Abstract
A rapid method for demonstration of gram-positive and gram-negative bacteria in milk is described. The technique is based on dilution of the sample in a medium, followed by filtration through a porous polysulfone membrane with a pore size retaining and concentrating bacteria from the sample. The bacteria concentrated on the surface of the membrane are stained with a cationic dye (toluidine blue) that can be visualized by the naked eye. After staining, the membrane is treated with ethanol-acetic acid (pH 2.8 to 3.0), which causes decolorization of gram-negative bacteria, whereas gram-positive bacteria retain the stain. The method does not require heat fixation, electrical power, microscopic examination, or specially trained personnel. The time needed to perform the test is approximately 5 min. The technique was applied to artificially infected milk and milk from cows with moderate or severe clinical mastitis for detection and differentiation of bacteria. The sensitivity of the filtration method was 92 and 100% for gram-positive and gram-negative bacteria, respectively, compared with traditional bacteriological culture of milk samples. The detection limit was 5 × 106 CFU/ml for Staphylococcus aureus and 1 × 106 CFU/ml for Escherichia coli in spiked milk samples. The overall specificity of the method was 86%. This diagnostic method can provide on-site guidance to the veterinarian to optimize use of antibiotics in mastitis therapy.
Gram staining of bacteria segregates bacteria into two categories based on cell wall composition. The cell wall of gram-positive bacteria consists of a cytoplasmic membrane, many polymeric layers of peptidoglycan connected by amino acid bridges, and a variable outer layer called the capsule (12). Gram-negative bacteria possess a bilayered outer membrane, a thin peptidoglycan layer, and a bilayered plasma membrane (7).
The Gram stain procedure, developed by Hans Christian Gram in 1884, relies on the differential cell wall staining properties of gram-positive and -negative bacteria. It remains today as a fundamental step in bacterial classification. The reaction is based on the retention of a dye (crystal violet complexed with iodine) within the cell wall of bacteria. The dye is retained in gram-positive organisms following an alcohol wash. The gram-negative bacteria, which lose the dye following the alcohol wash, may subsequently be counterstained with carbolfuchsin or safranin.
Alternative bacterial staining methods have been developed using fluorescein-labeled wheat germ agglutinin (22) or rhodamine 123 (a lipophilic cationic dye) (1, 9, 21). Furthermore, a staining technique for unfixed organisms in suspension has been developed (14) employing two fluorescent nucleic acid binding dyes, hexidium iodide and SYTO 13. These alternative staining methods are more expensive than traditional Gram staining and require expensive instruments such as epifluorescence microscopes or flow cytometers.
A method of membrane filtration of bacteria in raw milk, followed by staining with a fluorescent dye, e.g., acridine orange, and subsequent visualization by epifluorescence microscopy has been reported (18) and used with some modification by others (11). A similar procedure was developed for Gram stain characterization of mixed bacterial populations in environmental samples (8).
Mastitis, which is inflammation of the mammary gland, is considered the most costly disease of dairy cows worldwide (10). The most important microorganisms causing mastitis are staphylococci, streptococci, and coliform bacteria, although Enterococcus spp., Pseudomonas spp., Arcanobacterium pyogenes, and Pasteurella spp., are isolated frequently. From a therapeutic point of view, the differential diagnosis of clinical mastitis is essential, since the clinical signs of the cow are normally insufficient to predict the etiological agent (15) and since optimal treatments differ for each of these types of mastitis.
In this study, we developed a filter device system that retains bacteria from milk and determines their Gram staining classification. We evaluated the technique for the detection and differentiation of bacteria in artificially infected milk and milk from cows with moderate or severe clinical mastitis. The sensitivity and specificity of the technique were calculated, using bacteriological culture as the reference method.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains Staphylococcus aureus ATCC 25923, S. aureus 489/96, Staphylococcus intermedius 859/97, Streptococcus dysgalactiae 116/98, Escherichia coli 83 II, Listeria monocytogenes 92/99, Pseudomonas aeruginosa 114/98, Bacillus subtilis ATCC 27853, Staphylococcus epidermidis KS429, Staphylococcus haemolyticus DSM 20263, A. pyogenes 397/99, Enterococcus faecalis 332/99, Streptococcus agalactiae 356/99, and Klebsiella sp. strain 351/99 utilized in this study originated from the bacterial collection of the Bacteriology Laboratory, Norwegian School of Veterinary Science. With the exception of the type strains, all strains were isolated from animals. S. aureus 489/96, S. dysgalactiae 116/98, and E. coli 83 II were from milk of cows with mastitis. One colony from a fresh culture of each bacterial strain was inoculated into Luria-Bertani broth, into heart infusion broth (Difco, Detroit, Mich.), or on blood agar and incubated at 37°C overnight.
