Abstract
We have previously identified a genetic variant, rs34331122 in the 22q11.21 locus, as being associated with breast cancer risk in a genome-wide association study. This novel variant is located in the intronic region of the T-box transcription factor 1 (TBX1) gene. Cis-expression quantitative trait loci analysis showed that expression of TBX1 was regulated by the rs34331122 variant. In the current study, we investigated biological functions and potential molecular mechanisms of TBX1 in breast cancer. We found that TBX1 expression was significantly higher in breast cancer tumor tissues than adjacent normal breast tissues and increased with tumor stage (P < 0.05). We further knocked-down TBX1 gene expression in three breast cancer cell lines, MDA-MB-231, MCF-7 and T47D, using small interfering RNAs and examined consequential changes on cell oncogenicity and gene expression. TBX1 knock-down significantly inhibited breast cancer cell proliferation, colony formation, migration and invasion. RNA sequencing and flow cytometry analysis revealed that TBX1 knock-down in breast cancer cells induced cell cycle arrest in the G1 phase through disrupting expression of genes involved in the cell cycle pathway. Furthermore, survival analysis using the online Kaplan–Meier Plotter suggested that higher TBX1 expression was associated with worse outcomes in breast cancer patients, especially for estrogen receptor-positive breast cancer, with HRs (95% CIs) for overall survival (OS) and distant metastasis free survival (DMFS) of 1.5 (1.05–2.15) and 1.55 (1.10–2.18), respectively. In conclusion, our results suggest that the TBX1 gene may act as a putative oncogene of breast cancer through regulating expressions of cell cycle-related genes.
We investigated the biological functions and potential molecular mechanisms of TBX1 in breast cancer. Our results suggest that the TBX1 gene may act as a putative oncogene of breast cancer through regulating expressions of cell cycle-related genes.
Introduction
The latest global cancer burden statistics produced by the International Agency for Research on Cancer estimated 2.3 million (11.7%) new breast cancer cases in 2020, surpassing lung cancer and ranking as the most commonly diagnosed cancer (1). Breast cancer also remains the leading cause of cancer death among women worldwide (1–3). The increasing incidence and poor prognosis of patients with advanced breast cancer are still critical for global breast cancer control and treatment (4,5). Therefore, identifying unique expression patterns and regulatory mechanisms of novel breast cancer oncogenes will contribute to the discovery of potential intervention strategies and actionable drug targets for breast cancer patients.
We previously identified a genetic variant, rs34331122 in the 22q11.21 locus, to be associated with breast cancer risk in a genome-wide association study (GWAS) (6). This single nucleotide polymorphism (SNP) is located in the intronic region within the T-box (TBX) transcription factor 1 (TBX1) gene. Cis-expression quantitative trait loci (eQTL) analysis showed that expression of TBX1 was regulated by the rs34331122 variant. TBX1 belongs to the TBX transcription factor gene family, a group of evolutionarily conserved transcription factors with specific DNA-binding motifs, which have been shown to function as transcriptional regulators (7,8). TBX transcription factors play vital roles in embryonic development, cell fate specification and differentiation, and tissue organization in vertebrates and invertebrates (9,10). Dysregulation of TBX transcription factors was shown to be directly related to oncogenesis (11,12). The role of TBX1 in morphogenesis, as well as cardiac and vascular development, has been well studied (13,14). Recent studies have found that TBX1 could regulate tumorigenesis in skin, thyroid and prostate cancers (15–18). Other members of the TBX family, including TBX2 and TBX3, have been demonstrated to affect breast cancer progression (19–21). However, the associations and biological functions of TBX1 in breast cancer development and prognosis remain largely unknown.
In this study, we investigated the biological role and potential mechanisms of the TBX1 gene in human breast cancer cell lines using in vitro functional studies and further elucidated the clinical association of TBX1 expression with breast cancer prognosis.
Materials and methods
Cell culture
Three human breast cancer cell lines, including two hormone receptor positive cell lines (MCF-7 and T47D) and one triple-negative breast cancer cell line (MDA-MB-231), were used in this study. The cell lines MDA-MB-231 and MCF-7 were obtained from the American Type Culture Collection (ATCC), which the cells were authenticated by short tandem repeat. The T47D cell line was a kind gift from Dr. Jennifer Pietenpol from Vanderbilt University Medical Center and was authenticated by short tandem repeat assay by ATCC. All three cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Gibco) at 37 °C with 5% CO2 in a humidified incubator.
RNA interference
MDA-MB-231, T47D and MCF-7 cells were plated at 1.25 × 105 cells/well in 6-well plates and reverse-transfected with small interfering RNAs (siRNAs, Dharmacon) targeting TBX1 messenger RNA (mRNA) at 10 nM using RNAiMAX (Life Technologies), following the manufacturer’s protocol. Three siRNAs targeting different sites of TBX1 were used for transfections, and the target sequences for each siRNA have been provided in Supplementary Table S1, available at Carcinogenesis Online. A non-targeting control siRNA (AllStars Neg Control siRNA, Qiagen) and a positive control (PC) siRNA (AllStars Hs Cell Death Control siRNA, Qiagen) were used as the negative control (NC) and PC, respectively. Knock-down efficiency was assessed at 36–48 h post-transfection by quantitative real-time polymerase chain reaction (qPCR), and data were presented as relative values to cells transfected with the NC siRNA (Supplementary Figure S1, available at Carcinogenesis Online).
