Abstract
The new INNO-LiPA Mycobacteria (Innogenetics, Ghent, Belgium), a reverse-hybridization-based line probe assay, and the AccuProbe assay (Gen-Probe Inc., San Diego, Calif.) were applied to MB/BacT Alert 3D (MB/BacT) system (Organon Teknika, Boxtel, The Netherlands) culture bottles and evaluated for mycobacterial identification. From 2,532 respiratory and extrapulmonary specimens submitted for culture, 168 were flagged positive by the MB/BacT system and promptly evaluated for identification (within 24 h). Each of 163 vials grew one mycobacterial isolate, including Mycobacterium tuberculosis complex (n = 73), M. avium complex (n = 3), M. avium (n = 8), M. intracellulare (n = 5), M. kansasii (n = 15), M. gordonae (n = 8), M. malmoense (n = 3), M. chelonae (n = 13), M. abscessus (n = 2), M. xenopi (n = 11), M. scrofulaceum (n = 2), M. fortuitum (n = 7), M. terrae (n = 3), M. simiae (n = 2), M. celatum (n = 3), M. flavescens (n = 1), M. interjectum (n = 1), M. bohemicum (n = 1), and M. pulveris (n = 2). Five cultures yielded mixed growth of two mycobacterial species: M. tuberculosis complex plus M. gordonae (n = 2), M. tuberculosis complex plus M. chelonae (n = 1), M. tuberculosis complex plus M. xenopi (n = 1), and M. avium plus M. chelonae (n = 1). In testing of one-isolate vials, both systems showed excellent sensitivity and specificity for all species and complexes for which they are licensed (nine for INNO-LiPA Mycobacteria versus six for AccuProbe). There were minor discrepancies in results for two isolates identified by INNO-LiPA Mycobacteria as M. avium - M. intracellulare - M. scrofulaceum (MAIS) complex and by AccuProbe as M. intracellulare. In testing of two-isolate vials, INNO-LiPA Mycobacteria correctly identified all isolates, while the AccuProbe assay failed to identify three M. tuberculosis complex isolates and one M. avium isolate. The AccuProbe assay was completed within 2 h, while INNO-LiPA Mycobacteria required a 6-h period. In our opinion, INNO-LiPA Mycobacteria offers the following advantages: (i) it contains a genus-specific probe that, in addition to being used in genus identification, may be used as an internal control for both the amplification and hybridization steps; (ii) it simultaneously identifies M. tuberculosis complex, MAIS complex, and seven other mycobacterial species, even from mixed cultures; (iii) its mycobacterial DNA amplification ensures reliable results independent from the concentration of viable microorganisms; and (iv) it genotypically identifies M. kansasii and M. chelonae. In conclusion, even though INNO-LiPA Mycobacteria is considerably less easy to use than AccuProbe, requiring personnel skilled in molecular biology techniques, it represents an excellent approach for routine identification of frequently encountered mycobacteria.
In recent years members of the genus Mycobacterium underwent extensive research designed to develop new methods aimed at improving and expediting the diagnosis and treatment of tuberculosis and other mycobacterial infections. In this context, the suitability of newly developed methods for routine use in the clinical laboratory can be assessed by comparative evaluation with currently available technologies. Radiometric detection of mycobacteria coupled with AccuProbe DNA probe technology (Gen-Probe, San Diego, Calif.) dramatically improved conventional laboratory procedures, allowing a rapid and accurate diagnosis of mycobacterial infections. Recently, MB/BacT Alert 3D (MB/BacT) (Organon Teknika, Boxtel, The Netherlands), a nonradiometric, fully automated, continuously monitoring, walk-away system, has been introduced as an alternative to the radiometric system, currently considered the “gold standard.” The AccuProbe system, developed for the identification of M. tuberculosis complex (MTB), M. avium complex (MAC), M. avium, M. intracellulare, M. kansasii, and M. gordonae, is a simple and rapid assay featuring a single probe test whose choice can be oriented according to acid-fast bacillus (AFB) microscopic and/or colonial morphology. INNO-LiPA Mycobacteria (LiPA) (Innogenetics NV, Ghent, Belgium), a new DNA probe system for simultaneous identification of MTB, M. avium-M. intracellulare-M. scrofulaceum (MAIS) complex, and seven other mycobacterial species, has been introduced as an upgrade of AccuProbe technology. LiPA combines amplification of the 16S-23S rRNA spacer region of Mycobacterium species (1, 5) and reverse hybridization with membrane strip-associated probes.
