Abstract
Extracellular vesicles (EVs), especially exosomes, have been reported to play essential roles as extracellular messengers by transporting goods in various diseases, while their potential roles in bladder cancer (BC) still remain to be further studied. BC exhibits a high degree of chemoresistance and metastatic ability, which may be affected by cancer‐derived exosomes that carry proteins, lipids and RNA. To date, the most studied exosomal molecular cargo is long non‐coding RNA (lncRNA). Although there is increasing interest in its role and function, there is relatively little knowledge about it compared with other RNA transcripts. Nevertheless, in the past ten years, we have witnessed increasing interest in the role and function of lncRNA. For example, lncRNAs have been studied as potential biomarkers for the diagnosis of BC. They may play a role as a therapeutic target in precision medicine, but they may also be directly involved in the characteristics of tumour progression, such as metastasis, epithelial‐mesenchymal transition and drug resistance. Cancer cells are on chemotherapy acting. The function of lncRNA in various cancer exosomes has not yet been determined. In this review, we summarize the current studies about the prominent roles of exosomal lncRNAs in genome integrity, BC progression and carcinogenic features.
Keywords: biomarkers, bladder cancer, exosomes, long non‐coding RNAs, malignant progression
1. INTRODUCTION
Bladder cancer (BC) is one of the most common malignant tumours of the genitourinary system, accounting for the 9th most common malignant tumour in the world. 1 , 2 According to pathological classification, 90% of patients with BC have urothelial cancer. About one‐third of these patients are first diagnosed with muscle invasive bladder cancer (MIBC). 3 , 4 In some patients, even if the first diagnosis is non‐muscle invasive bladder cancer (NMIBC), 10%–30% of patients progress to MIBC. 4 , 5 BC has become a disease that seriously affects human health. 6 , 7 At present, its early diagnosis and treatment have made great progress, 8 , 9 but its specific mechanism of occurrence and development is still unclear.
In recent years, non‐coding RNAs (ncRNAs) have become a research hotspot. NcRNAs can be divided into housekeeping ncRNAs and regulatory ncRNAs. Among them, regulatory ncRNAs can be mainly divided into microRNA (miRNA), long noncoding RNA (lncRNA) and circular RNA (circRNA). 10 , 11 , 12 , 13 LncRNA is a general term for single‐stranded nucleotide sequences exceeding 200 bp. 14 Although it does not have the function of encoding proteins, it can participate in gene regulation at the epigenetic level, transcription level and post‐transcriptional level, 15 , 16 , 17 affect tumour occurrence, development, metastasis and malignant progression of drug resistance. 18 , 19 , 20 , 21 , 22 , 23 Based on the current research on the mechanism of lncRNA, the competitive endogenous ‘ceRNA’ mechanism is the most common type and a widely recognized regulatory mechanism, that is, some ncRNAs have binding sites with microRNAs. 24 , 25 The cell acts as a miRNA sponge, thereby releasing the inhibition of miRNA on the target gene, thereby increasing the expression level of the target gene. 26 , 27 , 28 , 29 For instances, elevated LINC00909 can promote tumour progression of ovarian cancer via regulating the miR‐23b‐3p/MRC2 axis. 30 Long noncoding RNA IL6‐AS1 is upregulated in chronic obstructive pulmonary disease and is interrelated to interleukin 6 via sponging miR‐149‐5p and regulating early B‐cell factor 1 expression 31 ; Long non‐coding RNA (lncRNA) DLEU2 can drive epithelial‐mesenchymal transition (EMT) genes and glycolysis in endometrial cancer through modulating the miR‐455/HK2 and EZH2/miR‐181a pathway. 32
Exosomes are small extracellular vesicles (EVs) (30–100 nm in diameter). 33 , 34 They are secretory vesicles containing cytoplasmic protein and RNA in a bilayer of phospholipids and exist in all eukaryotic and prokaryotic cells. 35 , 36 , 37 Through their role as transporters, exosomes form complex networks that connect tumour cells in the tumour microenvironment and play a crucial role in these networks. 38 , 39 , 40 Carrying substances through autocrine, paracrine, endocrine and other signalling pathways, exosomes transport specific proteins, DNA and RNA to recipient cells, thereby regulating the biological characteristics of the recipient cells. 41 , 42 , 43 , 44 For example, exosomes can promote tumour development, inhibit tumour cell apoptosis and immune escape, stimulate tumour angiogenesis and transfer genetic material. 45 , 46 , 47 , 48 , 49 , 50 Recently, it has been shown that the release of exosomes may act as a vital role in the chemotherapy resistance of cancer cells by mediating the transfer of drugs, nucleic acids and proteins. 51 , 52 , 53
In recent years, studies have shown that exosomal‐lncRNAs also play an indispensable role in the occurrence and progression of many cellular processes. Besides, exosomal lncRNAs can regulate the tumour microenvironment by modulating the expression of various key signalling pathways and molecular and play important regulatory roles in cancer metastasis. Moreover, due to their specificity and sensitivity, exosomal lncRNAs can also be released into tumour microenvironments and act as potential tumour markers. Ni et al. show that breast cancer‐derived exosomal lnc‐SNHG16 can enhance the activation of the TGF‐β1/SMAD5 pathway through the miR‐16‐5p/SMAD5 regulatory axis, thereby inducing the expression of CD73 in Vδ1 T cells and leading to malignant tumour progression. 54 Li et al. identify that the expression of Lnc‐FMR1‐AS1 is increased in the tissues of patients with oesophageal cancer and is related to the poor prognosis of patients. 55 Lnc‐FMR1‐AS1 can be packaged into exosomes and released into the tumour microenvironment, and maintain the dynamic interconversion state of tumour stem cells by activating TLR7‐NFκB signalling and up‐regulating c‐Myc levels in recipient cells. Guo et al. found that we can detect the occurrence of gastric cancer (GC) and predict the later progression of GC by detecting the expression of circulating exosomes lncRNA‐GC1. 56 Combining the detection of circulating exosomes lncRNA‐GC1 with endoscopy can improve the early diagnosis rate of GC. Lin et al. reveal that the AUC values of lncUEGC1 in distinguishing EGC patients from healthy individuals and patients with precancerous chronic atrophic gastritis were 0.8760 and 0.8406, respectively, which were higher than the diagnostic accuracy of carcinoembryonic antigen and were a good marker for early diagnosis of GC. 57
In this review, we focused on the latest evidence of major exosomal lncRNAs related to BC, and discussed the latent biological role of exosomal lncRNAs in the development, treatment and clinical applications of BC.
2. BIOGENESIS AND CHARACTERISTICS OF EXOSOME
2.1. Biochemical characteristics of exosomes
There are two main secretion mechanisms of exosomes: continuous secretion dependent on Golgi and induced secretion. 34 Different subtypes of exosomes may have different release mechanisms and carry different cargo components. A large number of proteins are enriched on and in the exosomal membrane, such as membrane transport and membrane fusion proteins (such as GTPases, Annexins and Flotillin), proteins required for the synthesis of multivesicles (such as tumour susceptibility gene 101), four Transmembrane proteins (such as CD9, CD63, CDS1), apoptosis‐linked gene 2 interacting protein X (ALIX), heat shock proteins (such as HSP70, HSP90). 45 , 58 , 59 , 60 Exosomes carry many nucleic acid molecules, such as miRNA, ncRNA and mRNA. 47 , 50 , 58 In addition, it also carries cytokines and growth factor proteins similar to the source cells. 60 , 61 The biological process of exosome biogenesis and release was showed in Figure 1.
FIGURE 1.