RNA isolation and qPCR
Total RNA was isolated from the three breast cancer cell lines using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qPCR amplification was performed on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) using Luna Universal qPCR Master Mix (New England BioLabs). Relative mRNA expression levels were calculated using the ∆∆Ct method, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene. Primer sequences are listed in Supplementary Table S2, available at Carcinogenesis Online.
Cell viability assay
Cell viability was determined using the alamarBlue™ Cell Viability Reagent (Thermo Fisher) assay, as described previously (22). MDA-MB-231, T47D and MCF-7 cells were plated at 5 × 103 cells/well in 96-well plates and reverse-transfected with TBX1, NC, and PC siRNAs. After 96 hours post-transfection, 10 µl of alamarBlue reagent was added to each well (1:10 dilution), incubated at 37°C for 4–6 h, and fluorescence (ex570 nm/em585 nm) was measured on a BioTek Synergy HT plate reader. Percent relative cell viability was calculated as: (mean siTBX1 value/ mean NC siRNA value) × 100. Figures represent data obtained from three independent experiments with six replicates per condition in each experiment.
Colony formation assay
After 16 h post-transfection, siRNA-transfected cells were harvested and re-seeded in 6-well plates at a density of 2000 cells per well in 2 ml of antibiotic-free culture media and allowed to proliferate for 10–14 days (22). Colonies, as defined to consist of ≥ 50 cells, were then fixed with 10% neutral buffered formalin (Sigma–Aldrich) for 30 min, stained with crystal violet (0.1% w/v in H2O) (Sigma–Aldrich) for 1 h and counted using ImageJ (National Institutes of Health, NIH). Colony formation efficiency (CFE) was normalized to the NC siRNA group and expressed as the percent (%) of NC. Figures represent data obtained from three independent experiments.
Cell migration and invasion assay
Cell migration and invasion assays were performed in 24-well plates using inserts with an 8 μm pore size (Millipore) and coated with (invasion assay) or without (migration assay) Corning Matrigel matrix, as described previously (23). MDA-MB-231 and MCF-7 cells were transfected with two TBX1 siRNAs and a NC siRNA for 24 h. Transfected cells were re-seeded into the upper chamber of the insert containing 200 µL serum-free media at 5 × 104 cells/well for MDA-MB-231 and 1 × 105 cells/well for MCF-7 cells. 750 µl of media containing 10% FBS was placed in the lower chamber as a chemoattractant. After incubation at 37°C for 48 and 72 h, cells migrating through the membrane were fixed in 10% neutral buffered formalin, stained with 0.1% crystal violet and observed under a microscope. Five visual fields from each insert were randomly selected for cell counting. To further quantify the cell density, crystal violet was eluted using 33% acetic acid, and absorbance at 590 nm was measured on a BioTek Synergy HT plate reader. Absorbance was normalized to the NC and expressed as relative absorbance. Figures represent data from three independent experiments. T47D cells were not assayed due to low migration ability.
RNA sequencing analysis
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. The integrity of RNA quality was confirmed by running an aliquot on the Agilent Bioanalyzer, and RNA concentration was measured using the Qubit RNA fluorometry assay. mRNA enrichment and cDNA library preparation were performed utilizing the stranded mRNA (polyA-selected) sample prep kit. RNA sequencing (RNA-Seq) was performed at paired-end 150 bp on the Illumina NovaSeq 6000. A minimum of 30M reads was obtained for each sample.
The RNA-Seq raw data quality control was analyzed by FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The STAR (24) two-pass method was used for raw data alignment to the human reference genome (hg38). Gene expression levels were determined from aligned BAM files using featureCounts (25). GENCODE v30 (26) was used for coding gene and noncoding RNA annotations in the human genome. Principal component analyses (PCA) were computed using all identified genes to visualize transcriptome variations between samples. DESeq2 (27) was used to quantify differential expression genes (DEGs) between NC and TBX1 knock-down samples. DEGs were identified with the false discovery rate (FDR) < 0.05 and fold change (FC) > 2. DEGs were used for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database pathway analyses by using WebGestalt (28). GO was classified as the cellular component (CC), biological process (BP) and molecular function (MF). Benjamini–Hochberg (BH) adjustment was used to identify enrichment GO categories and KEGG pathways, and only terms with FDR < 0.05 were considered significantly enriched. All statistical analyses were performed in R 3.6.0.
Cell cycle analysis
Propidium iodide staining was used to analyze cell cycle phase distributions. All three breast cancer cell lines (MDA-MB-231, MCF-7 and T47D) were transfected with TBX1 and NC siRNAs for 48 h. Transfected cells were trypsinized, washed with cold phosphate buffered saline (PBS) and then fixed with 70% ethanol at 4°C overnight. After another wash, cells were stained by FxCycleTM PI/RNase solution (Invitrogen) for 30 min at room temperature and then analyzed on a Fortessa cell analyzer (BD Biosciences). Cell cycle analyses were performed using FlowJo™ Software.
Statistical analysis
UALCAN is a publicly available online portal used to perform in-depth analyses of The Cancer Genome Atlas (TCGA) gene expression data (29). We used UALCAN analysis to compare TBX1 expression between breast cancer tumor tissues (n = 1097) and adjacent normal tissues (n = 114), as well as various subgroups, based on breast cancer stages and subtypes. Expression levels were normalized as transcripts per million reads (TPM). We further obtained gene expression profiles with the HTSeq fragments per kilobase million (FPKM) format of breast invasive carcinoma (BRCA) samples from the TCGA data portal (https://cancergenome.nih.gov/) and converted RNA-Seq data from the FPKM format to the TPM format. Expression differences of TBX1 in paired breast tumor tissues and normal tissues (n = 108) were further analyzed using the two-tailed paired t test.