Filtration and staining procedure.
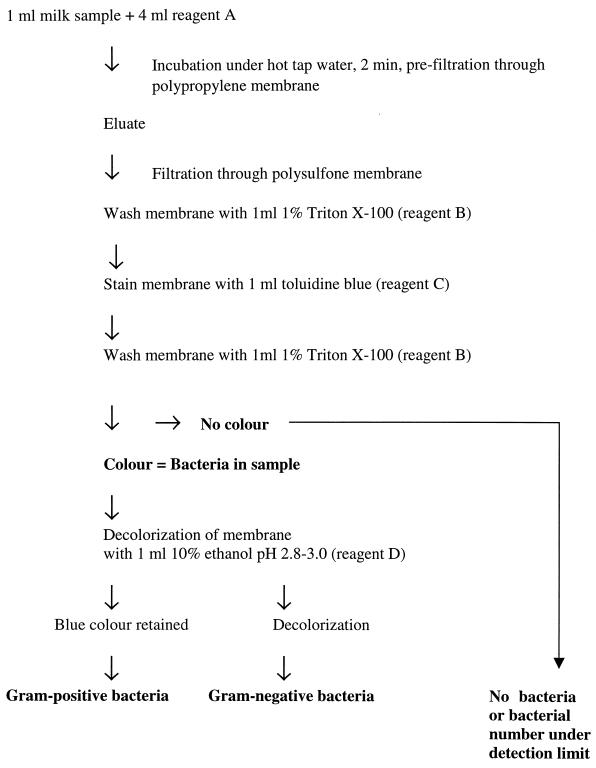
The procedure for filtration and staining is illustrated in Fig. 1. One milliliter of each stock bacterial culture, containing approximately 109 CFU/ml, was diluted in 9 ml of milk from healthy cows with no signs of mastitis, no bacterial growth in milk, and a somatic cell count (SCC) of <50,000/ml. Fivefold dilutions of these mixtures were performed in the same type of milk. The sample (1 ml of diluted culture) was added to 4 ml of reagent A (1 M NaCl, 0.5% Triton X-100, 0.1 M EDTA [pH 12.5]), which killed the bacteria in milk. The milk-reagent mixture was then prefiltered using a 25-mm-diameter polypropylene filter (Millipore Corporation, Bedford, Mass.) with an 80-μm pore size. The eluate was further filtered through a 25-mm-diameter circular polysulfone filter with a 0.8-μm pore size (Pall Gelman Sciences, Ann Arbor, Mich.). The filter membrane was subsequently washed by filtration of 1 ml of 1% Triton X-100 (reagent B), followed by staining with 1 ml of 133 μM toluidine blue (reagent C) (Sigma, St. Louis, Mo.) to stain any bacteria on the filter. After washing with 2 ml of reagent B the membrane was exposed to 1 ml of 10% ethanol adjusted to pH 2.8 to 3.0 with acetic acid (reagent D), which decolorized the gram-negative bacteria, whereas gram-positive bacteria retained the stain. In all stages, a filter holder (Millipore) connected to a 10-ml syringe was used to create a positive pressure over the filter membrane to promote filtration. Milk from cows with moderate or severe clinical mastitis was added directly to 4 ml of reagent A and incubated in hot tap water for 2 min. The polypropylene prefilter (25-mm diameter circular; 80-μm pore size) was mounted on a filter holder (Millipore) connected to a 10-ml syringe containing a 150- to 300-mg hydrophobic wad of polyurethane (Tamro, Vantaa, Finland). For mastitic milk samples with low liquid content, approximately 1 ml of milk samples was mixed with 1 ml of 0.9% NaCl and shaken for a few seconds prior to prefiltration and filtration. The prefiltration stage with the hydrophobic wad of polyurethane was omitted in the experiments involving analyses of milk from healthy cows supplemented with bacteria.