The aim of this study was to evaluate the performance of LiPA compared to that of AccuProbe. Both assays were applied to MB/BacT liquid medium soon after culture bottles were flagged as positive by the MB/BacT instrument.
MATERIALS AND METHODS
Specimen collection and processing.
This study included 2,532 clinical specimens, both respiratory (n = 1,511) and extrapulmonary (n = 1021), consecutively received for mycobacterial culture by two Italian microbiology laboratories: the Regional Mycobacteria Reference Centre, San Bortolo Hospital, Vicenza, and the Department of Clinical Microbiology, General Hospital Umberto I—Torrette, Ancona. Both labs share several years of experience in mycobacterial diagnostic procedures and amplification techniques. Clinical specimens collected from contaminated sites were digested and decontaminated with an equal volume of N-acetyl-l-cysteine and 2% NaOH (final concentration, 1%) according to standard decontamination procedures (4). After neutralization with phosphate-buffered saline (0.067 M, pH 6.8) and centrifugation at 3,500 × g for 20 min, the pellet was used for a smear, suspended in phosphate-buffered saline to a final volume of 2.0 ml, and cultured into liquid and solid media. Sterile specimens were inoculated into media without prior decontamination.
Culture systems.
MB/BacT bottles were supplemented with mycobacteria antibiotic supplement (MAS) (amphotericin B, azlocillin, nalidixic acid, polymyxin B, trimethoprim, and vancomycin) plus a vancomycin antibiotic mixture as recommended by the manufacturer. A volume of 0.5 ml of the sediment was randomly inoculated into liquid and solid media. All culture media were incubated at 37°C. Liquid cultures automatically monitored by the MB/BacT instrument (every 10 min) were incubated for 42 days, while solid ones were discarded after a 56-day incubation. Löwenstein-Jensen medium slants were visually inspected once a week for mycobacterial growth, and smears from suspect colonies were made. Liquid cultures were considered positive only when smears confirmed the presence of mycobacteria.
Microscopy.
Smears for AFB were stained with auramine-rhodamine stain, and positive slides were confirmed by the Ziehl-Neelsen method.
Identification of mycobacteria.
Isolates of MTB, MAC, M. avium, M. intracellulare, M. kansasii, and M. gordonae were identified by the AccuProbe assay (11). Other mycobacterial species were identified by high-performance liquid chromatography (HPLC) (13) and conventional biochemical tests (4, 6).
AccuProbe procedure.
The AccuProbe assay was performed on smear-positive MB/BacT bottles within the first working day following instrument detection. To reduce false-negative results associated with an insufficient mycobacterial cell mass, a 1.5-ml portion of liquid medium was centrifuged at 12,500 × g for 10 min in a sterile screw-cap microcentrifuge tube, and the pellet was used for a hybridization test. DNA probes were used according to the instructions of the kit package insert; samples producing signals greater than 30,000 relative light units (RLU) were considered positive. Selection of an appropriate probe(s) relied upon the microscopic appearance in liquid medium and pigmentation of the pellet (3, 10). When the initial test gave a negative result (<30,000 RLU) or mixed culture was suspected, additional probes were performed.
LiPA procedure. (i) Amplification.
For PCR, 0.2-ml aliquots of culture samples were centrifuged (15 min at 17,900 × g) and pellets were resuspended in 20 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8). Then samples were heat inactivated at 95°C for 30 min, centrifuged at 17,900 × g for 10 s, and frozen at −20°C for 30 min. Once defrosted, they were vortexed and centrifuged at 17,900 × g for 10 s. Ten microliters of the extracted DNA was added to 40 μl of the reagent mixture containing deoxyribonucleoside 5′ triphosphates, biotinylated primers complementary to regions flanking the 16S-23S rRNA spacer region, and Taq polymerase. Amplification was performed with a Perkin-Elmer model 9600 thermal cycler. The first step consisted of 1 min at 95°C and was followed by 40 cycles, each of which included three steps: denaturation at 95°C for 30 s, annealing of primers at 62°C for 30 s, and extension of primers at 72°C for 30 s. The presence of amplification products was checked by electrophoretic migration of the amplified sample (10 μl) through a 2% agarose gel, followed by ethidium bromide staining. The amplicon appeared as a single band with a length of 400 to 550 bp.
(ii) Hybridization.