The association of TBX1 expression with breast cancer prognosis for up to 1,809 patients was assessed using the Kaplan–Meier Plotter online tool (30). Breast cancer patients were divided into high- and low-expression groups by median mRNA expression levels. Differences were estimated and compared for overall survival (OS) and distant metastasis free survival (DMFS) of breast cancer patients by TBX1 mRNA expression using log-rank test and hazard ratios (HR) with 95% confidence intervals (CI), derived from univariate Cox regression models.
All in vitro functional assays were repeated in at least three experiments. Data are expressed as mean ± SD. Two-tailed t-tests were used to determine significant differences between the two groups. Comparisons between more than two groups were analyzed by one-way ANOVA, followed by Dunnett’s multiple comparison test. Statistics were performed by GraphPad Prism 8.0 statistical software. A P-value < 0.05 was considered as statistically significant unless otherwise specified.
Results
Knock-down of TBX1 inhibited breast cancer cell viability and colony formation efficiency
According to data from The Human Protein Atlas database (31), TBX1 was moderately expressed in breast cancer cell lines MCF-7 and T47D. siRNAs were further used to knock-down TBX1 transcript expression in three breast cancer cell lines, MDA-MB-231, MCF-7 and T47D. After transfection with three TBX1 siRNAs for 36–48 h, TBX1 expression was significantly decreased in all three breast cancer cell lines. qPCR results showed that compared with NC, TBX1 expression decreased by 55% to 90% with the three different siRNAs in all three cell lines (Supplementary Figure S1, available at Carcinogenesis Online; P-value < 0.01).
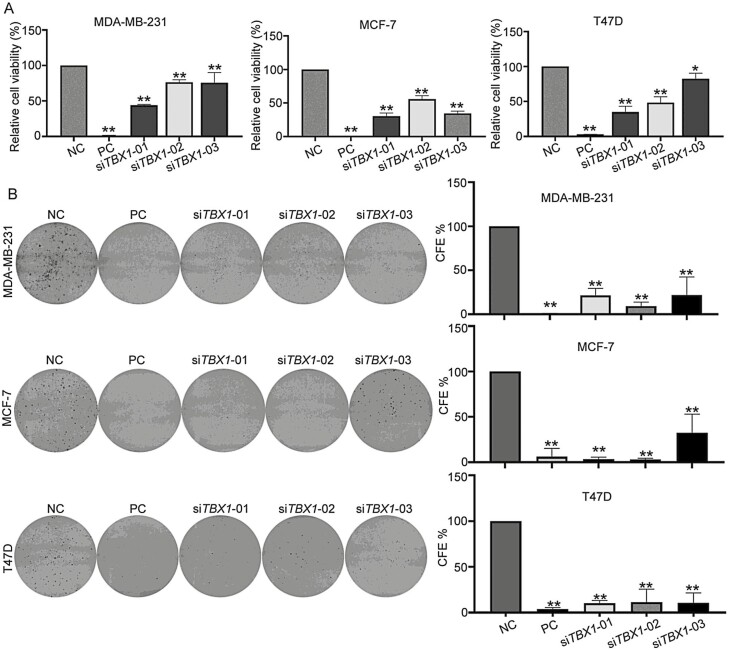
Cell viability and long-term colony formation assays were conducted following TBX1 knock-down. Compared with the NC siRNA transfection, TBX1 siRNA transfection significantly decreased cell viability in all three breast cancer cell lines (Figure 1). Compared with NC, normalized cell viabilities were 44.1% to 75.7%, 30.3% to 55.6%, and 35.0% to 82.5% in MDA-MB-231, MCF-7, and T47D cells transfected with three TBX1 siRNAs, respectively (Figure 1A, P-value < 0.05). CFE was further evaluated to detect the role of TBX1 in long-term cell proliferation and clonogenic survival. The results showed that CFE was also significantly decreased in all three breast cancer cells by all three TBX1 siRNAs. Compared with NC, normalized CFEs were 9.2% to 21.6%, 3.2% to 36.5%, and 10.5% to 11.3% for MDA-MB-231, MCF-7, and T47D cells transfected with three TBX1 siRNAs, respectively (Figure 1B, P-value < 0.01).
Figure 1.
Effect on cell viability and colony formation efficiency by silencing TBX1 expression in breast cancer cells. (A) MDA-MB-231, MCF-7 and T47D cells were transfected with three TBX1 siRNAs (siTBX1-01, siTBX1-02, siTBX1-03), negative control siRNA (NC) and positive control siRNA (PC) for 96 h. Relative cell viability was normalized to NC group and expressed as percent (%) of control. Bar chart presented as mean ± SD from three independent experiments. (B) Colony formation (CF) ability of MDA-MB-231, MCF-7 and T47D cells transfected with three TBX1 siRNAs (siTBX1-01, siTBX1-02, siTBX1-03), NC siRNA and PC siRNA, respectively. The CFE was normalized to NC siRNA group and expressed as percent (%) of control. All the experiments were repeated three times independently and presented as mean ± SD. P-values were determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. * P-value < 0.05, ** P-value < 0.01.
