules or receptors in AS patients vs. HCs. (C) RT‐qPCR analysis of gene expression fold changes in AS patients vs. HCs. (D) ROC curves were plotted to assess the ability of these five genes to differentiate between AS patients and HCs. (E) Representative flow cytometry plots showing GZMB+ NK cells. (F) Proportions of GZMB+ NK cells in total NK cells of HCs (n = 10) and AS patients (n = 10). (G) Top 10 biological processes for downregulated genes were shown in bubble plot according to gene ratio. (H) Use ClueGO plugin to analyse enriched KEGG pathways for downregulated genes. A gene involved in multiple pathways was presented with multiple colours. Horizontal lines and error bars show the mean ± SEM. *p < 0.05; **p < 0.01
We further studied the up‐ and downregulated DEGs by dropping them into GO and KEGG databases, respectively. As shown in Figure S2B, antigen processing and presentation, interferon‐γ response and T cell activation were significantly upregulated in patients with AS. By contrast, downregulated biological processes such as protein targeting to ER, protein targeting to membrane and translational initiation were identified (Figure 4G). Additionally, we evaluated affected KEGG pathways in AS patients. Consistent with GO analysis, the upregulated DEGs in NK cells from AS patients were enriched in antigen processing and presentation, T cell receptor signalling pathway and helper T cells differentiation (Figure S2C). For the downregulated DEGs, we noted that these genes were impactful in a number of pathways with immune or inflammatory context, and in particular, NK cell‐mediated cytotoxicity was hampered, which may result from impaired expression of cytotoxicity‐related genes encoding granzymes and NK cell receptors (Figure 4H).
3.5. Plasma levels of granzymes were declined in patients with AS
To learn more about secretory cytolytic products in peripheral blood, we examined the plasma levels of GZMA, GZMB and granulysin in AS patients and healthy controls using ELISA kits. Consistent with changes in NK cell transcriptomics, AS patients had significantly lower protein levels of GZMA (25.73 ± 3.5 vs. 42.62 ± 2.94 pg/ml, p < 0.001) and GZMB (25.05 ± 1.73 vs. 36.28 ± 3.08 pg/ml, p < 0.01), but not granulysin (1003 ± 125.1 vs. 1055 ± 108.4 pg/ml, p = 0.7559), compared to healthy controls (Figure 5A–C). For AS patients, we further analysed the association between plasma cytolytic molecule levels and disease activity by means of Pearson’s algorithm. Plasma levels of GZMA and GZMB in AS patients were negatively correlated with BASDAI; however, only GZMB showed statistical significance (r = −0.5275, p = 0.0358) (Figure 5D, E). We did not observe either an association between plasma granulysin levels and BASDAI (Figure 5F) or correlations between the three cytolytic products and biochemical indexes (ESR and CRP) (data not shown).
FIGURE 5.
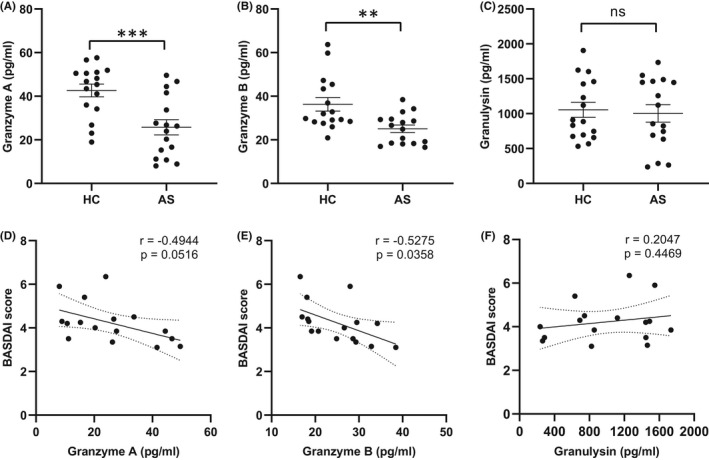
Expression of cytolytic molecules in plasma of HCs (n = 16) and AS patients (n = 16). The plasma levels of granzyme A (A), granzyme B (B) and granulysin (C) were determined by enzyme‐linked immunosorbent assay. Pearson correlation analysis was performed between granzyme A (D), granzyme B (E), granulysin (F) and disease activity (BASDAI). Horizontal lines and error bars show the mean ± SEM. **p < 0.01; ***p < 0.001; ns = not significant
4. DISCUSSION
In this study, we isolated PBMCs from peripheral blood and profiled them on the 10X Genomics platform. The primary clustering produced 20 clusters covering 10 cell types. Subsequently, we extracted NK cells from transcriptome data and performed secondary clustering to investigate the characteristics of NK subsets. Deep sequencing of RNA at single‐cell resolution uncovered remarkable heterogeneity in NK cell subsets and cytotoxic profiles between AS patients and healthy controls.