FIG. 1.
Flowchart of the filtration technique for detection and classification of bacteria in milk.
Clinical samples and bacteriological analysis.
Veterinary practitioners sampled milk from mammary gland quarters with moderate or severe clinical mastitis. All samples had high SCCs. In addition to high SCCs in milk, the moderate or severe clinical mastitis was accompanied by heat, pain, swelling, hardness, or abnormal secretion in the mammary gland. SCCs in milk were determined using the California mastitis test (20). Quarter milk samples were collected in 10-ml sterile plastic tubes using an aseptic collection technique. The tubes were numbered to identify the particular quarter, and information regarding the herds, cow, and clinical condition of the cow was recorded. The samples were transported fresh by mail to the Mastitis Laboratory of the National Veterinary Institute for identification of bacteria by culture. The milk samples were stored at 4°C in the laboratory for 24 to 48 h before analyzing by culturing and the filtration method. To determine whether mailing a milk sample alters the results of the filtration test compared with immediate mixing of the milk with reagent A, 21 milk samples from cows with moderate or severe clinical mastitis were sampled directly in reagent A by practicing veterinarians. An aliquot of the milk sample from the same quarter, without addition of reagent A, was also collected. Both samples were sent to the laboratory and analyzed by culture as well as by the filtration method. In all, 55 milk samples from 55 cows with moderate or severe clinical mastitis were analyzed. Milk samples from cows with moderate or severe clinical mastitis were examined according to the methods employed by the Mastitis Laboratory of the National Veterinary Institute (4), based on the recommendations of the International Dairy Federation (5). Fifty microliters of milk sample was plated on blood agar containing 5% washed animal erythrocytes. The plate was incubated at 37°C for 24 h. Bacteriological diagnosis was performed using standard biochemical, Gram staining, and morphological methods as deemed necessary. In parallel, an aliquot of the sample was analyzed by the method developed in this study.
Sensitivity and specificity of the test.
The detection limit of the filtration method was determined using fivefold dilutions of overnight cultures of S. aureus 489/96 and E. coli 83 II in milk from healthy, mastitis-free cows. To ensure that the filter test would work with a range of bacteria, the technique was applied to overnight cultures of clinically relevant bacteria listed above diluted in milk from healthy and mastitis-free cows in the medium/milk ratio of 1:5 to a final concentration between 1 × 107 and 5 × 108 bacteria/ml. Milk from mastitis-free cows without addition of bacteria was used as a negative control. Milk samples from cows with moderate or severe clinical mastitis were used to calculate the sensitivity and specificity of the filtration technique against the conventional culturing method. The sensitivity and specificity were determined, as described (23). Traditional Gram staining and microscopic examination was performed on 53 isolates of Streptococcus species from cows with subclinical or clinical mastitis.
RESULTS
The analytical technique developed in this study was applied to gram-negative and gram-positive bacteria (both cocci and rods) diluted in milk from mastitis-free cows, as well as milk from cows with moderate or severe clinical mastitis.
Overnight cultures of S. aureus 489/96 and E. coli 83 II were diluted in milk from a mastitis-free cow that was free of growth of any bacteria in order to determine the detection limit of the technique. The lowest detection limit of the technique was shown to be 5 × 106 bacteria/ml for S. aureus and 1 × 106 bacteria/ml for E. coli. The results of testing the bacterial stock cultures diluted to 1 × 107 to 5 × 106 CFU/ml in milk showed that the Gram stain identification found by the new method was in full agreement with the reference method. Fourteen samples containing bacteria were tested one time each and once for each bacterial type. No false-positive results occurred with unspiked (negative control) milk. No difference was observed when using Luria-Bertani or heart infusion broth.
Fifty-five milk samples from cows with moderate or severe clinical mastitis were tested for the presence of bacteria. A traditional culturing method was used to identify viable bacteria in milk. One species of bacteria was always dominant in the sample. Gram-positive bacteria were represented by staphylococci (28 samples), streptococci (8 samples), and A. pyogenes (1 sample). Gram-negative bacteria were represented by Pasteurella sp. (1 sample) and coliform bacteria (10 samples) including E. coli, Klebsiella sp., and Proteus sp. No bacteria were detected in seven milk samples. The test results compared with culturing results are presented in Table 1. A color photograph showing detection of gram-positive bacteria and gram-negative bacteria before and after decolorization is shown in Fig. 2. The ranges of viable bacteria in the milk samples from cows with moderate or severe clinical mastitis were between 104 and confluent growth, which corresponds to >106 bacteria/ml of milk.