Ten microliters of the amplified biotinylated product was added to the denaturation solution (placed into a disposable trough containing the test strip), carefully mixed by pipetting, and left for 5 min at room temperature. Two milliliters of the prewarmed hybridization solution was then added, and the solution was mixed by gentle shaking. The test strip was completely submerged by the solution, and the trough was placed into a 62°C shaking water bath (80 rpm) for a 30-min incubation. After hybridization, the test strip was washed twice at room temperature and then incubated at 62°C for 10 min with stringent wash solution. The subsequent procedure was carried out at room temperature using a shaker as follows. (i) After two washing steps (rinse solution for 1 min), the test strip was added to the conjugate solution (streptavidin labeled with alkaline phosphatase) for 30 min. (ii) The test strip was washed twice for 1 min using rinse solution and once again using substrate buffer. Then substrate solution was added to the trough, followed by incubation for 30 min. (iii) The test strip was washed twice for 3 min with distilled water to stop color development and then removed from the trough and placed on absorbent paper to dry.
(iii) Reading and interpretation.
The presence of a clearly visible line on the membrane-based test strip was considered a positive hybridization reaction. Visual comparison of test hybridization bands with the interpretation chart provided by the manufacturer allowed mycobacterial identification.
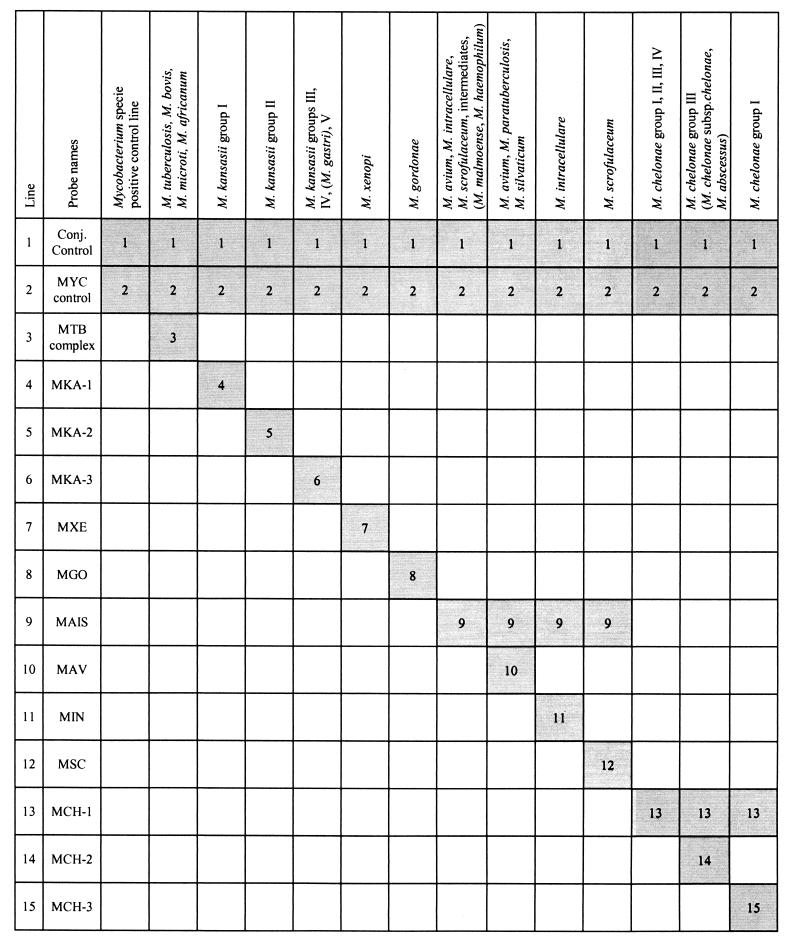
Moreover, at the end of the assay each tested strip was characterized by (i) a green line marking the correct position of the plastic reading chart; (ii) a positive line representing the conjugate control line, which must always be visible; (iii) another line (first probe) representing a Mycobacterium genus hybridization control, which should be positive when an appropriate amount of amplified mycobacterial DNA product has been tested for hybridization; and (iv) one or more species- or complex-specific probe lines (Fig. 1).
FIG. 1.
LiPA interpretation chart. Conj., Conjugated. For other abbreviations, see Table 1, footnote a.