Natural killer cells can be involved in the entire process of initiation, progression and remission of inflammatory response, hence its role in autoimmune diseases is of great interest. Szántó et al. 28 demonstrated an increase in NK cells in the peripheral circulation of AS patients; however, it was not replicated in our and other studies. 24 , 29 Our flow cytometry results supported a reduced frequency of total NK cells in patients with AS, accounting for 4.96% of PBMCs compared to 10.53% in healthy controls. This alteration was mainly attributed to the NK0 subset (CD56dim), as the percentage of NK1 subset (CD56bright) was actually elevated. Of note, inflammation in AS tends to be widespread and multiple, so the frequencies of NK cell subsets may vary among different specimens. Ciccia et al. 30 analyzed NK cell subsets on ileal samples from 15 AS patients and 15 controls by flow cytometry and revealed an enrichment of NKp44+ but not NKp46+ NK cells in patients’ gut, which resulted in an overproduction of IL‐22. In other autoimmune diseases, Liu et al. 31 revealed that CD56dim NK subset in SLE patients showed a trend towards decreased proportion of the total NK cells and correlated with disease activity. Complementarily, the fraction of CD56bright NK subset in peripheral blood of patients with active SLE was increased, accompanied by an accumulation of serum type I interferon levels. 32 To delve into the properties of NK cells, Cosan et al. 33 classified them into different types depending on cytokine secretion and found an increase in IFN‐γ+ NK cells in patients with Behcet's disease compared to controls, offset by a decrease in IL‐5+CD16+, IL‐17+CD16+ and IL‐10‐secreting regulatory NK subsets.
Previous microarray studies 34 , 35 demonstrated transcriptional heterogeneity between AS patients and healthy controls and captured a considerable number of DEGs, suggesting a multidimensional pathogenesis of this disease. However, these findings were based on whole blood profiling and failed to describe heterogeneity among distinct immune cell subset at a more precise level. Our scRNA‐seq provided novel insights by uncovering NK cell depletion in patients with AS, and then we asked whether this phenomenon was accompanied by altered transcripts. Compared with NK cells from healthy controls, we noted that the expression of cytotoxicity‐related molecules or receptors was hampered, including granzymes (GZMA, GZMB and GZMM) and NK cell receptors (such as NCR3 and KLRB1). Granzymes are a family of serine proteases containing five members in human, which was first proposed by Masson et al. in 1986. 36 As the most widely studied member, GZMB is understood as an important mediator of tissue healing, chronic inflammation and immune response. 37 An earlier study indicated that cytotoxic activity of NK cells and secretion of GZMB were reduced in systemic sclerosis patients, while proinflammatory cytokine secretion from NK cells was enhanced. 38 Likewise, the insufficient number of NK precursors and downregulation of genes encoding perforin and granzymes resulted in lower cytotoxicity and lymphokine‐activated killer activity of NK cells from SLE patients. 24 Additionally, the cytotoxicity of NK cells is regulated by a combination of signals from inhibitory and activating receptors. Jiao et al. 39 investigated the polymorphisms in genes encoding NK cell receptors and found that KIR2DL1 and KIR2DL5 were more common in AS patients than in controls, which may impede the ability of NK cells to recognize and lyse target cells in immune response, thus contributing to the development of AS. KLRB1, perhaps better known by its alias CD161, is a member of the killer lectin‐like receptor family and widely expressed on the surface of NK and certain T cells. Data from a flow cytometry study highlighted that SLE patients had decreased levels of CD161 expression in both NK and T cells, 40 and similar results were replicated by later investigators. 41
The reduced expression of cytotoxicity‐related genes and skewing of the NK cell compartment towards CD56bright phenotype (NK1) prompted us to question whether AS patients also exhibit altered circulating cytotoxicity. In our study, we found decreased plasma levels of granzymes (GZMA and GZMB) in AS patients using ELISA assay (no gross differences in granulysin). In agreement with our findings, Gracey et al. 13 compared the expression of granzymes and perforin‐1 in serum, synovial fluid and mononuclear cells from healthy controls and patients with AS, osteoarthritis and rheumatoid arthritis and concluded that AS patients showed reduced expression of cytotoxic genes. Intriguingly, in their research, the lower levels of serum cytotoxic molecules were limited to perforin‐1 but not granzymes. When considering the inconsistency of altered cytotoxic profiles, there are two potential reasons: differences in ethnicity and geographic region and distinct assay methods (cytokine bead array vs. ELISA).