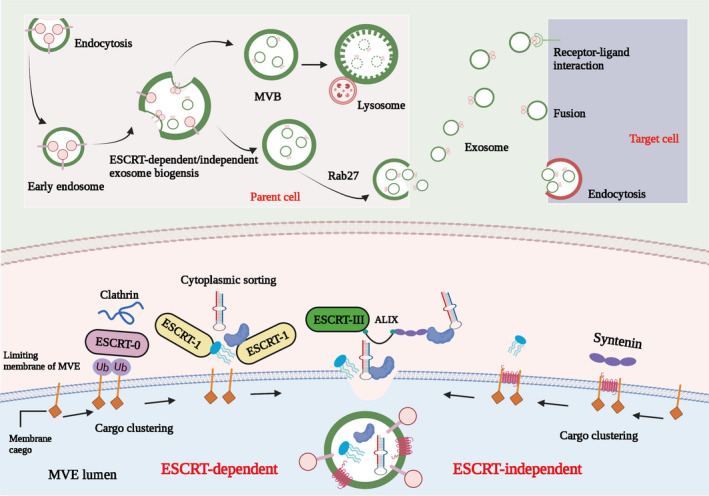
The biological process of exosome biogenesis and release. The biogenesis of exosomes begins with endosomes formed by plasma membrane endocytosis, and then early endosomes mature into multivesicular bodies (MVB). Exosomes are formed as intraluminal vesicles (ILV) in MVB through endosome‐sorting complexes required for transport (ESCRT)‐dependent or ‐independent pathways. MVB can be degraded by fusing with lysosomes or fused with the plasma membrane, leading to the secretion of ILV (exosomes). Target cells can take up exosomes through endocytosis, fusion with plasma membrane or ligand/receptor interaction. Cells can package different substances (including lncRNAs) into exosomes through ESCRT‐dependent manner and ESCRT‐independent manner
The separation methods of exosomes have not yet been unified, including sucrose gradient centrifugation, differential ultracentrifugation, filtration centrifugation, immunoaffinity capture technology, chromatography technology, microfluidic chip technology and PEG polymer precipitation. 62 , 63 , 64 The appropriate combination of these technologies may be effective. Even better, there are more commercial kits based on the above principles. The gold standard method is differential ultracentrifugation. 65 , 66 The exosomes obtained by the sucrose gradient centrifugation method have high purity, but the preliminary preparation is time‐consuming, the extraction process is very time‐consuming and the yield is low. Exosomes can be stored at 4°C for a short term (within 1–2 days) and stored at 80°C for a long term.
The identification of exosomes relies on morphological observation and protein composition analysis. 53 , 67 , 68 Observe the morphology of exosomes under an electron microscope. It can be seen that they are cup‐shaped or flat balloons. Nanoparticle tracking analysis (NTA) can also be used to measure their diameters; protein composition analysis usually uses Western blot to detect exosomes. The protein expression level of body enrichment, as usual, chooses to detect CD63 and CD81.
2.2. Exosomes and tumours
Exosomes were first discovered to participate in antigen presentation and immune activation and suppression. 69 , 70 Mast cells transport their mRNA and miRNA to recipient cells through the released exosomes, and translate proteins in the recipient cells, thus proving that exosomes have the function of transporting substances. 71 , 72 The lipid bilayer membrane of exosomes reduces the degradation of exosomes by proteases and ribonuclease, and is shed from the cell membrane through autocrine, paracrine and endocrine secretion pathways. 73 , 74 , 75 The membrane carries proteins and nucleic acid signal molecules. Body–ligand interaction, direct membrane fusion and endocytosis (or phagocytosis) are 3 ways to transfer signals from exosomes to recipient cells, and participate in intercellular communication, angiogenesis, immune response and tumour growth physiology and pathology process. 76
Exosomes participate in the composition of the tumor microenvironment and promote soluble proteins, nucleic acids, functional transmembrane proteins, chemokine receptors, epidermal growth factor receptors to mediate tumorigenesis, growth, tumor vascular growth, tumor metastasis, tumor immune escape, formation of tumor microenvironment. 77 , 78 Tumour cells release exosomes, and the signal molecular characteristics they carry can reflect the phenotype of tumour cells, such as tumour‐specific antigen proteins and RNA. They have great potential as tumour diagnostic markers. 79 , 80 At the same time, tumour cells can excrete anti‐tumour drugs by secreting exosomes, resulting in multiple tumours. Exosomes are closely related to tumours, participating in tumour formation, metastasis, drug resistance, evading immune surveillance and can also assist in diagnosis and treatment. 81 , 82
3. EXOSOMAL‐LNCRNA IN BLADDER CANCER
3.1. The potential biological role of exosomal lncRNAs in BC
Previous studies have shown that not only exosomal miRNAs serve as a vital role in the occurrence and progression of tumours, 83 , 84 , 85 , 86 but also lncRNAs in exosomes have important biological effects. We summarized the biological role of exosomal‐lncRNAs in BC (Figure 2; Table 1).
FIGURE 2.
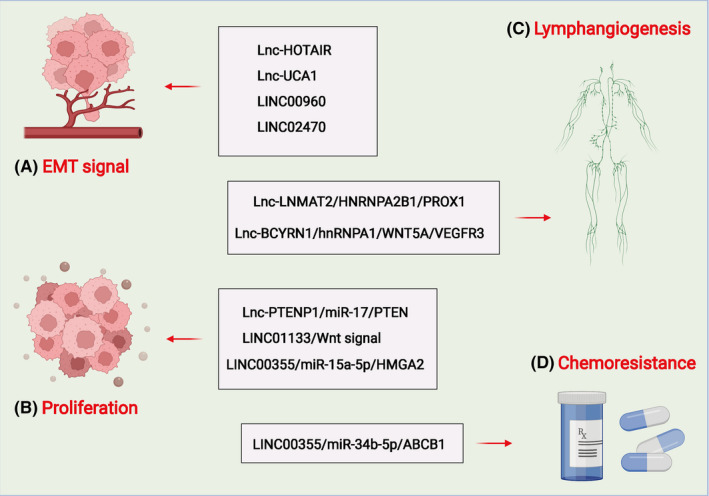
Role and functions of deregulated exosomal lncRNAs in BC progression. Exosomal lncRNAs can participate in the progression of BC by regulating the EMT (A), cell proliferation (B), lymphangiogenesis (C) and chemoresistance (D). Long noncoding RNA, lncRNA; bladder cancer, BC; HOX transcript antisense RNA, HOTAIR; epithelial‐to‐mesenchyme transition, EMT; urothelial cancer‐associated 1, UCA1; phosphatase and tensin homologue deleted on chromosome ten, PTEN; micro RNA, miRNA; lymph node metastasis‐associated transcript 2, LNMAT2; heterogeneous nuclear ribonucleoprotein A2B1, hnRNPA2B1; prospero homeobox 1, PROX1; ATP binding cassette subfamily B member 1, ABCB1; brain cytoplasmic RNA 1, BCYRN1; integration site family member 5A, Wnt5a; Vascular endothelial growth factor receptor 3, VEGF3; cancer‐associated fibroblasts, CAFs; human lymphatic endothelial cells, HLECs. Long noncoding RNA, lncRNA; bladder cancer, BC; orthodenticle homeobox 2 antisense 1, OTX2‐AS1; metastasis‐associated lung adenocarcinoma transcript 1, MALAT1; prostate cancer‐associated transcript 1, PCAT1; SPRY4 intronic transcript 1, SPRY4‐IT1; phosphatase and tensin homolog pseudogene 1, PTENP1; small nucleolar RNA host gene 16, SNHG16; H19 imprinted maternally expressed transcript, H19
TABLE 1.