RESULTS
One hundred sixty-eight out of 2,532 specimens grew mycobacteria by the MB/BacT system and were subjected to LiPA and AccuProbe assays for identification. All mycobacteria subjected to LiPA exhibited a positive conjugate control line as well as a Mycobacterium genus line. In testing of bottles yielding one isolate, both assays correctly identified all species for which the kits are licensed, showing discrepant results for only two isolates identified by LiPA as MAIS complex (reactive lines 1, 2, and 9) and by AccuProbe as M. intracellulare (Table 1).
TABLE 1.
Identification of mycobacterial species from MB/BacT bottles yielding one strain by the LiPA and AccuProbe assay
| Isolates
|
LiPA
|
AccuProbe
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total | Identification | No. of isolates identified | Reactive lines | Identificationa | No. of isolates identified | Avg RLU | Range (RLU) | Identificationa |
| 73 | MTB | 73 | 1, 2, 3 | MTB | 73 | 306,768 | 31,754–690,336 | MTB |
| 3 | MAC | 3 | 1, 2, 9 | MAIS | 3 | 440,097 | 310,815–584,290 | MAC |
| 2 | MAC | 2 | 1, 2, 9 | MAIS | 2 | 241,729 | 214,134–269,324 | MIN |
| 8 | M. avium | 8 | 1, 2, 9, 10 | MAV | 8 | 275,025 | 36,645–660,218 | MAV |
| 3 | M. intracellulare | 3 | 1, 2, 9, 11 | MIN | 3 | 334,655 | 214,135–589,165 | MIN |
| 15 | M. kansasii | 9 | 1, 2, 4 | MKA-1 | 15 | 415,618 | 41,122–1,330,490 | MKA |
| 6 | 1, 2, 5 | MKA-2 | ||||||
| 8 | M. gordonae | 8 | 1, 2, 8 | MGO | 8 | 304,398 | 39,169–831,084 | MGO |
| 3 | M. malmoense | 3 | 1, 2, 9 | MAIS | 3 | 2,354 | 2,821–4,944 | NTM |
| 13 | M. chelonae | 8 | 1, 2, 13, 15 | MCH-1 | 13 | 2,467 | 891–4,703 | NTM |
| 5 | 1, 2, 13, 14 | MCH-3 | ||||||
| 2 | M. abscessus | 2 | 1, 2, 13, 14 | MCH-3 | 2 | 1,955 | 1,522–2,388 | NTM |
| 11 | M. xenopi | 11 | 1, 2, 7 | MXE | 11 | 3,716 | 890–16,077 | NTM |
| 2 | M. scrofulaceum | 2 | 1, 2, 9, 12 | MSC | 2 | 3,714 | 1,003–14,598 | NTM |
| 7 | M. fortuitum | 7 | 1, 2 | NTM | 7 | 2,914 | 1,122–7,494 | NTM |
| 3 | M. terrae | 3 | 1, 2 | NTM | 3 | 1,745 | 1,129–2,987 | NTM |
| 2 | M. simiae | 2 | 1, 2 | NTM | 2 | 1,752 | 1,345–2,987 | NTM |
| 3 | M. celatum | 3 | 1, 2 | NTM | 3 | 3,945 | 1,992–6,432 | NTM |
| 1 | M. flavescens | 1 | 1, 2 | NTM | 1 | 1,632 | 991–2,421 | NTM |
| 1 | M. interjectum | 1 | 1, 2 | NTM | 1 | 1,938 | 1,231–4,494 | NTM |
| 1 | M. bohemicum | 1 | 1, 2 | NTM | 1 | 1,428 | 998–2,439 | NTM |
| 2 | M. pulveris | 2 | 1, 2 | NTM | 2 | 1,824 | 2,220–4,323 | NTM |
MAV, M. avium; MIN, M. intracellulare; MKA-1, M. kansasii group I; MKA-2, M. kansasii group II; MGO, M. gordonae; MCH-1, M. chelonae group I; MCH-3, M. chelonae group III; MXE, M. xenopi; MSC, M. scrofulaceum NTM, nontuberculous mycobacteria.
Three MAC isolates, when tested by LiPA, showed positive hybridization bands with the MAIS complex probes and negative results with the M. avium, M. intracellulare, and M. scrofulaceum probes. These three isolates, when tested by AccuProbe, hybridized with MAC probes but not with probes specific for M. avium and M. intracellulare. Moreover, three other isolates, when tested by LiPA, showed positive hybridization with the MAIS complex probes and negative results with the M. avium, M. intracellulare, and M. scrofulaceum probes. When tested by AccuProbe, they did not hybridize either with MAC probes or with M. avium- and M. intracellulare-specific probes. The former strains were correctly identified as MAC, and the latter strains were identified as M. malmoense by HPLC and standard biochemical tests.