Our study has some limitations. We mainly focused on NK cells and cytotoxic profiles without detailing the transcriptome features of other members (e.g. T cells and monocytes) as well as changes in inflammatory cytokines. We are also aware of the limitations in sample source. Due to the scarcity of human skeletal and ligamentous specimens, we relied on peripheral blood throughout the study.
In conclusion, we demonstrate the differences in NK cell subsets and cytotoxic profiles between AS patients and healthy controls by 10X scRNA‐seq technology and experimental validation. Depletion of NK cells and impaired expression of cytotoxic molecules distinguish AS patients from healthy controls and are associated with disease activity (BASDAI), which may offer new insights for disease diagnosis and therapeutic intervention.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Conglin Ren: Conceptualization (equal); methodology (equal); writing – original draft (equal). Mingshuang Li: Methodology (equal); writing – original draft (equal). Yang Zheng: Data curation (equal); software (equal). Bingbing Cai: Methodology (equal). Weibin Du: Funding acquisition (equal). Helou Zhang: Methodology (equal). Fengqing Wu: Validation (equal). Mengsha Tong: Validation (equal). Fu Lin: Validation (equal). Jinfu Wang: Conceptualization (equal); writing – review and editing (equal). Renfu Quan: Conceptualization (equal); supervision (equal); writing – review and editing (equal).
Supporting information
Fig S1‐S2
Table S1‐S8
ACKNOWLEDGMENTS
We thank Academy of Chinese Medical Sciences, Zhejiang Chinese Medical University for the professional technical support to this work. This study was financially supported by the National Natural Science Foundation of China (No. 81904053).
Ren C, Li M, Zheng Y, et al. Single‐cell RNA‐seq reveals altered NK cell subsets and reduced levels of cytotoxic molecules in patients with ankylosing spondylitis. J Cell Mol Med. 2022;26:1071–1082. doi: 10.1111/jcmm.17159
Conglin Ren and Mingshuang Li contributed equally to this work.
Contributor Information
Jinfu Wang, Email: quanrenfu@st.btbu.edu.cn.
Renfu Quan, Email: wjfu@zju.edu.cn.
DATA AVAILABILITY STATEMENT
The raw data of single‐cell RNA sequencing in this study have been deposited with the Sequence Read Archive (SRA) under accession number PRJNA749866.
REFERENCES
- 1. Rudwaleit M, van der Heijde D, Landewé R, et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann Rheum Dis. 2009;68:777‐783. doi: 10.1136/ard.2009.108233 [DOI] [PubMed] [Google Scholar]
- 2. van den Berg R, de Hooge M, Rudwaleit M, et al. ASAS modification of the Berlin algorithm for diagnosing axial spondyloarthritis: results from the SPondyloArthritis Caught Early (SPACE)‐cohort and from the Assessment of SpondyloArthritis international Society (ASAS)‐cohort. Ann Rheum Dis. 2013;72:1646‐1653. doi: 10.1136/annrheumdis-2012-201884 [DOI] [PubMed] [Google Scholar]
- 3. Appel H, Maier R, Loddenkemper C, et al. Immunohistochemical analysis of osteoblasts in zygapophyseal joints of patients with ankylosing spondylitis reveal repair mechanisms similar to osteoarthritis. J Rheumatol. 2010;37:823‐828. doi: 10.3899/jrheum.090986 [DOI] [PubMed] [Google Scholar]
- 4. Tyrrell JS, Redshaw CH. Physical activity in ankylosing spondylitis: evaluation and analysis of an eHealth tool. J Innov Health Inform. 2016;23:169. doi: 10.14236/jhi.v23i2.169 [DOI] [PubMed] [Google Scholar]
- 5. van der Heijde D, Song IH, Pangan AL, et al. Efficacy and safety of upadacitinib in patients with active ankylosing spondylitis (SELECT‐AXIS 1): a multicentre, randomised, double‐blind, placebo‐controlled, phase 2/3 trial. Lancet. 2019;394:2108‐2117. doi: 10.1016/s0140-6736(19)32534-6 [DOI] [PubMed] [Google Scholar]
- 6. Poddubnyy D, Sieper J. Similarities and differences between nonradiographic and radiographic axial spondyloarthritis: a clinical, epidemiological and therapeutic assessment. Curr Opin Rheumatol. 2014;26:377‐383. doi: 10.1097/bor.0000000000000071 [DOI] [PubMed] [Google Scholar]