Potential role and mechanism of exosomal lncRNA in BC
| LncRNAs | Parent cell/source | Target cell | Mechanism | Biological function | Reference |
|---|---|---|---|---|---|
| HOTAIR | Urine | T24 and TCC‐SUP | Regulate EMT signalling pathway | Promote cell migration and invasion | [91] |
| UCA1 | 5637 | UMUC2 | Regulate EMT signalling pathway | Promote tumor growth and progression | [92] |
| PTENP1 | 293A | J82 and EJ | PTENP1/miR‐17/PTEN | Increase cell apoptosis and reduce the ability to invade and migrate | [97] |
| LNMAT2 | UMUC3/5637 | HLEC | LNMAT2/HNRNPA2B1/PROX1 | Stimulated HLEC tube formation and migration enhanced tumour lymphangiogenesis | [102] |
|
LINC00960 LINC02470 |
T24 and J82 | TSGH‐8301 | Regulate EMT | Enhance the cell viability, migration, invasion and clonogenicity | [93] |
| LINC00355 | CAFs | T24 and 5637 | LINC00355/miR‐34b‐5p/ABCB1 | promotes BC cell resistance to cisplatin | [107] |
| LINC01133 | SV‐HUC‐1 | T24 and J82 | Regulate Wnt signalling pathway | Restrain cell viability, proliferation, migration, | [98] |
| BCYRN1 | UMUC3/5637 | HLEC | BCYRN1/hnRNPA1/WNT5A/VEGFR3 | Promote tube formation and migration of HLECs, lymphangiogenesis and LN metastasis of BCa‐HOTAIR | [103] |
| LINC00355 | CAFs | T24 and 5637 | LINC00355/miR‐15a‐5p/HMGA2 | Promote BC cell proliferation and invasion | [107] |
Abbreviations: ABCB1, ATP‐binding cassette subfamily B member 1; BC, bladder cancer; BCYRN1, brain cytoplasmic RNA 1; CAFs, cancer‐associated fibroblasts; EMT, epithelial–mesenchyme transition; HLECs, human lymphatic endothelial cells; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; HOTAIR, HOX transcript antisense RNA; lncRNA, Long noncoding RNA; LNMAT2, lymph node metastasis‐associated transcript 2; miRNA, micro RNA; PROX1, prospero homeobox 1; PTEN, phosphatase and tensin homologue deleted on chromosome ten; UCA1, urothelial cancer‐associated 1; VEGF3, Vascular endothelial growth factor receptor 3; Wnt5a, integration site family member 5A.
3.1.1. Exosomal lncRNAs and epithelial‐mesenchymal transition (EMT) in BC
Epithelial‐mesenchymal transition is intimately interrelated to the development of tissues or organs during embryogenesis. Additionally, this phenomenon is significantly associated with tumour development 87 and is a trigger for invasion, migration and acquisition of stem cell‐like phenotype in cells of diverse cancers, including BC. 88 EMT can promote the gain of epithelial stem cell properties, association with stem‐like cell markers and generation of cancer stem cells. 89 EMT is established by EMT‐inducible transcription factors, such as ZEB1, ZEB2, Snail, Slug and Twist. These transcription factors can inhibit the expression of epithelial marker E‐cadherin and increase the mesenchymal marker N‐cadherin expression to promote EMT. 90 Besides, EMT can also be regulated by several diverse upstream regulators, including signalling molecules and exosomal lncRNAs by various mechanisms.
Studies have shown that exosomal lncRNAs can regulate the EMT of BC (Figure 2A). Berondo et al. showed that HOX transcribed antisense RNA (HOTAIR) and several tumour‐related lncRNAs were rich in biological fluids, such as urine from EU patients with urothelial bladder cancer (UBC) with highly aggressive muscle diseases (HGMI pT2‐pT4) (EU). 91 Inhibition of HOTAIR in UBC cell lines could reduce cell migration and invasion. Besides, the loss of HOTAIR expression in UBC cell lines altered the expression of EMT‐related genes. They also utilized RNA sequencing to identify four 4 EU‐rich ncRNAs from UBC patients. Xue et al. proved that hypoxic BC cells could reshape the tumour microenvironment to promote tumour growth and progression, and secrete carcinogenic exosomes rich in lncRNA‐UCA1. Besides, exosomal lnc‐UCA1 might be used as a latent diagnostic biomarker for BC. 92 Huang and colleagues reported that LINC00960 and LINC02470 from high‐grade BC cell exosomes could promote the malignant behaviour of receptor low‐grade BC cells and induce EMT by up‐regulating receptor β‐catenin, Notch and Smad2/3 signalling. 93
3.1.2. Exosomal lncRNAs and cell proliferation in BC
Cell proliferation is a precision control process, which is vital for embryonic and postnatal development. 94 Under pathological conditions, abnormal cell proliferation is a central mechanism attributing to disease progressions. Abnormal cell proliferation includes both abnormal cell division and abnormal cell differentiation. 95 Besides, cell proliferation is also a main characteristic of cancer cells and the base of metastasis. 96
Studies have shown that exosomal lncRNAs can regulate the cell proliferation of BC (Figure 2B). Zheng and colleagues showed that exosomal PTENP1 was a novel biomarker that could be applied for clinical detection of BC. 97 Exosomes secreted by normal cells could transfer PTENP1 to BC cells and suppress cell growth and metastasis. The results indicated that exosomal PTENP1 might participate in the communication between normal cells and BC cells in the process of BC carcinogenesis. In addition, exosome‐mediated could transfer LINC01133 inhibits the progression of BC by regulating the Wnt signalling pathway. 98
3.1.3. Exosomal lncRNAs and lymphangiogenesis in BC
Tumour‐induced lymphangiogenesis plays an essential role in promoting tumour growth and metastasis. 99 Tumour‐associated lymphatic vessel density is closely correlated with sentinel lymph node metastasis, distant metastasis and patient survival. 100 In addition, lymph endothelial cells can interact with various immune cells to modulate immune cell activity. 101 By the above‐mentioned means, lymphatic vessels can act as vital roles in the malignant progression of tumours.
Studies have shown that exosomal lncRNAs can regulate the lymphangiogenesis of BC (Figure 2C). Chen et al. identified that lncRNA lymph node metastasis‐associated transcript 2 (LNMAT2) could stimulate the tube formation and migration of human lymphatic endothelial cell (HLEC) and strengthen lymphatic vessels of tumour generation and LN metastasis. 102 Mechanically, exosomal LNMAT2 could be secreted by BC cells and interact with the heterogeneous ribonucleoprotein A2B1 (hnRNPA2B1). Subsequently, the expression of prospero homeobox 1 (PROX1) was up‐regulated by the recruitment of hnRNPA2B1, leading to lymphangiogenesis and lymphatic metastasis. Zheng et al. discovered through which exosomal BCYRN1 synergistically enhances lymphatic metastasis induced by VEGF‐C/VEGFR3 signalling from BCa, indicating that BCYRN1 may serve as an encouraging therapeutic target for BCa patients. 103
3.1.4. Exosomal lncRNAs and chemoresistance in BC
Chemoresistance can be divided into primary drug resistance and multiple drug resistance (MDR). The former refers to cancer cells that are resistant to induced drugs, whereas the latter refers to cancer cells that develop resistance to induced drugs or other chemotherapeutic agents. 104 The establishment of chemoresistance in cancer cells involves various mechanisms, including downregulation of apoptosis, increased DNA repair, altered drug targets and overexpression of MDR proteins. 105 , 106
Studies have shown that exosomal lncRNAs can regulate the chemoresistance of BC ( Figure 2D ). Luo et al. proved that the CAF‐derived exosome LINC00355 could promote the resistance of BC cells to cisplatin by regulating the miR‐34b‐5p/ABCB1 axis. 107
3.2. Exosomal lncRNAs could act as diagnostic and prognostic Biomarkers in BC
Previous studies have shown that exosomal lncRNAs play a crucial role in the early diagnosis and prognostic evaluation of tumours. 108 , 109 , 110 , 111 We summarized the diagnostic and prognostic value of exosomal‐lncRNAs in BC (Table 2).
TABLE 2.
Potential of exosomal lncRNA as diagnostic and prognostic tool in BC
| LncRNA | Source of exosome | Exosome isolation techniques | Biomarker potential | References |
|---|---|---|---|---|
| HYMA1, LINC00477, LOC100506688 and OTX2‐AS1 | Urine | Ultracentrifugation | Biomarkers for BC prognosis | [91] |
| UCA1 | Serum | ExoQuick solution | A biomarker for BC diagnosis | [92] |
| MALAT1, PCAT1 and SPRY4‐IT1 | Urine | Ultracentrifugation | Biomarkers for BC diagnosis and prognosis | [112] |
| PTENP1 | Plasma | ExoQuick solution | A biomarker for BC diagnosis | [97] |
| PCAT‐1, UBC1 and SNHG16 | Serum | ExoQuick solution | Biomarkers for BC diagnosis and prognosis | [113] |
| H19 | Serum | ExoQuick solution | A biomarker for BC diagnosis and prognosis | [114] |
| UCA1‐201, HOTAIR, HYMA1 and MALAT1 | Urine | Ultracentrifugation | Biomarkers for BC diagnosis | [115] |
Abbreviations: BC, bladder cancer; H19, H19 imprinted maternally expressed transcript; HOTAIR, HOX transcript antisense RNA; lncRNA, Long noncoding RNA; MALAT1, metastasis associated lung adenocarcinoma transcript 1; OTX2‐AS1, orthodenticle homeobox 2 antisense 1; PCAT1, prostate cancer associated transcript 1; PTENP1, phosphatase and tensin homolog pseudogene 1; SNHG16, small nucleolar RNA host gene 16; SPRY4‐IT1, SPRY4 intronic transcript 1; UCA1, urothelial cancer associated 1.