Knock-down of TBX1 inhibited breast cancer cell migration and invasion
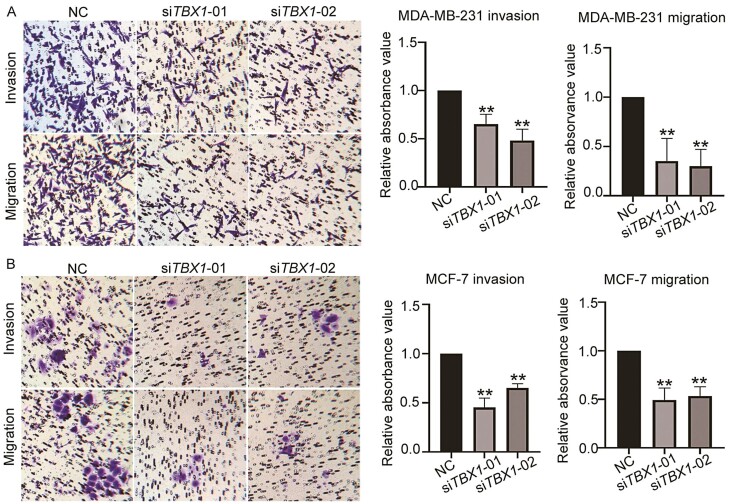
We further investigated the effects of TBX1 knock-down on breast cancer cell migration and invasion by conducting transwell assays in MDA-MB-231 and MCF-7 cell lines. As shown in Figure 2, knock-down of TBX1 expression significantly decreased cell migration and invasion ability in both breast cancer cell lines. When normalized to NC transfected cells, 35.2% (siTBX1-01) and 29.9% (siTBX1-02) of MDA-MB-231 cells had migrated; 65.1% (siTBX1-01) and 48.2% (siTBX1-02) of transfected MDA-MB-231 cells had invaded the membrane (P-value < 0.01). In MCF-7 cells, 49.3% (siTBX1-01) and 53.3% (siTBX1-02) of transfected cells had migrated compared with NC transfected cells. 45.3% (siTBX1-01) and 65.1% (siTBX1-02) of transfected cells had invaded the membrane when compared with NC transfected cells (P-value < 0.01).
Figure 2.
Effect on cell migration and invasion ability by silencing TBX1 expression in breast cancer cells. Representative images and quantitative analysis of migration and invasion of MDA-MB-231(A) and MCF-7 (B) cells transfected with two TBX1 siRNAs (siTBX1-01, siTBX1-02) or NC siRNA, respectively. Relative migrated and invaded cells were normalized to the NC siRNA group and expressed as percent (%) of control. All the experiments were repeated three times independently and presented as mean ± SD. P-values were determined by one-way ANOVA followed by Dunnett’s multiple comparisons test: ** P-value < 0.01.
Differential gene expression profiles in TBX1 knock-down MDA-MB-231 cells
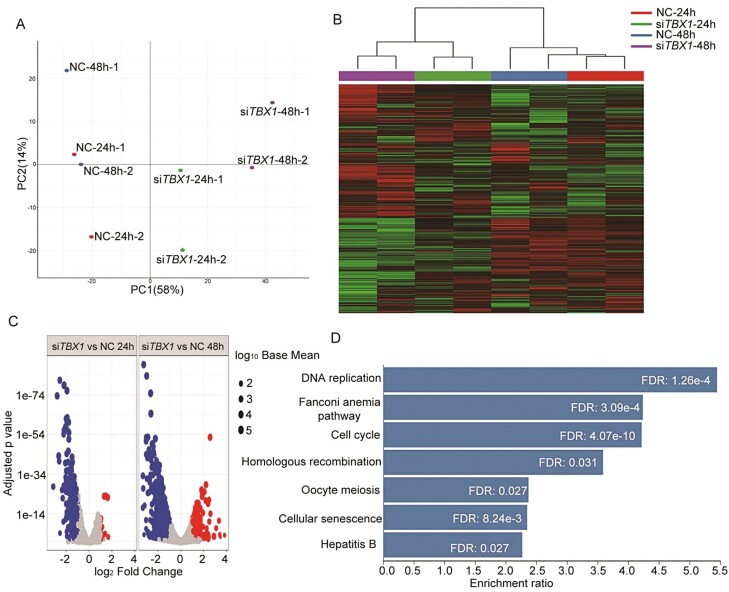
Transcriptomic analysis using RNA-Seq data was performed following siRNA-mediated knock-down of TBX1- and NC-transfected MDA-MB-231 cells to identify downstream genes and signaling pathways regulated by the TBX1 gene. MDA-MB-231 cells were treated with TBX1 siRNA (siTBX1-01) and NC siRNA for 24 and 48 h in independent duplicates. Sequencing reads were aligned to a human reference genome (hg38), with over 88% of the reads being uniquely aligned (Supplementary Table S3, available at Carcinogenesis Online).
Expressions for each gene were compared between NC and TBX1 knock-down samples. Genes with a FC > 2 and FDR < 0.05 were considered as DEGs. There were a total 310 and 1195 DEGs in TBX1 knock-down, compared with NC, 24- and 48-h treatment groups, respectively. Clustering and PCA of gene expression patterns in our PCA plots indicated that duplicate samples of each group clustered together very well (Figure 3A). TBX1 knock-down and NC samples segregated well, indicating that gene expression differences between the two groups were from inter-group variability rather than individual heterogeneity. The heat map also showed significantly different expression patterns between TBX1 knock-down and NC groups (Figure 3B). Volcano plots showed the distributions of different genomic profiles between the compared groups (Figure 3C).
Figure 3.