- 7. DeLay ML, Turner MJ, Klenk EI, Smith JA, Sowders DP, Colbert RA. HLA‐B27 misfolding and the unfolded protein response augment interleukin‐23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009;60:2633‐2643. doi: 10.1002/art.24763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu D, Liu B, Lin C, Gu J. Imbalance of peripheral lymphocyte subsets in patients with ankylosing spondylitis: a meta‐analysis. Front Immunol. 2021;12:696973. doi: 10.3389/fimmu.2021.696973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith JA, Colbert RA. Review: The interleukin‐23/interleukin‐17 axis in spondyloarthritis pathogenesis: Th17 and beyond. Arthritis Rheumatol. 2014;66:231‐241. doi: 10.1002/art.38291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L, Li YG, Li YH, et al. Increased frequencies of Th22 cells as well as Th17 cells in the peripheral blood of patients with ankylosing spondylitis and rheumatoid arthritis. PLoS One. 2012;7:e31000. doi: 10.1371/journal.pone.0031000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cortes A, Hadler J, Pointon JP, et al. Identification of multiple risk variants for ankylosing spondylitis through high‐density genotyping of immune‐related loci. Nat Genet. 2013;45:730‐738. doi: 10.1038/ng.2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Z, Haynes K, Pennisi DJ, et al. Epigenetic and gene expression analysis of ankylosing spondylitis‐associated loci implicate immune cells and the gut in the disease pathogenesis. Genes Immun. 2017;18:135‐143. doi: 10.1038/gene.2017.11 [DOI] [PubMed] [Google Scholar]
- 13. Gracey E, Yao Y, Qaiyum Z, Lim M, Tang M, Inman RD. Altered cytotoxicity profile of CD8+ T cells in ankylosing spondylitis. Arthritis Rheumatol. 2020;72:428‐434. doi: 10.1002/art.41129 [DOI] [PubMed] [Google Scholar]
- 14. Penkava F, Velasco‐Herrera MDC, Young MD, et al. Single‐cell sequencing reveals clonal expansions of pro‐inflammatory synovial CD8 T cells expressing tissue‐homing receptors in psoriatic arthritis. Nat Commun. 2020;11:4767. doi: 10.1038/s41467-020-18513-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lefferts AR, Regner EH, Stahly A, et al. Circulating mature granzyme B+ T cells distinguish Crohn's disease‐associated axial spondyloarthritis from axial spondyloarthritis and Crohn's disease. Arthritis Res Ther. 2021;23:147. doi: 10.1186/s13075-021-02531-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garrett S, Jenkinson T, Kennedy LG, Whitelock H, Gaisford P, Calin A. A new approach to defining disease status in ankylosing spondylitis: the Bath Ankylosing Spondylitis Disease Activity Index. J Rheumatol. 1994;21:2286‐2291. [PubMed] [Google Scholar]
- 17. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single‐cell data. Cell. 2019;177:1888‐1902.e1821. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aran D, Looney AP, Liu L, et al. Reference‐based analysis of lung single‐cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163‐172. doi: 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284‐287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug‐in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091‐1093. doi: 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ziegler‐Heitbrock L, Ancuta P, Crowe S, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74‐80. doi: 10.1182/blood-2010-02-258558 [DOI] [PubMed] [Google Scholar]
- 22. Liao HT, Lin YF, Tsai CY, Chou CT. Regulatory T cells in ankylosing spondylitis and the response after adalimumab treatment. Joint Bone Spine. 2015;82:423‐427. doi: 10.1016/j.jbspin.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 23. Surdacki A, Sulicka J, Korkosz M, et al. Blood monocyte heterogeneity and markers of endothelial activation in ankylosing spondylitis. J Rheumatol. 2014;41:481‐489. doi: 10.3899/jrheum.130803 [DOI] [PubMed] [Google Scholar]
- 24. Park YW, Kee SJ, Cho YN, et al. Impaired differentiation and cytotoxicity of natural killer cells in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1753‐1763. doi: 10.1002/art.24556 [DOI] [PubMed] [Google Scholar]