Berondo et al. detected the expression level of lncRNAs in the original 8 patients plus two additional patient UEs (n = 10 UBC patient UEs) compared to the original 3 HV UEs and additional 4 HV UEs (n = 7 HVs) ‐seq data by qRT‐PCR. 91 The results showed that UE of patients with HGMI disease (pT2‐pT4) is rich in lncRNA HYMA1, LINC00477, LOC100506688 and OTX2‐AS1. It identified that UEs from UBC patients contain ncRNA and might eventually play a role in the prognosis of BC. Xue et al. utilized the ROC curve to analyse the diagnostic value of exosomal UCA1 in BC patients’ serum. The results showed that exosomal UCA1 could be a promising potential biomarker for BC. 92 Moreover, three differently expressed lncRNAs (MALAT1, PCAT‐1 and SPRY4‐IT1) were established to diagnose BC, and they are identified to be potential biomarkers. 112 In addition, Kaplan–Meier analysis showed that the up‐regulation of PCAT‐1 and MALAT1 was interrelated to poor recurrence‐free survival (RFS) of NMIBCs, and more the variable Cox proportional hazard regression analysis showed that the overexpression of PCAT‐1 in exosomes was an independent prognostic factor of NMIBC RFS. Zheng and colleagues found that exosomal PTENP1 could distinguish BC patients from healthy controls. 97 In addition, a set of three lncRNAs (PCAT‐1, UBC1 and SNHG16) were finally identified by a multiple logistic regression model to provide BC with high diagnostic accuracy. 113 Additionally, Kaplan–Meier analysis revealed that NMIBC patients with high UBC1 expression had a significantly lower recurrence‐free survival rate. Cox multivariate analysis showed that UBC1 was independently correlated to tumour recurrence in NMIBC. Wang et al. showed that the detection of exosomal H19 serum clarifies the use of exosomal lncRNA as a non‐invasive diagnostic and prognostic biomarker for BC patients. 114 Yu et al. selected four lncRNAs, namely UCA1‐201, HOTAIR, HYMA1 and MALAT1, to form a set of urine biomarkers of BC. 115 With the help of this panel, BC patients could be distinguished from patients with allantoicitis, with sensitivity and specificity reaching 95.7% and 94.3%, respectively. Finally, they confirmed the applicability of the four lncRNA combinations in an independent validation study involving 60 patients with BC and 60 patients with allantoicitis.
4. FUTURE EXPECTATIONS OF EXOSOME IN BC
No matter how hard the current multidisciplinary treatment is, the high recurrence rate of BC is still the biggest obstacle for treatment. 116 , 117 , 118 The important role of exosomes‐mediated signal transduction in cancer progression makes exosomes a potential new therapeutic target, which focuses on inhibiting the key components of the tumour cell communication network. Exosomes are expected to play an important role in the treatment of BC patients, help early diagnosis and monitoring and provide accurate predictive markers.
4.1. Exosomes are used to develop carriers for the transportation of anti‐cancer drugs
The lipid bilayer membrane of exosomes can protect nucleic acids and proteins in the membrane from being degraded. At the same time, there are recognition molecules on the membrane, and exosomes can become a good carrier for targeted drug delivery. It can accurately transport interfering RNA, suicide mRNA, protein, miRNA and drugs. Despite the huge therapeutic potential of exosomes, the field still needs new in vivo models and powerful imaging systems to track the pathways of the synthesis, release, transportation and function of single‐cell exosomes.
4.2. Inhibition of tumour progression and metastasis by targeting tumour‐derived exosomes
Exosomes participate in the formation of the tumour microenvironment, and the signal transduction between tumour cells can inhibit the occurrence and development of tumours. There are currently several potential strategies. By interfering with the pathway components involved in the formation of exosomes (such as ESCRT, neural Amide) or release (such as Rab27, ARF6, RhoA) to inhibit the biogenesis or release of exosomes. Remove exosomes from the circulation through extracorporeal hemofiltration. Block those exosomes involved in exosome binding or internalization. Exosomal ligands (such as four transmembrane proteins) or cell surface receptors (such as HSPG) inhibit the uptake of exosomes by recipient cells.
4.3. Future prospects of exosomal lncRNAs in BC
At present, studies have found that exosomal lncRNA has an important biological role in BC, but more research is still needed to explore the clinical translational value of exosomal lncRNAs in BC. Many studies have confirmed that exosomal lncRNAs can promote the malignant progression of tumours by promoting angiogenesis. 119 , 120 , 121 In addition, exosomal lncRNA can also mediate immunosuppressive microenvironments, 122 , 123 regulate cell radioresistance 124 and mediate metabolic reprogramming, 125 but there is no relevant research report in BC. In the future, more research should be done to explore the role of exosomal lncRNAs in the above aspects and the prospects of clinical application. The role of Exosomal lncRNAs in tumour liquid biopsy has been confirmed by research. The current research on exosomal lncRNAs in the diagnosis of BC is mostly focused on the experience of a single centre, and the diagnostic potential of exosomal lncRNAs can only be tested by further verification in a multi‐centre joint study.
5. CONCLUSION
Due to its very aggressive nature, BC has the lower survival rate of urology cancers. 126 , 127 , 128 This extremely high mortality rate is primarily the result of its early asymptomatic development, so it is diagnosed as late. 129 , 130 Therefore, there is an urgent need for new diagnostic tools and new treatment strategies. In recent years, the potential role of lncRNAs as biomarkers, therapeutic targets and therapeutics in cancer research have attracted increasing interest. However, the pathophysiological function of lncRNAs still remains unknown, and whether they are the cause or consequence of cancer remains to be determined. Additionally, the same lncRNA can play completely distinct roles in various cancer environments, which makes the characterization of lncRNA particularly difficult. Although most studies are still in the preclinical stage, the diagnostic and prognostic applications of lncRNA related to exosomes are very promising for BC treatment. New advances in lncRNAs‐related studies in specific fields, such as bioinformatics, pharmacokinetics, and improved nanotechnology to deliver lncRNAs‐containing exosomes to the tumour microenvironment, will lay the foundation for future clinical applications. Understanding the function and role of lncRNAs is essential for their effective use as biomarkers, precision medicine or therapeutic targets.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Qiang Liu: Original draft preparation, allocation, revision, supplement and edition.
ACKNOWLEDEGMENTS
We thank the generous support by Liaoning Cancer Hospital & Institute (Shenyang).
Liu Q. The emerging roles of exosomal long non‐coding RNAs in bladder cancer. J Cell Mol Med. 2022;26:966–976. doi: 10.1111/jcmm.17152
DATA AVAILABILITY STATEMENT
The data in the current study are available from the corresponding authors on reasonable request.