TABLE 1.
Comparison of results from the filtration technique and a conventional culturing method using milk from cows with moderate or severe clinical mastitis
| Result according to reference method (culturing) | Filtration technique result
|
Total no. of samples tested | |
|---|---|---|---|
| Correct diagnosis | Misdiagnosis | ||
| Presence of bacteria | |||
| Gram positive | 34a | 3b | 37 |
| Gram negative | 11 | 0 | 11 |
| No bacteria presentc | 6 | 1d | 7 |
| Total | 51 | 4 | 55 |
One Streptococcus strain was found to be Gram stain variable and was partially decolorized after the test was performed.
Filtration method misidentified these samples containing S. aureus as bacterium free milk.
Bacteria were not detected by conventional culturing (<50 CFU/ml).
Identified as gram positive by filtration method.
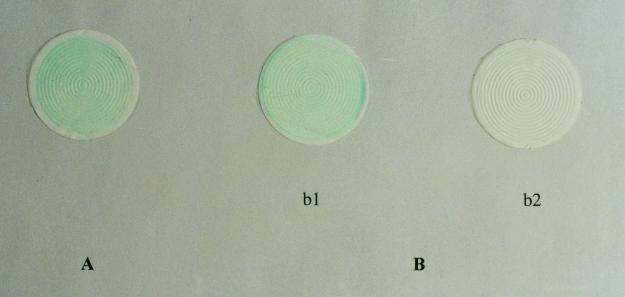
FIG. 2.
The photograph shows results of the filtration method derived from a milk sample from a cow with mastitis containing approximately 107 S. aureus organisms/ml (A) and a milk sample from a cow with mastitis containing approximately 107 E. coli organisms/ml (B), before decolorization (b1) and after decolorization (b2).
The polysulfone filter used to analyze one milk sample containing a Streptococcus sp. was partially decolorized after being washed with 10% ethanol adjusted to pH 2.9 with acetic acid. To determine the likelihood of occurrence of Gram stain-variable streptococci in mastitis, streptococcal strains isolated from 53 cases of clinical and subclinical mastitis were stained by traditional Gram staining. Gram staining followed by microscopic examination of the bacteria was performed after plating of milk samples on blood agar. Gram-variable streptococci were found in 8 of 53 samples (15%).
Thirty-four (n = 37) milk samples from cows with moderate or severe clinical mastitis containing gram-positive bacteria were identified correctly by the filtration method, corresponding to a sensitivity of 92%. All milk samples (n = 11) containing gram-negative bacteria were identified correctly, corresponding to a sensitivity of 100%. Therefore, the filtration method identified 45 (of 48) milk samples containing gram-positive and gram-negative bacteria correctly, corresponding to a sensitivity of 94%. The test also correctly identified six (of seven) milk samples without the presence of bacteria, which corresponds to 86% specificity. The filtration method was approximately 93% (= 45 + 6/55 × 100) accurate in predicting culture results.
Bacterial growth was not observed in samples mixed with reagent A since the reagent kills the bacteria. No differences were observed in test results by the filtration method from milk samples collected directly in reagent A and milk samples to which reagent A was added just prior to analysis (data not shown).
DISCUSSION
Traditional methods for detection and characterization of bacteria in milk require special equipment and are too slow for application in on-site diagnosis and treatment of moderate or severe clinical mastitis.