Deregulated genes expression profile after TBX1 knock-down in MDA-MB-231 cells. MDA-MB-231 cells were transfected with TBX1 (siTBX1-01) and NC siRNA for 24 and 48 h, respectively. RNA samples from duplicate treatments were collected and analyzed by RNA sequencing. (A) Principal components analysis performed on RNA sequencing data obtained from eight treatment samples. Principal Component 1 (PC1, x-axis) represents 58% and PC2 (y-axis) represents 24% of total variation in the data. (B) Heat map demonstrated the top 100 differentially expressed genes identified with RNA sequencing in negative control siRNA (NC) and TBX1 siRNA (siTBX1) transfected MDA-MB-231 cell samples. Red color indicates relative high expression and green color indicates relative low expression. (C) Volcano plot for comparison of RNA expression profiles between duplicate TBX1 siRNA (siTBX1) and NC samples. The x-axis indicates the differential expression profiles, plotting the fold-change in a log scale; the y-axis indicates the statistical significance of difference in expression. Blue color dots represent downregulated genes and red ones represent upregulated genes with FC > 2 and FDR < 0.05. (D) KEGG pathway analysis using DEGs between the siTBX1 and NC treatment group for 48 h, the significantly enriched pathways were presented with FDR < 0.05.
To identify pathways that are regulated by the TBX1 gene, GO and KEGG enrichment analyses were further performed using the list of DEGs. The most significant GO biological processes included cell cycle phase transition and nuclear division after a 48-hour treatment with the TBX1 siRNA (Supplementary Figure S2, available at Carcinogenesis Online). KEGG analysis indicated that cell cycle was the most significantly changed pathway in TBX1 knock-down cells (Figure 3D), with 30 downregulated and 2 upregulated genes (Supplementary Figure S3, available at Carcinogenesis Online).
Knock-down of TBX1 induced cell cycle arrest by regulating downstream gene expressions
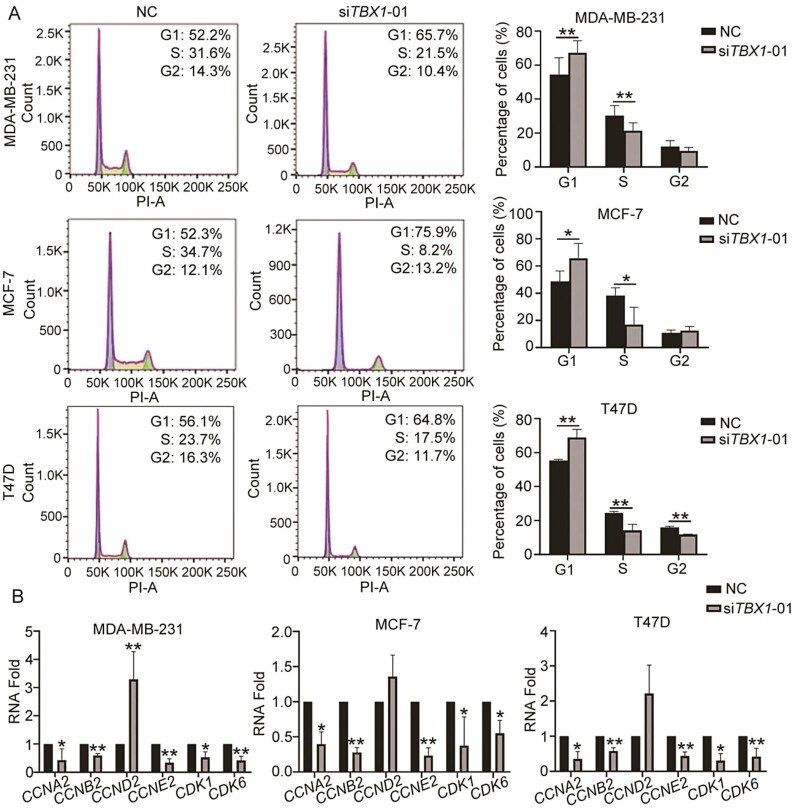
We further examined the effects of TBX1 on the breast cancer cell cycle by using propidium iodide staining and flow cytometry analysis. TBX1 knock-down cells showed an accumulation in the G1 phase and decreased S phase in all three breast cancer cell lines when compared to the NC (Figure 4A, P-value < 0.05). The differential expressions of key genes in the cell cycle pathway, validated by qPCR, further revealed that knocking-down TBX1 expression resulted in significantly lower levels of cyclin A2 (CCNA2), cyclin B2 (CCNB2), cyclin E2 (CCNE2), cyclin-dependent kinase 1 (CDK1) and cyclin-dependent kinase 6 (CDK6) in all three breast cancer cell lines (Figure 4B). In addition, knocking-down TBX1 expression resulted in a significantly higher mRNA level of cyclin D2 (CCND2) in MDA-MB-231 cells. The similar trend of CCND2 differential expression was also found in MCF-7 and T47D cells, but the difference was not statistically significant (Figure 4B). Furthermore, using the JASPAR database (32) to predict the TBX1 binding motif, we found that TBX1 may bind to the promoter regions of the above cell cycle related genes (Supplementary Table S4, available at Carcinogenesis Online).
Figure 4.