REFERENCES
- 1. Li R, Zhang J, Gilbert SM, Conejo‐Garcia J, Mule JJ. Using oncolytic viruses to ignite the tumour immune microenvironment in bladder cancer. Nat Rev Urol. 2021;18(9):543‐555. 10.1038/s41585-021-00483-z [DOI] [PubMed] [Google Scholar]
- 2. Witjes JA, Bruins HM, Cathomas R, et al. European association of urology guidelines on muscle‐invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur Urol. 2021;79(1):82‐104. 10.1016/j.eururo.2020.03.055 [DOI] [PubMed] [Google Scholar]
- 3. Jain P, Kathuria H, Momin M. Clinical therapies and nano drug delivery systems for urinary bladder cancer. Pharmacol Ther. 2021;226:107871. 10.1016/j.pharmthera.2021.107871 [DOI] [PubMed] [Google Scholar]
- 4. Jiang DM, Gupta S, Kitchlu A, et al. Defining cisplatin eligibility in patients with muscle‐invasive bladder cancer. Nat Rev Urol. 2021;18(2):104‐114. 10.1038/s41585-020-00404-6 [DOI] [PubMed] [Google Scholar]
- 5. van Puffelen JH, Keating ST, Oosterwijk E, et al. Trained immunity as a molecular mechanism for BCG immunotherapy in bladder cancer. Nat Rev Urol. 2020;17(9):513‐525. 10.1038/s41585-020-0346-4 [DOI] [PubMed] [Google Scholar]
- 6. Meeks JJ, Al‐Ahmadie H, Faltas BM, et al. Genomic heterogeneity in bladder cancer: challenges and possible solutions to improve outcomes. Nat Rev Urol. 2020;17(5):259‐270. 10.1038/s41585-020-0304-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schneider AK, Chevalier MF, Derre L. The multifaceted immune regulation of bladder cancer. Nat Rev Urol. 2019;16(10):613‐630. 10.1038/s41585-019-0226-y [DOI] [PubMed] [Google Scholar]
- 8. Wu J, Abraham SN. The roles of t cells in bladder pathologies. Trends Immunol. 2021;42(3):248‐260. 10.1016/j.it.2021.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heath EI, Rosenberg JE. The biology and rationale of targeting nectin‐4 in urothelial carcinoma. Nat Rev Urol. 2021;18(2):93‐103. 10.1038/s41585-020-00394-5 [DOI] [PubMed] [Google Scholar]
- 10. Kan RL, Chen J, Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2021; 10.1016/j.tig.2021.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang XW, Liu CX, Chen LL, Zhang QC. RNA structure probing uncovers RNA structure‐dependent biological functions. Nat Chem Biol. 2021;17(7):755‐766. 10.1038/s41589-021-00805-7 [DOI] [PubMed] [Google Scholar]
- 12. Mohapatra S, Pioppini C, Ozpolat B, Calin GA. Non‐coding RNAs regulation of macrophage polarization in cancer. Mol Cancer. 2021;20(1):24. 10.1186/s12943-021-01313-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang Z, Xu J, Zhang B, et al. The promising role of noncoding RNAs in cancer‐associated fibroblasts: an overview of current status and future perspectives. J Hematol Oncol. 2020;13(1):154. 10.1186/s13045-020-00988-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mirzadeh Azad F, Polignano IL, Proserpio V, Oliviero S. Long noncoding RNAs in human stemness and differentiation. Trends Cell Biol. 2021;31(7):542‐555. 10.1016/j.tcb.2021.02.002 [DOI] [PubMed] [Google Scholar]
- 15. Zhang H, Liu B, Shi X, Sun X. Long noncoding RNAs: Potential therapeutic targets in cardiocerebrovascular diseases. Pharmacol Ther. 2021;221:107744. 10.1016/j.pharmthera.2020.107744 [DOI] [PubMed] [Google Scholar]
- 16. Yang S, Lim KH, Kim SH, Joo JY. Molecular landscape of long noncoding RNAs in brain disorders. Mol Psychiatry. 2021;26(4):1060‐1074. 10.1038/s41380-020-00947-5 [DOI] [PubMed] [Google Scholar]
- 17. Guo CJ, Xu G, Chen LL. Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci. 2020;45(11):947‐960. 10.1016/j.tibs.2020.07.001 [DOI] [PubMed] [Google Scholar]
- 18. Hu Q, Egranov SD, Lin C, Yang L. Long noncoding RNA loss in immune suppression in cancer. Pharmacol Ther. 2020;213:107591. 10.1016/j.pharmthera.2020.107591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schulte C, Barwari T, Joshi A, Zeller T, Mayr M. Noncoding RNAs versus protein biomarkers in cardiovascular Disease. Trends Mol Med. 2020;26(6):583‐596. 10.1016/j.molmed.2020.02.001 [DOI] [PubMed] [Google Scholar]
- 20. Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19(1):77. 10.1186/s12943-020-01188-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng J, Meng J, Zhu L, Peng Y. Exosomal noncoding RNAs in Glioma: Biological functions and potential clinical applications. Mol Cancer. 2020;19(1):66. 10.1186/s12943-020-01189-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu K, Gao L, Ma X, et al. Long non‐coding RNAs regulate drug resistance in cancer. Mol Cancer. 2020;19(1):54. 10.1186/s12943-020-01162-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang L, Xu X, Su X. Noncoding RNAs in cancer immunity: functions, regulatory mechanisms, and clinical application. Mol Cancer. 2020;19(1):48. 10.1186/s12943-020-01154-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee H, Zhang Z, Krause HM. Long Noncoding RNAs and repetitive elements: junk or intimate evolutionary partners? Trends Genet. 2019;35(12):892‐902. 10.1016/j.tig.2019.09.006 [DOI] [PubMed] [Google Scholar]
- 25. Wei L, Wang X, Lv L, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18(1):147. 10.1186/s12943-019-1086-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramnarine VR, Kobelev M, Gibb EA, et al. The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur Urol. 2019;76(5):546‐559. 10.1016/j.eururo.2019.07.040 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Du W, Yang B. Long non‐coding RNAs as new regulators of cardiac electrophysiology and arrhythmias: Molecular mechanisms, therapeutic implications and challenges. Pharmacol Ther. 2019;203:107389. 10.1016/j.pharmthera.2019.06.011 [DOI] [PubMed] [Google Scholar]
- 28. Zhang L, Meng X, Zhu XW, et al. Long non‐coding RNAs in Oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol Cancer. 2019;18(1):102. 10.1186/s12943-019-1021-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y, Cheng Z, Pang Y, et al. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J Hematol Oncol. 2019;12(1):51. 10.1186/s13045-019-0734-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang X, Wu G, Yang F, et al. Elevated LINC00909 promotes tumor progression of ovarian cancer via regulating the miR‐23b‐3p/MRC2 axis. Oxid Med Cell Longev. 2021;2021:5574130. 10.1155/2021/5574130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yi E, Zhang J, Zheng M, et al. Long noncoding RNA IL6‐AS1 is highly expressed in chronic obstructive pulmonary disease and is associated with interleukin 6 by targeting miR‐149‐5p and early B‐cell factor 1. Clin Transl Med. 2021;11(7):e479. 10.1002/ctm2.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong P, Xiong Y, Konno Y, et al. Long non‐coding RNA DLEU2 drives EMT and glycolysis in endometrial cancer through HK2 by competitively binding with miR‐455 and by modulating the EZH2/miR‐181a pathway. J Exp Clin Cancer Res. 2021;40(1):216. 10.1186/s13046-021-02018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shirejini SZ, Inci F. The Yin and Yang of exosome isolation methods: Conventional practice, microfluidics, and commercial kits. Biotechnol Adv. 2022;54:107814. 10.1016/j.biotechadv.2021.107814 [DOI] [PubMed] [Google Scholar]