Of the 15 M. kansasii isolates, all positive by AccuProbe assay, 9 were identified by LiPA as M. kansasii group I and 6 were identified as M. kansasii group II.
Of the 15 M. chelonae isolates, all of which were negative by AccuProbe, 8 were identified by LiPA as M. chelonae group I and 7 were identified as M. chelonae group III, which includes M. chelonae subsp. chelonae and M. abscessus. Two isolates from the latter group identified as M. abscessus by HPLC and standard biochemical tests were shown to react with M. chelonae group III probes (lines 1, 2, 13, and 14).
In our study, selection of the appropriate probe according to microscopic appearance and pellet color was always successful. Similarly, RLU values were clearly positive except for 14 isolates (5 of these were MTB strains) showing results slightly above the cutoff (range, 31,754 to 49,922 RLU).
Five cultures yielded two mycobacterial species (Table 2). Two cultures from a patient reporting a history of repeatedly treated pulmonary tuberculosis, grew MTB and M. gordonae. One culture from a patient with leukemia grew MTB and M. chelonae, another from a patient with lung cancer grew MTB and M. xenopi, and the last culture from an AIDS patient grew M. avium and M. chelonae. These species were correctly identified by LiPA, which exhibited specific bands for all coupled strains. When probed by the AccuProbe assay, mixed cultures gave positive results only for three strains out of seven expected. Both cultures growing MTB and M. gordonae and the culture growing MTB and M. chelonae gave negative results with the MTB probe. Similarly, culture growing M. avium and M. chelonae was negative for MAC.
TABLE 2.
Identification of mycobacterial species from MB/BacT bottles yielding mixed cultures by the LiPA and AccuProbe assaya
| Mycobacterial culture
|
LiPA
|
AccuProbe
|
|||||
|---|---|---|---|---|---|---|---|
| No. of isolates | Identification | No. of isolates | Reactive lines | Identification | Probe | RLU | Identification |
| 1 | MTB + MGO | 2 | 1, 2, 3, 8 | MTB + MGO | MGO | 366,577 | MGO |
| MTB | 13,416 | ||||||
| MK | 1,679 | ||||||
| MAC | 2,134 | ||||||
| 1 | MTB + MGO | 2 | 1, 2, 3, 8 | MTB + MGO | MGO | 297,467 | MGO |
| MTB | 15,725 | ||||||
| MAC | 1,929 | ||||||
| MK | 1,761 | ||||||
| 1 | MTB + MCH | 2 | 1, 2, 3, 13, 14 | MTB + MCH-3 | MTB | 7,933 | |
| MAC | 2,524 | ||||||
| MK | 1,187 | ||||||
| MGO | 2,416 | ||||||
| 1 | MTB + MXE | 2 | 1, 2, 3, 7 | MTB + MXE | MTB | 89,829 | MTB |
| MAC | 2,518 | ||||||
| MK | 892 | ||||||
| MGO | 1,014 | ||||||
| 1 | MAV + MCH | 2 | 1, 2, 9, 10, 13, 14 | MAV + MCH-3 | MAC | 11,134 | |
| MTB | 2,412 | ||||||
| MK | 1,826 | ||||||
| MGO | 1,361 | ||||||
MAV, M. avium; MK, M. kansasii; MGO, M. gordonae; MCH, M. chelonae; MCH-3, M. chelonae group III; MXE, M. xenopi.
DISCUSSION
Technological advances in molecular biology have provided new nucleic acid-based amplification strategies for a rapid and more effective detection of MTB from clinical specimens. Presently, however, the most recommended tool to achieve laboratory diagnosis of tuberculosis and other mycobacterial infections is culture. In this context, new culture systems and identification assays have been developed to make AFB isolation and identification faster and more reliable.