Effect on cell cycle distribution by silencing TBX1 expression in breast cancer cells. MDA-MB-231, MCF-7 and T47D cells were transfected with TBX1 siRNA (siTBX1-01) and NC siRNA for 48 h. (A) Cell cycle distribution was analyzed by propidium iodide staining and subsequent analysis by flow cytometry. The percentage of different cell cycle phase were presented as mean ± SD and compared with NC group from three independent experiments. P-values were determined by two-tailed paired t-test. (B) RNA expression of cell cycle genes was determined by qPCR. Relative RNA fold of each gene was compared to NC and showed as mean ± SD from three independent experiments. P-values were determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. * P-value < 0.05, ** P-value < 0.01.
High TBX1 mRNA expression predicted worse prognosis in breast cancer patients
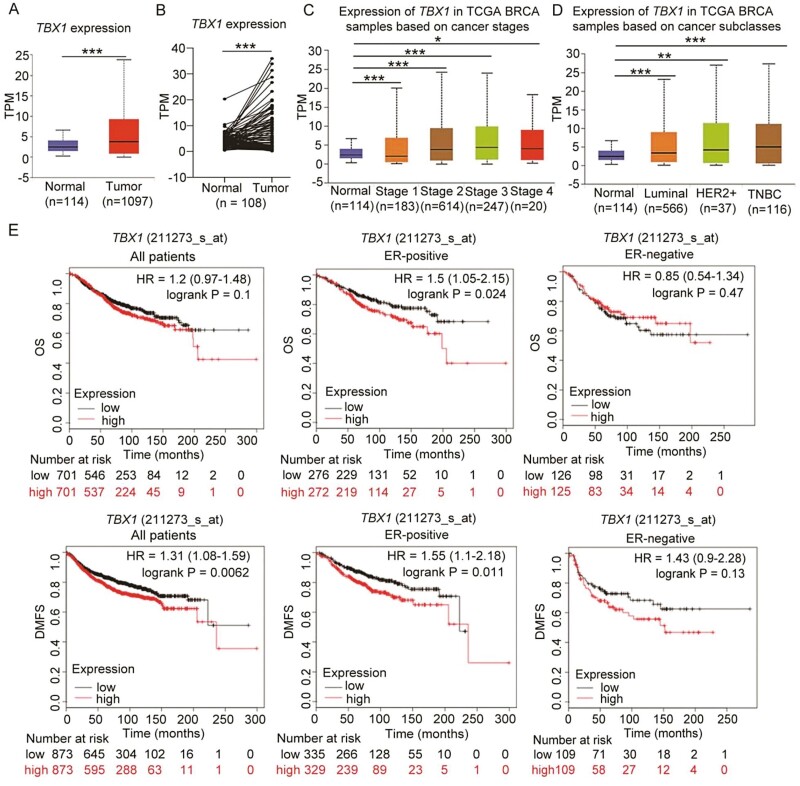
To evaluate potential functions of TBX1 in vivo, TBX1 transcript expression was further analyzed in TCGA-BRCA samples. Using UALCAN analysis (29), we found that TBX1 expression levels were higher in breast cancer primary tumor tissues (n = 1097) compared with adjacent normal tissues (n = 114) (Figure 5A, P-value = 1.62E-12). In the paired TCGA-BRCA data, TBX1 expression was also increased in tumor tissues than adjacent normal tissues (n = 108) (Figure 5B, P-value < 0.001). Furthermore, TBX1 expression levels increased significantly with tumor stage (Figure 5C, P-value < 0.05), and upregulated TBX1 expression was also observed in different cancer subtypes, including luminal, human epidermal growth factor receptor 2 (HER2)-positive, and triple-negative breast cancer. The highest expression was found in invasive triple-negative breast cancer tissues when compared with normal tissues (Figure 5D, P-value < 0.01).
Figure 5.
High TBX1 gene expression predicted worse prognosis in breast cancer patients. (A) TBX1 gene expression in breast cancer tissue (primary tumor) and adjacent normal tissue were obtained from UALCAN database by analyzing the TCGA-BRCA samples. (B) TBX1 expression in the tumors and adjacent normal tissues of paired TCGA-BRCA data as analyzed by two-tailed paired t-test. TBX1 expression in different breast cancer stage (C), and various breast cancer subclasses. (D) were obtained from UALCAN database by analyzing the TCGA-BRCA samples. * P-value < 0.05, ** P-value < 0.01, *** P-value < 0.001. (E) Kaplan–Meier curves for overall survival (OS) and distant metastasis free survival (DMFS) were created by using the Kaplan–Meier Plotter online analysis with all breast cancer patients, ER-positive and ER-negative breast cancer patients according to high and low TBX1 gene expression (Affymetrix probe IDs: 211273_s_at). Hazard ratio (HR) with 95% confidence interval and log-rank P values were calculated. A log-rank P value < 0.05 was considered as statistically significant.
Associations between TBX1 mRNA expression and clinical outcomes of breast cancer patients were analyzed using the Kaplan-Meier Plotter online survival analysis (30). We included the top two probes for TBX1 (236926_at, 211273_s_at) presented on Affymetrix platforms, based on the average expression in 1,809 breast cancer patients (Figure 5E; Supplementary Figure S4, available at Carcinogenesis Online). For all patients, results showed that TBX1 expression (probe: 211273_s_at) was positively associated with a significantly decreased DFMS, while no significant association was found with OS (Figure 5E). Furthermore, stratified analysis indicated that higher TBX1 expression was significantly associated with poor OS and DMFS in estrogen receptor (ER)-positive breast cancer patients, while similar associations were also identified in ER-negative breast cancer patients, though results were not significant (Figure 5E).