- 34. Li YJ, Wu JY, Liu J, et al. Artificial exosomes for translational nanomedicine. J Nanobiotechnology. 2021;19(1):242. 10.1186/s12951-021-00986-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yao Y, Fu C, Zhou L, Mi QS, Jiang A. DC‐derived exosomes for cancer immunotherapy. Cancers (Basel). 2021;13(15):3667. 10.3390/cancers13153667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fu P, Zhang J, Li H, Mak M, Xu W, Tao Z. Extracellular vesicles as delivery systems at nano‐/micro‐scale. Adv Drug Deliv Rev. 2021;179:113910. 10.1016/j.addr.2021.113910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng S, Li Y, Yan H, et al. Advances in microfluidic extracellular vesicle analysis for cancer diagnostics. Lab Chip. 2021;21(17):3219‐3243. 10.1039/D1LC00443C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tian Y, Fu C, Wu Y, Lu Y, Liu X, Zhang Y. Central nervous system cell‐derived exosomes in neurodegenerative diseases. Oxid Med Cell Longev. 2021;2021:9965564. 10.1155/2021/9965564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu J, Zhang Y, Liang J, Diao J, Liu P, Zhao H. Role of exosomal microRNAs and their crosstalk with oxidative stress in the pathogenesis of osteoporosis. Oxid Med Cell Longev. 2021;2021:6301433. 10.1155/2021/6301433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang XH, Guo T, Gao XY, et al. Exosome‐derived noncoding RNAs in gastric cancer: functions and clinical applications. Mol Cancer. 2021;20(1):99. 10.1186/s12943-021-01396-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng W, Ji D, Zhou Y, et al. Exosomal non‐coding RNAs in hepatobiliary cancer: A rising star. Mol Cancer Ther. 2021;20(10):1777‐1788. 10.1158/1535-7163.MCT-21-0363 [DOI] [PubMed] [Google Scholar]
- 42. Burgos‐Ravanal R, Campos A, Diaz‐Vesga MC, et al. Extracellular vesicles as mediators of cancer disease and as nanosystems in theranostic applications. Cancers (Basel). 2021;13(13):3324. 10.3390/cancers13133324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reed SL, Escayg A. Extracellular vesicles in the treatment of neurological disorders. Neurobiol Dis. 2021;157:105445. 10.1016/j.nbd.2021.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhong Y, Li X, Wang F, et al. Emerging potential of exosomes on adipogenic differentiation of mesenchymal stem cells. Front Cell Dev Biol. 2021;9:649552. 10.3389/fcell.2021.649552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen J, Zhang Q, Liu D, Exosomes LZ. Advances, development and potential therapeutic strategies in diabetic nephropathy. Metabolism. 2021;122:154834. 10.1016/j.metabol.2021.154834 [DOI] [PubMed] [Google Scholar]
- 46. Sharma S, Masud MK, Kaneti YV, et al. Extracellular vesicle nanoarchitectonics for novel drug delivery applications. Small. 2021;17(42):e2102220. 10.1002/smll.202102220 [DOI] [PubMed] [Google Scholar]
- 47. Uddin MH, Al‐Hallak MN, Philip PA, et al. Exosomal microRNA in pancreatic cancer diagnosis, prognosis, and treatment: from bench to bedside. Cancers (Basel). 2021;13(11):2777. 10.3390/cancers13112777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruan S, Greenberg Z, Pan X, Zhuang P, Erwin N, He M. Extracellular vesicles as an advanced delivery biomaterial for precision cancer immunotherapy. Adv Healthc Mater. 2021:e2100650. 10.1002/adhm.202100650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parada N, Romero‐Trujillo A, Georges N, Alcayaga‐Miranda F. Camouflage strategies for therapeutic exosomes evasion from phagocytosis. J Adv Res. 2021;31:61‐74. 10.1016/j.jare.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He X, Kuang G, Wu Y, Ou C. Emerging roles of exosomal miRNAs in diabetes mellitus. Clin Transl Med. Jun 2021;11(6):e468. 10.1002/ctm2.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grieco GE, Fignani D, Formichi C, et al. Extracellular vesicles in immune system regulation and Type 1 diabetes: Cell‐to‐cell communication mediators, disease biomarkers, and promising therapeutic tools. Front Immunol. 2021;12:682948. 10.3389/fimmu.2021.682948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khalaf K, Hana D, Chou JT, Singh C, Mackiewicz A, Kaczmarek M. Aspects of the tumor microenvironment involved in immune resistance and drug resistance. Front Immunol. 2021;12:656364. 10.3389/fimmu.2021.656364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiong H, Huang Z, Yang Z, et al. Recent progress in detection and profiling of cancer cell‐derived exosomes. Small. 2021;17(35):e2007971. 10.1002/smll.202007971 [DOI] [PubMed] [Google Scholar]
- 54. Ni C, Fang QQ, Chen WZ, et al. Breast cancer‐derived exosomes transmit lncRNA SNHG16 to induce CD73+gammadelta1 Treg cells. Signal Transduct Target Ther. 2020;5(1):41. 10.1038/s41392-020-0129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li W, Zhang L, Guo B, et al. Exosomal FMR1‐AS1 facilitates maintaining cancer stem‐like cell dynamic equilibrium via TLR7/NFkappaB/c‐Myc signaling in female esophageal carcinoma. Mol Cancer. 2019;18(1):22. 10.1186/s12943-019-0949-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guo X, Lv X, Ru Y, et al. Circulating exosomal gastric cancer‐associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: A multiphase study. JAMA Surg. 2020;155(7):572. 10.1001/jamasurg.2020.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lin LY, Yang L, Zeng Q, et al. Tumor‐originated exosomal lncUEGC1 as a circulating biomarker for early‐stage gastric cancer. Mol Cancer. 2018;17(1):84. 10.1186/s12943-018-0834-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feng J, Zhang Y, Zhu Z, Gu C, Waqas A, Chen L. Emerging exosomes and exosomal MiRNAs in spinal cord injury. Front Cell Dev Biol. 2021;9:703989. 10.3389/fcell.2021.703989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ghafouri‐Fard S, Niazi V, Hussen BM, Omrani MD, Taheri M, Basiri A. The emerging role of exosomes in the treatment of human disorders with a special focus on mesenchymal stem cells‐derived exosomes. Front Cell Dev Biol. 2021;9:653296. 10.3389/fcell.2021.653296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mittal R, Bencie N, Langlie J, Mittal J, Eshraghi AA. Exosomes as drug delivery vehicles and biomarkers for neurological and auditory systems. J Cell Physiol. 2021;236(12):8035‐8049. 10.1002/jcp.30484 [DOI] [PubMed] [Google Scholar]
- 61. Benjamin‐Davalos S, Koroleva M, Allen CL, Ernstoff MS, Shu S. Co‐isolation of cytokines and exosomes: implications for immunomodulation studies. Front Immunol. 2021;12:638111. 10.3389/fimmu.2021.638111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang S, Ji X, Jackson KK, et al. Rapid separation of blood plasma exosomes from low‐density lipoproteins via a hydrophobic interaction chromatography method on a polyester capillary‐channeled polymer fiber phase. Anal Chim Acta. 2021;1167:338578. 10.1016/j.aca.2021.338578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Boer C, Calder B, Blackhurst D, et al. Analysis of the regenerative capacity of human serum exosomes after a simple multistep separation from lipoproteins. J Tissue Eng Regen Med. 2021;15(1):63‐77. 10.1002/term.3155 [DOI] [PubMed] [Google Scholar]
- 64. An M, Wu J, Zhu J, Lubman DM. Comparison of an optimized ultracentrifugation method versus size‐exclusion chromatography for isolation of exosomes from human serum. J Proteome Res. 2018;17(10):3599‐3605. 10.1021/acs.jproteome.8b00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guan S, Yu H, Yan G, Gao M, Sun W, Zhang X. Characterization of urinary exosomes purified with size exclusion chromatography and ultracentrifugation. J Proteome Res. 2020;19(6):2217‐2225. 10.1021/acs.jproteome.9b00693 [DOI] [PubMed] [Google Scholar]
- 66. Cao F, Gao Y, Chu Q, et al. Proteomics comparison of exosomes from serum and plasma between ultracentrifugation and polymer‐based precipitation kit methods. Electrophoresis. 2019;40(23–24):3092‐3098. 10.1002/elps.201900295 [DOI] [PubMed] [Google Scholar]