In this study, in order to reduce identification time as much as possible, we compared LiPA and the AccuProbe assay, which were run directly from AFB-positive MB/BacT bottles within the first working day following flagging by the instrument. An early identification of mycobacteria may provide patients with faster treatment and a better prognosis. Evaluations of the AccuProbe assay in identifying MTB and nontuberculous mycobacteria directly from 12B broth and MB/BacT bottles have been reported in the literature (2, 3, 9, 10). It is now well established that AccuProbe's major drawback is represented by the number of viable mycobacterial organisms, which may be responsible for false-negative results. In fact, while positive B460 vials usually require additional incubation after radiometric detection to be successfully probed, evidence of considerable AFB growth in MB/BacT bottles since the first positive signal allows a very early tentative identification. According to our previous experience (9), we centrifuged 1.5 ml of MB/BacT liquid medium at 12,500 × g for 10 min and used the pellet for hybridization testing. With all samples growing one AFB isolate (even though 14 of them produced signals slightly above the cutoff) AccuProbe's sensitivity was 100%. Although it has been reported that centrifugation of MB/BacT medium is not necessary to obtain a positive AccuProbe reaction (2), we recommend this step, which in our opinion is essential to obtain a reliable performance. LiPA includes probes for M. xenopi, M. scrofulaceum, and M. chelonae, which are not covered by the AccuProbe system. Both assays showed a 100% sensitivity and an excellent specificity. Full agreement with conventional identification tests was shown for all AFB isolates, including two strains identified as being of the MAIS group by LiPA and M. intracellulare by AccuProbe. Although this discrepancy was related to the different genetic targets of AccuProbe (16S rRNA region) and LiPA (16S-23S rRNA spacer) and could be resolved only by sequencing (5), it did not demonstrate any relevance from a clinical standpoint. Two evaluations of LiPA have been reported in the literature, and this discrepancy was observed in both (7, 12). A positive result with probe 9 identified the MAIS group, which includes different species. Some of these are identifiable by the simultaneous hybridization of probes 10, 11, and 12. Other species, such as M. malmoense, that show negative MAC, M. avium, and M. intracellulare hybridizations by AccuProbe can be discriminated only by HPLC and conventional biochemical tests. Moreover, strains belonging to the M. avium-M. intracellulare-X group (14) (otherwise defined as “intermediate”) showed a MAC-positive but M. avium- and M. intracellulare-negative probe pattern in the AccuProbe assay and a MAIS-positive and M. avium-, M. intracellulare-, M. scrofulaceum-negative probe pattern in the LiPA.
Identification of M. chelonae and M. xenopi represents an important improvement. Moreover, LiPA exhibits also a genotypic identification of M. chelonae isolates, whose correlation with putative pathogenicity is not clear. In this study, 15 isolates were identified as genotypes I and III, but only two isolates of the last genotype were identified as M. abscessus by HPLC and standard biochemical tests. The existence of five well-defined genotypic clusters within M. kansasii and their different clinical relevance were previously reported (1, 8). Of these, only subtypes I and II (Table 2), which represent the most frequent isolates of human origin, were detected in our study. The AccuProbe assay allowed correct identification of all M. kansasii isolates. Mixed mycobacterial cultures (confirmed by subculture onto solid media of five specimens) could be correctly detected only by LiPA (100% sensitivity), showing the simultaneous presence of specific bands for both species on the test strip (Table 3). Mixed cultures represent a further drawback of the AccuProbe technology, not only for a restricted identification coverage in comparison with that of LiPA, but also for a considerably reduced sensitivity (42.8%), even in presence of identifiable strains. Possible explanations of these false-negative results can be related to different growth kinetics and competition for medium nutrients which do not permit coupled strains to achieve a sufficient cell mass. This failure can cause possible delays in diagnosis and treatment of mycobacterial infections.
In pure and mixed cultures, both assays' specificity was 100%, except with MAIS (LiPA) and M. intracellulare (AccuProbe) probes, whose specificity according to the above-reported unresolved discrepancies was 98.8%.
Advantages of the LiPA can be summarized as follows. (i) It uses a general probe that reacts with the whole genus Mycobacterium. (ii) It identifies MTB complex, MAIS complex, and seven other mycobacterial species in a single assay from pure and mixed cultures. This considerable number of mycobacterial DNA probes allowed correct identification of the majority (88.4%) of mycobacterial isolates recovered in our laboratories. In contrast, the AccuProbe system allowed a correct identification of only 68.8% of AFB isolates. (iii) Amplification gets rid of any cell mass-related false-negative result. (iv) It genotypically identifies M. kansasii and M. chelonae. As minor disadvantages, we report that it requires a stringent hybridization temperature, whose control is essential to avoid development of nonspecific bands, and 6 h to perform the procedure, including preliminary PCR amplification. AccuProbe procedures are completed in about 2 h, but when a negative result is obtained, additional time is required for testing new probes. In addition, the LiPA is a complex procedure that should be performed in a mycobacteriology lab by personnel skilled in molecular biology techniques. In conclusion, the LiPA allows a rapid and accurate species identification of most commonly encountered mycobacteria directly from positive liquid media and therefore represents an important technological improvement in clinical mycobacteriology.