- 25. Pfefferle A, Jacobs B, Haroun‐Izquierdo A, Kveberg L, Sohlberg E, Malmberg KJ. Deciphering natural killer cell homeostasis. Front Immunol. 2020;11:812. doi: 10.3389/fimmu.2020.00812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hashemi E, Malarkannan S. Tissue‐resident NK cells: development, maturation, and clinical relevance. Cancers. 2020;12(6):1553. doi: 10.3390/cancers12061553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fogel LA, Yokoyama WM, French AR. Natural killer cells in human autoimmune disorders. Arthritis Res Ther. 2013;15:216. doi: 10.1186/ar4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Szántó S, Aleksza M, Mihály E, et al. Intracytoplasmic cytokine expression and T cell subset distribution in the peripheral blood of patients with ankylosing spondylitis. J Rheumatol. 2008;35:2372‐2375. doi: 10.3899/jrheum.070839 [DOI] [PubMed] [Google Scholar]
- 29. Kim TJ, Lee SJ, Cho YN, et al. Immune cells and bone formation in ankylosing spondylitis. Clin Exp Rheumatol. 2012;30:469‐475. [PubMed] [Google Scholar]
- 30. Ciccia F, Accardo‐Palumbo A, Alessandro R, et al. Interleukin‐22 and interleukin‐22‐producing NKp44+ natural killer cells in subclinical gut inflammation in ankylosing spondylitis. Arthritis Rheum. 2012;64:1869‐1878. doi: 10.1002/art.34355 [DOI] [PubMed] [Google Scholar]
- 31. Liu M, Liu J, Zhang X, Xiao Y, Jiang G, Huang X. Activation status of CD56(dim) natural killer cells is associated with disease activity of patients with systemic lupus erythematosus. Clin Rheumatol. 2021;40:1103‐1112. doi: 10.1007/s10067-020-05306-x [DOI] [PubMed] [Google Scholar]
- 32. Schepis D, Gunnarsson I, Eloranta ML, et al. Increased proportion of CD56bright natural killer cells in active and inactive systemic lupus erythematosus. Immunology. 2009;126:140‐146. doi: 10.1111/j.1365-2567.2008.02887.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cosan F, Aktas Cetin E, Akdeniz N, Emrence Z, Cefle A, Deniz G. Natural killer cell subsets and their functional activity in Behçet's disease. Immunol Invest. 2017;46:419‐432. doi: 10.1080/08820139.2017.1288240 [DOI] [PubMed] [Google Scholar]
- 34. Pimentel‐Santos FM, Ligeiro D, Matos M, et al. Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue‐destructive disease aspects. Arthritis Res Ther. 2011;13:R57. doi: 10.1186/ar3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gracey E, Yao Y, Green B, et al. Sexual dimorphism in the Th17 signature of ankylosing spondylitis. Arthritis Rheumatol. 2016;68:679‐689. doi: 10.1002/art.39464 [DOI] [PubMed] [Google Scholar]
- 36. Masson D, Nabholz M, Estrade C, Tschopp J. Granules of cytolytic T‐lymphocytes contain two serine esterases. EMBO J. 1986;5:1595‐1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turner CT, Hiroyasu S, Granville DJ. Granzyme B as a therapeutic target for wound healing. Expert Opin Ther Targets. 2019;23:745‐754. doi: 10.1080/14728222.2019.1661380 [DOI] [PubMed] [Google Scholar]
- 38. Horikawa M, Hasegawa M, Komura K, et al. Abnormal natural killer cell function in systemic sclerosis: altered cytokine production and defective killing activity. J Invest Dermatol. 2005;125:731‐737. doi: 10.1111/j.0022-202X.2005.23767.x [DOI] [PubMed] [Google Scholar]
- 39. Jiao YL, Zhang BC, You L, et al. Polymorphisms of KIR gene and HLA‐C alleles: possible association with susceptibility to HLA‐B27‐positive patients with ankylosing spondylitis. J Clin Immunol. 2010;30:840‐844. doi: 10.1007/s10875-010-9444-z [DOI] [PubMed] [Google Scholar]
- 40. Lin YL, Lin SC. Analysis of the CD161‐expressing cell quantities and CD161 expression levels in peripheral blood natural killer and T cells of systemic lupus erythematosus patients. Clin Exp Med. 2017;17:101‐109. doi: 10.1007/s10238-015-0402-1 [DOI] [PubMed] [Google Scholar]
- 41. Park Y, Lim J, Kim SY, Kwon GC, Koo SH, Kim J. Changes of frequency and expression level of CD161 in CD8(+) T cells and natural killer T cells in peripheral blood of patients with systemic lupus erythematosus. Microbiol Immunol. 2020;64:532‐539. doi: 10.1111/1348-0421.12798 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2
Table S1‐S8
Data Availability Statement
The raw data of single‐cell RNA sequencing in this study have been deposited with the Sequence Read Archive (SRA) under accession number PRJNA749866.