- 67. Ke W, Afonin KA. Exosomes as natural delivery carriers for programmable therapeutic nucleic acid nanoparticles (NANPs). Adv Drug Deliv Rev. 2021;176:113835. 10.1016/j.addr.2021.113835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu L, Sun HT, Wang S, et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13(1):152. 10.1186/s13045-020-00987-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Whiteside TL. Exosomes and tumor‐mediated immune suppression. J Clin Invest. 2016;126(4):1216‐1223. 10.1172/JCI81136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bai K, Li X, Zhong J, et al. Placenta‐derived exosomes as a modulator in maternal immune tolerance during pregnancy. Front Immunol. 2021;12:671093. 10.3389/fimmu.2021.671093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rincon‐Riveros A, Lopez L, Villegas EV, Antonia Rodriguez J. Regulation of antitumor immune responses by exosomes derived from tumor and immune cells. Cancers (Basel). 2021;13(4):847. 10.3390/cancers13040847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xiong YY, Gong ZT, Tang RJ, Yang YJ. The pivotal roles of exosomes derived from endogenous immune cells and exogenous stem cells in myocardial repair after acute myocardial infarction. Theranostics. 2021;11(3):1046‐1058. 10.7150/thno.53326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kugeratski FG, Kalluri R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021;288(1):10‐35. 10.1111/febs.15558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Giordano C, La Camera G, Gelsomino L, et al. The biology of exosomes in breast cancer progression: Dissemination, immune evasion and metastatic colonization. Cancers (Basel). 2020;12(8):2179. 10.3390/cancers12082179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yan W, Jiang S. Immune cell‐derived exosomes in the cancer‐immunity cycle. Trends Cancer. 2020;6(6):506‐517. 10.1016/j.trecan.2020.02.013 [DOI] [PubMed] [Google Scholar]
- 76. Li Q, Wang H, Peng H, Huyan T, Cacalano NA. Exosomes: Versatile nano mediators of immune regulation. Cancers. 2019;11(10):1557‐ 10.3390/cancers11101557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jena BC, Mandal M. The emerging roles of exosomes in anti‐cancer drug resistance and tumor progression: An insight towards tumor‐microenvironment interaction. Biochim Biophys Acta Rev Cancer. 2021;1875(1):188488. 10.1016/j.bbcan.2020.188488 [DOI] [PubMed] [Google Scholar]
- 78. Vafaei S, Roudi R, Madjd Z, Aref AR, Ebrahimi M. Potential theranostics of circulating tumor cells and tumor‐derived exosomes application in colorectal cancer. Cancer Cell Int. 2020;20:288. 10.1186/s12935-020-01389-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tian X, Shen H, Li Z, Wang T, Wang S. Tumor‐derived exosomes, myeloid‐derived suppressor cells, and tumor microenvironment. J Hematol Oncol. 2019;12(1):84. 10.1186/s13045-019-0772-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheng L, Zhang X, Tang J, Lv Q, Liu J. Gene‐engineered exosomes‐thermosensitive liposomes hybrid nanovesicles by the blockade of CD47 signal for combined photothermal therapy and cancer immunotherapy. Biomaterials. 2021;275:120964. 10.1016/j.biomaterials.2021.120964 [DOI] [PubMed] [Google Scholar]
- 81. Lee NK, Kothandan VK, Kothandan S, Byun Y, Hwang SR. Exosomes and cancer stem cells in cancer immunity: current reports and future directions. Vaccines (Basel). 2021;9(5):441. 10.3390/vaccines9050441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ma YS, Yang XL, Xin R, Liu JB, Fu D. Power and promise of exosomes as clinical biomarkers and therapeutic vectors for liquid biopsy and cancer control. Biochim Biophys Acta Rev Cancer. 2021;1875(1):188497. 10.1016/j.bbcan.2020.188497 [DOI] [PubMed] [Google Scholar]
- 83. Di W, Zhang W, Zhu B, Li X, Tang Q, Zhou Y. Colorectal cancer prompted adipose tissue browning and cancer cachexia through transferring exosomal miR‐146b‐5p. J Cell Physiol. 2021;236(7):5399‐5410. 10.1002/jcp.30245 [DOI] [PubMed] [Google Scholar]
- 84. Kim DH, Park H, Choi YJ, et al. Exosomal miR‐1260b derived from non‐small cell lung cancer promotes tumor metastasis through the inhibition of HIPK2. Cell Death Dis. 2021;12(8):747. 10.1038/s41419-021-04024-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li J, Sun L, Chen Y, et al. Gastric cancer‐derived exosomal miR‐135b‐5p impairs the function of Vgamma9Vdelta2 T cells by targeting specificity protein 1. Cancer Immunol Immunother. 2021. 10.1007/s00262-021-02991-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao Q, Huang L, Qin G, et al. Cancer‐associated fibroblasts induce monocytic myeloid‐derived suppressor cell generation via IL‐6/exosomal miR‐21‐activated STAT3 signaling to promote cisplatin resistance in esophageal squamous cell carcinoma. Cancer Lett. 2021;518:35‐48. 10.1016/j.canlet.2021.06.009 [DOI] [PubMed] [Google Scholar]
- 87. Graziani V, Rodriguez‐Hernandez I, Maiques O, Sanz‐Moreno V. The amoeboid state as part of the epithelial‐to‐mesenchymal transition programme. Trends Cell Biol. 2021. 10.1016/j.tcb.2021.10.004 [DOI] [PubMed] [Google Scholar]
- 88. Taki M, Abiko K, Ukita M, et al. Tumor immune microenvironment during epithelial‐mesenchymal transition. Clin Cancer Res. 2021;27(17):4669‐4679. 10.1158/1078-0432.CCR-20-4459 [DOI] [PubMed] [Google Scholar]
- 89. Katsuno Y, Derynck R. Epithelial plasticity, epithelial‐mesenchymal transition, and the TGF‐beta family. Dev Cell. 2021;56(6):726‐746. 10.1016/j.devcel.2021.02.028 [DOI] [PubMed] [Google Scholar]
- 90. Greco L, Rubbino F, Morelli A, et al. Epithelial to mesenchymal transition: A challenging playground for translational research. current models and focus on TWIST1 relevance and gastrointestinal cancers. Int J Mol Sci. 2021;22(21):11469. 10.3390/ijms222111469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Berrondo C, Flax J, Kucherov V, et al. Expression of the long non‐coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS One. 2016;11(1):e0147236. 10.1371/journal.pone.0147236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xue M, Chen W, Xiang A, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non‐coding RNA‐UCA1. Mol Cancer. 2017;16(1): 10.1186/s12943-017-0714-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Huang CS, Ho JY, Chiang JH, Yu CP, Yu DS. Exosome‐derived LINC00960 and LINC02470 promote the epithelial‐mesenchymal transition and aggressiveness of bladder cancer cells. Cells. 2020;9(6):1419. 10.3390/cells9061419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xin Y, Li J, Wu W, Liu X. Mitofusin‐2: A new mediator of pathological cell proliferation. Front Cell Dev Biol. 2021;9:647631. 10.3389/fcell.2021.647631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Feola T, Centello R, Sesti F, et al. Neuroendocrine carcinomas with atypical proliferation index and clinical behavior: A systematic review. Cancers (Basel). 2021;13(6):1247. 10.3390/cancers13061247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pandey M, Mukhopadhyay A, Sharawat SK, Kumar S. Role of microRNAs in regulating cell proliferation, metastasis and chemoresistance and their applications as cancer biomarkers in small cell lung cancer. Biochim Biophys Acta Rev Cancer. 2021;1876(1):188552. 10.1016/j.bbcan.2021.188552 [DOI] [PubMed] [Google Scholar]