ACKNOWLEDGMENTS
We thank Innogenetics Italia (Pomezia, Italy) and Organon Teknika (Rome, Italy) for providing instrumentation and reagents for this study.
This study is part of the scientific activity undertaken by the AMCLI (Italian Association of Clinical Microbiology) Committee of Mycobacteriology.
REFERENCES
- 1.Alcaide F, Richter I, Bernasconi C, Sprinter B, Hagenau C, Schulze-Röbbecke R, Tortoli E, Martin R, Böttger E C, Talenti A. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implication for epidemiological and pathogenicity studies. J Clin Microbiol. 1997;35:1959–1964. doi: 10.1128/jcm.35.8.1959-1964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badak F Z, Goksel S, Sertoz R, Nafile B, Ermertcan S, Cavusogliu C, Bilgic A. Use of nucleic acid probes for identification of Mycobacterium tuberculosis directly from MB/BacT bottles. J Clin Microbiol. 1999;37:1602–1605. doi: 10.1128/jcm.37.5.1602-1605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaminski D A, Hardy D J. Selective utilization of DNA probes for identification of Mycobacterium species on the basis of cord formation in primary BACTEC 12B cultures. J Clin Microbiol. 1995;33:1548–1550. doi: 10.1128/jcm.33.6.1548-1550.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent P T, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. Atlanta, Ga: Department of Health and Human Services, Centers for Disease Control and Prevention; 1985. [Google Scholar]
- 5.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metchock B, Nolte F S, Wallace R J., Jr . Mycobacterium. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 399–437. [Google Scholar]
- 7.Miller N, Infante S, Cleary T. Evaluation of the LiPA MYCOBACTERIA assay for identification of mycobacterial species from BACTEC 12B bottles. J Clin Microbiol. 2000;38:1915–1919. doi: 10.1128/jcm.38.5.1915-1919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picardeau M, Prod'Hom G, Raskine L, Lepennec M P, Vincent V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol. 1997;35:25–32. doi: 10.1128/jcm.35.1.25-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piersimoni C, Scarparo C, Callegaro A, Passerini Tosi C, Nista D, Bornigia S, Scagnelli M, Rigon A, Ruggiero G, Goglio A. Comparison of MB/BacT ALERT 3D system with radiometric BACTEC system and Löwenstein-Jensen medium for recovery and identification of mycobacteria from clinical specimens: a multicenter study. J Clin Microbiol. 2001;39:651–657. doi: 10.1128/JCM.39.2.651-657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reisner B S, Gatson A M, Wood G L. Use of Gen-Probe AccuProbes to identify Mycobacterium avium complex, Mycobacterium tuberculosis complex, Mycobacterium kansasii, and Mycobacterium gordonae directly from BACTEC TB broth cultures. J Clin Microbiol. 1994;32:2995–2998. doi: 10.1128/jcm.32.12.2995-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stockman L. DNA probes for the identification of mycobacteria. In: Isenberg H D, editor. Clinical microbiology procedures handbook. Washington, D.C.: American Society for Microbiology; 1992. pp. 3.15.1–3.15.4. [Google Scholar]
- 12.Tortoli E, Nannetti A, Piersimoni C, Cichero P, Farina C, Mucignat G, Scarparo C, Bortolini L, Valentini R, Nista D, Gesu G, Passerini Tosi C, Crovato M, Brusarosco G. Performance assessment of new multiplex probe assay for identification of mycobacteria. J Clin Microbiol. 2001;39:1079–1084. doi: 10.1128/JCM.39.3.1079-1084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tortoli E, Bartoloni A. High-performance liquid chromatography and identification of mycobacteria. Rev Med Microbiol. 1996;7:207–219. [Google Scholar]
- 14.Viljanen MK, Olkkonen L, Katila M L. Conventional identification characteristics, mycolate and fatty acid composition, and clinical significance of MAIX AccuProbe-positive isolates of Mycobacterium avium complex. J Clin Microbiol. 1993;31:1376–1378. doi: 10.1128/jcm.31.5.1376-1378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]