Discussion
In this study, we found that knocking-down of TBX1 expression by siRNA could inhibit cell viability, colony formation, migration, and invasion and induce a G1 phase arrest in both ER-positive and ER-negative breast cancer cell lines. These effects were confirmed by three siRNAs targeted to different TBX1 mRNA sequences, supporting the oncogenic role of TBX1 in breast cancer. Furthermore, using TCGA data, we found higher expression levels of the TBX1 gene in breast cancer tumor tissues compared with adjacent normal breast tissue. Kaplan-Meier analysis also showed evidence for the association between higher TBX1 expression and worse DMFS in breast cancer patients, and the association was more pronounced in ER-positive than ER-negative breast cancer patients (Figure 5E). A similar trend was observed with the probe 236926_st (Supplementary Figure S4, available at Carcinogenesis Online); however, this did not reach statistical significance, most likely due to the small sample size.
Previous studies have shown that TBX1 regulates tumor growth and metastasis in several cancer types. Wang et al. reported that overexpression of TBX1 inhibited tumor growth and invasiveness through PI3K/AKT and MAPK/ERK signaling pathways in thyroid cancer (15). Verdelli et al. found that TBX1 silencing exerted cell cycle arrest in parathyroid tumors (16). A recent study reported that TBX1 could promote prostate cancer growth through epigenetic control, thereby increasing ribosomal RNA (rRNA) gene transcription (18). Therefore, the role of TBX1 in cancer may be tissue-specific or cell-type dependent. The TBX gene family is an evolutionarily conserved transcription factor family, and studies have shown that TBX genes, including TBX2, TBX3 and TBX21, are involved in the genesis and progression of breast cancer (33,34). TBX2 was shown to be upregulated in BRCA1/2-associated breast cancers, and TBX3 overexpression was associated with advanced stage disease in ER-positive breast cancer (35). To the best of our knowledge, no studies investigating TBX1 function in breast cancer have been reported.
To identify possible oncogenic mechanisms of TBX1 in breast cancer, we performed RNA-Seq analysis after knocking-down of TBX1 mRNA levels. DEGs and pathway enrichment analyses indicated that the cell cycle pathway was significantly regulated by TBX1 in breast cancer. qPCR results validated changes in expressions of genes regulating cell cycle following TBX1 knock-down. Among these genes, our analysis included key cell cycle checkpoint genes including CCNA2, CCNB2, CCNE2, CDK6 and CDK1 (Figure 4). CCND could form active complexes with either CDK4 or CDK6, which in turn phosphorylate the retinoblastoma protein (Rb) and drive G1 to S phase progression (36). The decreased CDK6 level could inhibit the activation of the CCND/CDK6 complex and prevent G1 phase cells from entering the S phase even with higher CCND2 levels. These results indicate that silencing of TBX1 expression induced G1 phase arrest and inhibited cell cycle transition from the G1 to the S phase in breast cancer cells. We used the triple-negative breast cancer cell line, MDA-MB-231, in the RNA-Seq experiment. Additional RNA-Seq experiments using other breast cancer cell lines are warranted to explore potential differential regulating effects of TBX1 on gene expressions in other breast cancer cells.
As a transcription factor, TBX1 was shown to regulate expression by interacting with the SWI-SNF-like BAF complex and histone methyltransferases, with either transcriptional activator or repressor activity (7,15,37). Using JASPAR, an open-access database of curated, non-redundant binding profiles from published and experimentally defined transcription factor eukaryotic binding sites (32), we showed that TBX1 was predicted to bind with promoter regions of the above cell cycle-related genes, including CCNA2, CCNB2, CCNE2, CCND2, CDK6 and CDK2. However, whether TBX1 directly regulates the expressions of the above genes needs further validation.
In summary, our study indicates that TBX1 could promote cell proliferation, migration, invasion, and cell cycle progression in breast cancer. TBX1 gene expression was associated with worse prognosis in breast cancer patients. One major mechanism underpinning TBX1’s activity as a potential oncogene in breast cancer is through regulating the transcription of cell cycle genes. Our study identifies TBX1 as a putative oncogene for breast cancer and a potential molecular target for breast cancer therapies.
Supplementary Material
Acknowledgements
We thank the staffs at The Vanderbilt Technologies for Advanced Genomics (VANTAGE) for their help with the RNA sequencing assay. We thank Dr. Mary Shannon Byers for assistance with editing and manuscript preparation and Ms. Regina Courtney for laboratory assistance.
Glossary
Abbreviations
- OS
overall survival
- DMFS
distant metastasis free survival
- GWAS
genome-wide association study
- SNP
single nucleotide polymorphism
- ATCC
American Type Culture Collection
Funding
This work was supported, in part, by UL1 TR002243 from the National Center for Advancing Translational Sciences. X.S. was partially supported by a grant from National Cancer Institute (K99/R00CA230205), J.B. was partially supported by a grant from National Cancer Institute (R50CA211206), and Q.C. was partially support by a grant from National Cancer Institute (R01CA235553). The in vitro functional studies were conducted at the Survey and Biospecimen Shared Resources, which is supported in part by the Vanderbilt-Ingram Cancer Center (P30CA068485).
Conflict of Interest Statement
The authors declare no conflicts of interest.