Preliminary testing was performed to determine the procedures and concentrations arrived at in the final filtration method (data not shown). The reagent developed to treat milk samples contains the nonionic detergent Triton X-100, which facilitates the passage of milk through the filter. Addition of salts (NaCl) to the reagent increased solubilization of samples and augmented the effect of the detergent in the sample solution. EDTA also increased solubilization of milk samples. The final pH (10.3 to 10.8) of the mixture also promoted the filtration process. The solubilization of milk samples was more rapid at higher temperatures, i.e., when the milk-reagent mixture was kept under hot tap water for approximately 2 min. Toluidine blue (reagent C) was used to stain the bacteria captured on the filter. This compound is an amine-substituted phenothiazine, which has the ability to bind to gram-positive as well as gram-negative bacteria. The bacterial surface has a net negative charge at pHs above 2 (16). Toluidine blue binds to the anionic sites on the surface of bacteria. The binding between toluidine blue and the anionic sites on the surface of bacteria is suggested to be stronger in gram-positive than gram-negative bacteria (17). The combination of toluidine blue for staining and polysulfonic filters constitutes a unique combination that allows specific bacterial staining with negligible background staining of the filters.
Milk samples from cows with moderate or severe clinical mastitis frequently have a high number of somatic cells (up to several million per milliliter of milk), fibrin particles, and substances causing problems in filtration process. Such substances are not readily dissolved, even after the described reagent (reagent A) is applied. Such material may, however, be removed by a prefiltration step applying a hydrophobic filter made from polyurethane or polypropylene. The hydrophobic materials used in the prefiltration stage allow bacteria to pass through, whereas the solid material, including the SCC, is retained. Some bacteria might be trapped in the inflammatory debris and therefore do not enter the eluate. This may influence the detection limit of the test in mastitic milk compared with spiked milk. The recovery of viable bacteria after the prefiltration stage was not possible since mastitic milk was always added to reagent A (pH 12.5) and no bacteria were alive following prefiltration.
The technique developed in this study correctly identified the bacteria in 51 of 55 milk samples (93%). Three milk samples containing S. aureus tested negative, however. All these samples contained a high degree of solid particles, and it is possible that the bacteria were retained with particles and inflammatory debris in milk during the prefiltration stage. One milk sample found to be free of bacteria by culture was identified as weakly positive for the presence of gram-positive bacteria. This sample contained a visible amount of blood, which may account for the weakly false-positive result. One milk sample containing Streptococcus sp. was partially decolorized after application of 10% ethanol, pH 2.9. Traditional Gram staining of this isolate as well as streptococci isolated from other cows with mastitis confirmed the Gram stain-variable property of some strains isolated from cows with mastitis in Norwegian dairy herds.
Staphylococci, streptococci, and coliform bacteria caused, respectively, 46, 22, and 13.5% of cases of moderate or severe clinical mastitis in Norway, whereas 14% of samples from quarters of mammary glands of cows with clinical mastitis contained no bacteria (2). According to our knowledge, moderate or severe clinical mastitis is typically associated with a high number of a single species of bacteria, normally above 106 bacteria/ml of milk. Consequently, the sensitivity established for the new test should be sufficient for detection of most cases of moderate or severe clinical mastitis caused by bacteria. In a study performed in cows with moderate or severe clinical mastitis, caused by coliform bacteria, the number of viable bacteria was estimated to be >106 CFU/ml of milk sample in 78% of the tested cows (13). Since the low detection limit of the test is >106/ml of milk, the number of bacteria (viable as well as unviable) should be at least 106/ml of milk when the test result is positive. Mastitic glands with bacterial counts below 106 bacteria/ml often indicate chronic or mild infection. Such infections should be treated differently from an acute infection based on the evaluation of the history of infection before deciding the treatment strategy. Since the filtration method detects viable as well as nonviable bacteria in mastitic milk, the results obtained by the filtration method may not always correlate to the results obtained by culture.