- 97. Zheng R, Du M, Wang X, et al. Exosome‐transmitted long non‐coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17(1):143. 10.1186/s12943-018-0880-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lin H, Wu LJ, Guo SQ, et al. Dynamic monitoring of serum liver function indexes in patients with COVID‐19. World J Clin Cases. 2021;9(7):1554‐1562. 10.12998/wjcc.v9.i7.1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Md Yusof K, Rosli R, Abdullah M, A. Avery‐Kiejda K The roles of non‐coding RNAs in tumor‐associated lymphangiogenesis. Cancers (Basel). 2020;12(11):3290. 10.3390/cancers12113290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Roy S, Banerjee P, Ekser B, et al. Targeting lymphangiogenesis and lymph node metastasis in liver cancer. Am J Pathol. 2021;191(12):2052‐2063. 10.1016/j.ajpath.2021.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang L, Li L, Zhu G. Role of Extracellular Vesicles on Cancer Lymphangiogenesis and Lymph Node Metastasis. Front Oncol. 2021;11:721785. 10.3389/fonc.2021.721785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen C, Luo Y, He W, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404‐421. 10.1172/JCI130892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zheng H, Chen C, Luo Y, et al. Tumor‐derived exosomal BCYRN1 activates WNT5A/VEGF‐C/VEGFR3 feedforward loop to drive lymphatic metastasis of bladder cancer. Clin Transl Med. 2021;11(7):e497. 10.1002/ctm2.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li J, Gao N, Gao Z, et al. The Emerging Role of Exosomes in Cancer Chemoresistance. Front Cell Dev Biol. 2021;9:737962. 10.3389/fcell.2021.737962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chai JY, Sugumar V, Alshanon AF, Wong WF, Fung SY, Looi CY. Defining the Role of GLI/Hedgehog Signaling in Chemoresistance: Implications in Therapeutic Approaches. Cancers. 2021;13(19):4746‐ 10.3390/cancers13194746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Campos A, Sharma S, Obermair A, Salomon C. Extracellular Vesicle‐Associated miRNAs and Chemoresistance: A Systematic Review. Cancers. 2021;13(18):4608. 10.3390/cancers13184608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Luo G, Zhang Y, Wu Z, Zhang L, Liang C, Chen X. Exosomal LINC00355 derived from cancer‐associated fibroblasts promotes bladder cancer cell resistance to cisplatin by regulating miR‐34b‐5p/ABCB1 axis. Acta Biochim Biophys Sin (Shanghai). 2021;53(5):558‐566. 10.1093/abbs/gmab023 [DOI] [PubMed] [Google Scholar]
- 108. Hashemipour M, Boroumand H, Mollazadeh S, et al. Exosomal microRNAs and exosomal long non‐coding RNAs in gynecologic cancers. Gynecol Oncol. 2021;161(1):314‐327. 10.1016/j.ygyno.2021.02.004 [DOI] [PubMed] [Google Scholar]
- 109. Yao Z, Jia C, Tai Y, et al. Serum exosomal long noncoding RNAs lnc‐FAM72D‐3 and lnc‐EPC1‐4 as diagnostic biomarkers for hepatocellular carcinoma. Aging (Albany NY). 2020;12(12):11843‐11863. 10.18632/aging.103355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zheng P, Zhang H, Gao H, et al. Plasma exosomal long noncoding RNA lnc‐SLC2A12‐10:1 as a novel diagnostic biomarker for gastric cancer. Onco Targets Ther. 2020;13:4009‐4018. 10.2147/OTT.S253600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cai C, Zhang H, Zhu Y, et al. Serum exosomal long noncoding RNA pcsk2‐2:1 as a potential novel diagnostic biomarker for gastric cancer. Onco Targets Ther. 2019;12:10035‐10041. 10.2147/OTT.S229033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhan Y, Du L, Wang L, et al. Expression signatures of exosomal long non‐coding RNAs in urine serve as novel non‐invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. 2018;17(1):142. 10.1186/s12943-018-0893-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang S, Du L, Wang L, et al. Evaluation of serum exosomal LncRNA‐based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23(2):1396‐1405. 10.1111/jcmm.14042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang J, Yang K, Yuan W, Gao Z. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307‐9316. 10.12659/MSM.912018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yu X, Wang R, Han C, Wang Z, Jin X. A panel of urinary long non‐coding RNAs differentiate bladder cancer from urocystitis. J Cancer. 2020;11(4):781‐787. 10.7150/jca.37006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gouin KH 3rd, Ing N, Plummer JT, et al. An N‐Cadherin 2 expressing epithelial cell subpopulation predicts response to surgery, chemotherapy and immunotherapy in bladder cancer. Nat Commun. 2021;12(1):4906. 10.1038/s41467-021-25103-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Miyata Y, Tsurusaki T, Hayashida Y, et al. Intravesical MMC and MMC+Ara‐C for non‐muscle invasive bladder cancer: A randomized clinical trial. BJU Int. 2021. 10.1111/bju.15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kobayashi M, Fujiyama N, Tanegashima T, et al. Effect of HLA genotype on intravesical recurrence after bacillus Calmette‐Guerin therapy for non‐muscle‐invasive bladder cancer. Cancer Immunol Immunother. 2021. 10.1007/s00262-021-03032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yi Y, Wu M, Zeng H, et al. Tumor‐derived exosomal non‐coding RNAs: The emerging mechanisms and potential clinical applications in breast cancer. Front Oncol. 2021;11:738945. 10.3389/fonc.2021.738945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Li Y, Lin S, Xie X, Zhu H, Fan T, Wang S. Highly enriched exosomal lncRNA OIP5‐AS1 regulates osteosarcoma tumor angiogenesis and autophagy through miR‐153 and ATG5. Am J Transl Res. 2021;13(5):4211‐4223. [PMC free article] [PubMed] [Google Scholar]
- 121. Han W, Sulidankazha Q, Nie X, Yilidan R, Len K. Pancreatic cancer cells‐derived exosomal long non‐coding RNA CCAT1/microRNA‐138‐5p/HMGA1 axis promotes tumor angiogenesis. Life Sci. 2021;278:119495. 10.1016/j.lfs.2021.119495 [DOI] [PubMed] [Google Scholar]
- 122. Guo X, Qiu W, Liu Q, et al. Immunosuppressive effects of hypoxia‐induced glioma exosomes through myeloid‐derived suppressor cells via the miR‐10a/Rora and miR‐21/Pten Pathways. Oncogene. 2018;37(31):4239‐4259. 10.1038/s41388-018-0261-9 [DOI] [PubMed] [Google Scholar]
- 123. Guo X, Qiu W, Wang J, et al. Glioma exosomes mediate the expansion and function of myeloid‐derived suppressor cells through microRNA‐29a/Hbp1 and microRNA‐92a/Prkar1a pathways. Int J Cancer. 2019;144(12):3111‐3126. 10.1002/ijc.32052 [DOI] [PubMed] [Google Scholar]
- 124. Abels ER, Maas SLN, Nieland L, et al. Glioblastoma‐associated microglia reprogramming is mediated by functional transfer of extracellular miR‐21. Cell Rep. 2019;28(12):3105‐3119 e7. 10.1016/j.celrep.2019.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhao M, Xu J, Zhong S, et al. Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radio‐resistant glioma cells. Oncol Rep. 2019;41(3):1893‐1900. 10.3892/or.2019.6972 [DOI] [PubMed] [Google Scholar]
- 126. Kartolo A, Kassouf W, Vera‐Badillo FE. Adjuvant immune checkpoint inhibition in muscle‐invasive bladder cancer: Is it ready for prime time? Eur Urol. 2021;80(6):679‐681. 10.1016/j.eururo.2021.07.019 [DOI] [PubMed] [Google Scholar]
- 127. Wigner P, Bijak M, Saluk‐Bijak J. The green anti‐cancer weapon. The role of natural compounds in bladder cancer treatment. Int J Mol Sci 2021;22(15):7787. 10.3390/ijms22157787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Yang N, Gao J, Hou R, Xu X, Yang N, Huang S. Grape seed proanthocyanidins inhibit migration and invasion of bladder cancer cells by reversing emt through suppression of TGF‐beta signaling pathway. Oxid Med Cell Longev. 2021;2021:5564312. 10.1155/2021/5564312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Eldh M, Mints M, Hiltbrunner S, et al. Proteomic profiling of tissue exosomes indicates continuous release of malignant exosomes in urinary bladder cancer patients, even with pathologically undetectable tumour. Cancers. 2021;13(13):3242. 10.3390/cancers13133242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wang P, Zhou R, Thomas P, et al. Epithelial‐to‐mesenchymal transition enhances cancer cell sensitivity to cytotoxic effects of cold atmospheric plasmas in breast and bladder cancer systems. Cancers (Basel). 2021;13(12):2889. 10.3390/cancers13122889 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data in the current study are available from the corresponding authors on reasonable request.