References
- 1. Sung, H., et al. (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin., 71, 209–249. [DOI] [PubMed] [Google Scholar]
- 2. Siegel, R.L., et al. (2019) Cancer statistics, 2019. CA. Cancer J. Clin., 69, 7–34. [DOI] [PubMed] [Google Scholar]
- 3. Bray, F., et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin., 68, 394–424. [DOI] [PubMed] [Google Scholar]
- 4. Nagini, S. (2017) Breast cancer: current molecular therapeutic targets and new players. Anticancer. Agents Med. Chem., 17, 152–163. [DOI] [PubMed] [Google Scholar]
- 5. Karaman, S., et al. (2014) Mechanisms of lymphatic metastasis. J. Clin. Invest., 124, 922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shu, X., et al. (2020) Identification of novel breast cancer susceptibility loci in meta-analyses conducted among Asian and European descendants. Nat. Commun., 11, 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baldini, A., et al. (2017) Tbx1: transcriptional and developmental functions. Curr. Top. Dev. Biol., 122, 223–243. [DOI] [PubMed] [Google Scholar]
- 8. Naiche, L.A., et al. (2005) T-box genes in vertebrate development. Annu. Rev. Genet., 39, 219–239. [DOI] [PubMed] [Google Scholar]
- 9. Papaioannou, V.E. (2014) The T-box gene family: emerging roles in development, stem cells and cancer. Development, 141, 3819–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takashima, Y., et al. (2013) Regulation of organogenesis and stem cell properties by T-box transcription factors. Cell. Mol. Life Sci., 70, 3929–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abrahams, A., et al. (2010) The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life, 62, 92–102. [DOI] [PubMed] [Google Scholar]
- 12. Wan, Z., et al. (2016) T-box transcription factor brachyury promotes tumor cell invasion and metastasis in non-small cell lung cancer via upregulation of matrix metalloproteinase 12. Oncol. Rep., 36, 306–314. [DOI] [PubMed] [Google Scholar]
- 13. Tsuchihashi, T., et al. (2016) Modification of cardiac phenotype in Tbx1 hypomorphic mice. In Nakanishi, T., et al. (eds.), Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology. Springer, Tokyo, pp. 215–217. [PubMed] [Google Scholar]
- 14. Cioffi, S., et al. (2014) Tbx1 regulates brain vascularization. Hum. Mol. Genet., 23, 78–89. [DOI] [PubMed] [Google Scholar]
- 15. Wang, N., et al. (2019) TBX1 Functions as a tumor suppressor in thyroid cancer through inhibiting the activities of the PI3K/AKT and MAPK/ERK pathways. Thyroid, 29, 378–394. [DOI] [PubMed] [Google Scholar]
- 16. Verdelli, C., et al. (2017) Expression, function, and regulation of the embryonic transcription factor TBX1 in parathyroid tumors. Lab. Invest., 97, 1488–1499. [DOI] [PubMed] [Google Scholar]
- 17. Caprio, C., et al. (2020) TBX1 and basal cell carcinoma: expression and interactions with Gli2 and Dvl2 signaling. Int J Mol Sci., 21, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui, J., et al. (2020) TBX1 functions as a tumor activator in prostate cancer by promoting ribosome RNA gene transcription. Front. Oncol., 10, 616173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crawford, N.T., et al. (2019) TBX2 interacts with heterochromatin protein 1 to recruit a novel repression complex to EGR1-targeted promoters to drive the proliferation of breast cancer cells. Oncogene, 38, 5971–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu, P., et al. (2019) Long non-coding RNA TTN-AS1 promotes the metastasis in breast cancer by epigenetically activating DGCR8. Eur. Rev. Med. Pharmacol. Sci., 23, 10835–10841. [DOI] [PubMed] [Google Scholar]
- 21. Krstic, M., et al. (2019) TBX3 promotes progression of pre-invasive breast cancer cells by inducing EMT and directly up-regulating SLUG. J. Pathol., 248, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu, L., et al. ; PRACTICAL, CRUK, BPC3, CAPS, PEGASUS Consortia. (2019) Identification of novel susceptibility loci and genes for prostate cancer risk: a transcriptome-wide association study in over 140,000 European descendants. Cancer Res., 79, 3192–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu, H., et al. (2016) MiR-374a suppresses lung adenocarcinoma cell proliferation and invasion by targeting TGFA gene expression. Carcinogenesis, 37, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dobin, A., et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao, Y., et al. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- 26. Frankish, A., et al. (2019) GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res., 47(D1), D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Love, M.I., et al. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao, Y., et al. (2019) WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res., 47(W1), W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chandrashekar, D.S., et al. (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia, 19, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Györffy, B., et al. (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat., 123, 725–731. [DOI] [PubMed] [Google Scholar]
- 31. Uhlén, M., et al. (2015) Proteomics. Tissue-based map of the human proteome. Science, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 32. Fornes, O., et al. (2020) JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res., 48(D1), D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu, H., et al. (2014) T-box transcription factor 21 expression in breast cancer and its relationship with prognosis. Int. J. Clin. Exp. Pathol., 7, 6906–6913. [PMC free article] [PubMed] [Google Scholar]
- 34. Chang, F., et al. (2016) The role of T-box genes in the tumorigenesis and progression of cancer. Oncol. Lett., 12, 4305–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Douglas, N.C., et al. (2013) The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. J. Mammary Gland Biol. Neoplasia, 18, 143–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. VanArsdale, T., et al. (2015) Molecular pathways: targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin. Cancer Res., 21, 2905–2910. [DOI] [PubMed] [Google Scholar]
- 37. Pane, L.S., et al. (2018) Tbx1 represses Mef2c gene expression and is correlated with histone 3 deacetylation of the anterior heart field enhancer. Dis Model Mech, 11, dmm029967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.