For effective treatment of clinical mastitis, reliable and rapid diagnosis of the nature of the inflammation is essential. The most accepted method for diagnosis of a case of moderate or severe clinical mastitis is observation of the clinical symptoms. A quarter of mammary gland with moderate or severe clinical mastitis shows visible changes and a high score of somatic cells in the milk, as shown by the California mastitis test. However, high SCCs are not necessarily correlated to the presence of bacteria in the udder. Optimal use of antibiotics in treatment of mastitis requires far better differential diagnostic tools that can provide on-site guidance to the veterinarian. Broad-spectrum antibiotics should be avoided unless the nature of the infection and the general health condition of a diseased animal definitely indicate such treatment. In Norway, it is recommended that mastitis caused by coliform bacteria should not be treated by antibiotics, whereas streptococcal and staphylococcal mastitis should be treated with penicillin (3). The self-cure rate of mastitis caused by coliform bacteria is close to 90%, and it has been suggested that therapy of E. coli mastitis should focus on anti-inflammatory therapy, whereas antibiotics should be used rationally (19). Therefore, a test aiming to differentiate whether a case of mastitis is caused by bacterial infection, and if so whether the infection is caused by either staphylococci and streptococci or coliform bacteria, is essential. The frequency of resistance against penicillin among staphylococci isolated from mastitis is low in Norwegian dairy herds (2). S. aureus causes approximately 38% of moderate or severe clinical mastitis in Norway, but only 4.2% demonstrate resistance against penicillin. In a herd with penicillin-resistant staphylococcus isolates, most of the staphylococci in the herd also show resistance to this drug. This indicates that the clinician is likely to know which herds exhibit resistance problems.
Most of the previous filtration studies (8, 11, 18) for detection of bacteria in milk were based on filtration of milk through a polycarbonate filter, subsequent staining by acridine orange, and visualization of stained bacteria with a microscope. The prerequisite for all these techniques was that the samples were treated so that bacteria remained sufficiently intact to keep their DNA intracellular. In one version of these methods, the bacteria were visualized with a monoclonal antibody conjugated to alkaline phosphatase (6). However, none of these studies tested mastitic milk samples, which may contain solid particles that render filtration of the sample difficult.
Wallis and Melnick (24) have presented a method in which bacteria in urine were treated with a solution causing their retention in a filter with a net positive electrostatic surface charge. In contrast to this method, we have found that bacteria and bacterial fragments could be retained in certain filters with neutral net charge and that gram-negative bacteria lost their stain after treatment with ethanol at acidic pH. Neither the method developed by Wallis and Melnick nor the other methods (9, 11, 14, 18, 22) have, however, provided a rapid procedure by which bacteria could be detected in mastitic milk and the results could be read by the naked eye.
The Limast-test (Maybec Vettech, Hägersten, Sweden) is the only method on the market used as a cow side test for detection of bacteria in milk from cows with moderate or severe clinical mastitis. The test takes up to 15 min and detects only gram-negative bacteria. The lack of classification properties leaves a negative result inconclusive as to the choice between an infection with gram-positive bacteria and the absence of bacterial infection.
The filtration test presented herein may be used in order to make a quick decision for treatment of moderate or severe clinical mastitis. The milk sample should, however, also be cultured to make prognostic and control decisions. In conclusion, a technique was developed for detection and classification of bacteria in milk from cows with moderate or severe clinical mastitis. The effective time to perform the technique is approximately 5 min. We are working to adapt the method as a cow side test.
ACKNOWLEDGMENTS
This work was supported by The National Center for Veterinary Contract Research and Commercial Services Ltd. (VESO), Oslo, Norway; FORNY Østlandet, Oslo Research Park, Norway; and Procaryo AS, Oslo, Norway.
We thank the Mastitis Laboratory in Molde, Norway, and The Mastitis Laboratory, National Veterinary Institute, Oslo, Norway for providing milk samples.
REFERENCES
- 1.Allman R, Manchee R, Lloyd D. Flow cytometric analysis of heterogenous bacterial populations. In: Lloyd D, editor. Flow cytometry in microbiology. London, England: Springer-Verlag; 1993. pp. 27–47. [Google Scholar]
- 2.Anonymous. Annual report of mastitis laboratories in Norway. Oslo, Norway: National Veterinary Institute; 1999. . (In Norwegian.) [Google Scholar]
- 3.Anonymous. Strategies to reduce the usage of antibacterial agents in cattle production, especially for treatment of mastitis. Guidelines of the Norwegian Cattle Health Service. Aas, Norway: Norwegian Cattle Health Service; 1996. . (In Norwegian.) [Google Scholar]
- 4.Anonymous. Routines for mastitis diagnostics. Oslo, Norway: State Veterinary Laboratory of Norway; 1993. . (In Norwegian.) [Google Scholar]
- 5.Anonymous. Laboratories methods for use in mastitis work. Document 132. Brussels, Belgium: International Dairy Federation; 1981. [Google Scholar]
- 6.Batina P, Arnold F, Bedouet L, Robreau G, Talbot F, Malcoste R. Monoclonal antibody detection of Clostridium microcolonies directly on membrane used for milk filtration. J Appl Microbiol. 1997;82:619–624. [PubMed] [Google Scholar]
- 7.Beveridge T J, Graham A A. Surface-layers of bacteria. Microbiol Rev. 1991;55:684–705. doi: 10.1128/mr.55.4.684-705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binnerup S J, Højberg O, Sørensen J. Gram characterisation determined on single cells and the microcolony level of bacteria immobilized on polycarbonate membrane filters. J Microbiol Methods. 1998;31:185–192. [Google Scholar]
- 9.Davey H M, Kell D B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single cell analyses. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Oliveria A P, Watts J L, Salmon S A, Aarestrup F M. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and United States. J Dairy Sci. 2000;83:855–862. doi: 10.3168/jds.S0022-0302(00)74949-6. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Astorga A, Hijarrubia M J, Lázaro B, Barcina I. Effect of the pre-treatments for milk samples filtration on direct viable cell counts. J Appl Bacteriol. 1996;80:511–516. doi: 10.1111/j.1365-2672.1996.tb03250.x. [DOI] [PubMed] [Google Scholar]
- 12.Jawets E, Melnick J L, Adelberg E A, editors. Review of medical microbiology. Los Altos, Calif: Appelton & Lange; 1987. Cell structure; pp. 1–29. [Google Scholar]
- 13.Katholm J, Haubro Andersen P. Acute coliforms mastitis in dairy cows: endotoxin and biochemical changes in plasma and colony-forming units in milk. Vet Rec. 1992;131:513–514. doi: 10.1136/vr.131.22.513. [DOI] [PubMed] [Google Scholar]
- 14.Mason D J, Shanmuganathan S, Mortimer F C, Gant V A. A fluorescent gram stain for flow cytometry and epifluorescence microscopy. Appl Environ Microbiol. 1998;7:2681–2685. doi: 10.1128/aem.64.7.2681-2685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin D E, Constable P D, McCoy G C. Use of clinical parameters for differentiation of gram-positive and gram-negative mastitis in dairy cows vaccinated against lipopolysaccharide core antigens. J Am Vet Med Assoc. 1998;212:1423–1431. [PubMed] [Google Scholar]
- 16.Noda Y, Kanemasa Y. Determination of surface charge of some bacteria by colloid titration. Physiol Chem Phys Med NMR. 1984;16:263–274. [PubMed] [Google Scholar]
- 17.Noda Y, Toei K. A new bacterial staining method involving Gram stain with theoretical consideration of the staining mechanism. Microbios. 1992;70:49–55. [PubMed] [Google Scholar]
- 18.Pettipher G, Mansell R, McKinnon C H, Cousins C M. Rapid membrane filtration-epifluorescent microscopy technique for direct enumeration of bacteria in raw milk. Appl Environ Microbiol. 1980;39:423–429. doi: 10.1128/aem.39.2.423-429.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandholm M, Pyörälä S. Coliform mastitis, endotoxins mastitis—endotoxin shock. In: Sandholm M, Honkanen-Buzalski T, Kaartinen L, Pyörälä M, editors. The bovine udder and mastitis. Helsinki, Finland: Faculty of Veterinary Medicine, University of Helsinki; 1995. pp. 149–160. [Google Scholar]
- 20.Schalm O W, Caroll E J, Jain N C. Bovine mastitis. Philadelphia, Pa: Lea & Febiger; 1971. [Google Scholar]
- 21.Shapiro H M. Practical flow cytometry. 3rd ed. New York, N.Y: Alan R. Liss, Inc.; 1995. pp. 367–425. [Google Scholar]
- 22.Sizemore R K, Caldwell J J, Kendrick A S. Alternate Gram staining technique using a fluorescent lectin. Appl Environ Microbiol. 1990;56:2245–2247. doi: 10.1128/aem.56.7.2245-2247.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thrusfield M, editor. Veterinary epidemiology. 2nd ed. Oxford, England: Blackwell Science Ltd.; 1995. Diagnostic testing; pp. 266–285. [Google Scholar]
- 24.Wallis, C., and J. L. Melnick. June 1982. Detection of bacteria. U.S. patent 4,336,337.