Abstract
In Ahmedabad, a major city in the state of Gujarat, India, an outbreak of acute secretory diarrhea caused by Vibrio cholerae O1 Ogawa El Tor, V. cholerae O139, and multiple serotypes of enterotoxigenic Escherichia coli (ETEC) occurred in January 2000. All of the representative V. cholerae O1 and O139 isolates examined harbored the ctxA gene (encoding the A subunit of cholera toxin) and the El Tor variant of the tcpA gene (encoding toxin-coregulated pilus). ETEC isolates of different serotypes were positive for the elt gene, encoding heat-labile enterotoxin. To further understand the molecular characteristics of the pathogens, representative isolates were examined by ribotyping and pulsed-field gel electrophoresis (PFGE). Ribotyping showed that the isolates of V. cholerae O1 Ogawa exhibited a pattern identical to that of the prevailing clone of O1 in areas where cholera is endemic in India, and all of the O139 isolates were identical to the BII clone of V. cholerae O139. PFGE of the representative O1 Ogawa isolates exhibited an identical pattern, comparable to the H pattern of the new clone of O1 reported in Calcutta, India. PFGE analysis of the V. cholerae O139 isolates showed identical patterns, but these differed from the PFGE patterns of O139 isolates reported during 1992 to 1997 in Calcutta. ETEC isolates showed genetic heterogeneity among isolates belonging to the same serotype, although the identical PFGE pattern was also observed among ETEC isolates of different serotypes. Antibiograms of the isolates were unusual, because all of the O139 isolates were resistant to nalidixic acid. Likewise, all of the E. coli isolates showed resistance to ciprofloxacin, norfloxacin, and nalidixic acid. This is a unique outbreak, and we believe that it is the first in which V. cholerae and ETEC were concomitantly involved.
Acute diarrheal diseases have been recognized as one of the major causes of morbidity and mortality in developing and underdeveloped countries. The common pathogens associated with diarrhea in developing countries are diarrheagenic Escherichia coli, Vibrio cholerae, Salmonella spp., and Shigella spp., etc. Cholera is caused by toxigenic strains of V. cholerae belonging to the O1 or O139 serogroup, which have the potential to cause epidemics (4, 25). It is estimated that tens of thousands of people in the world are affected every year due to cholera outbreaks and epidemics. Outbreaks of cholera are generally due to lack of sanitation or contamination of drinking water (28, 30). The etiologic agent, enterotoxigenic E. coli (ETEC), causes nearly 400 million diarrheal episodes and 700,000 deaths annually among children less than 5 years old (15). The present investigation highlights an association of three pathogens associated with a large outbreak of diarrhea in a metropolitan city of Gujarat state, India.
MATERIALS AND METHODS
Description of the outbreak.
From 1 to 17 January 2000, a total of 809 patients reported to three different hospitals, namely, I. D. Hospital, V. S. General Hospital, and L. G. Hospital, in Ahmedabad, India, with acute watery diarrhea. Cases of diarrhea were reported from at least 40 wards. The total population served by these three hospitals is in the range of 800,000 to 900,000. Bacteriological culture was performed on 734 of the 809 hospitalized patients. Pathogens were not isolated from all patients. Only the patients with acute illness accompanied by severe dehydration were hospitalized. Of the 734 stool specimens tested, 72 were positive for V. cholerae O1, 31 were positive for V. cholerae O139, and 24 were positive for E. coli. The attack rate of this outbreak was about 0.2%. Data on the background prevalence of these pathogens during this outbreak are not available, since routine surveillance for diarrheal etiologies is not maintained. However, in Ahmedabad, the seasonal peak of cholera is generally recorded between summer and early monsoon season, i.e., from April to August (J. S. Deokule, unpublished observation).
Identification of bacterial isolates.
One hundred three isolates of V. cholerae and 24 isolates of E. coli from this outbreak were received at the National Institute of Cholera and Enteric Diseases (NICED). For confirmation of identity, the V. cholerae isolates were plated on thiosulfate-citrate-bile salts-sucrose agar (Eiken, Tokyo, Japan) and the E. coli isolates were plated on MacConkey agar (Difco, Detroit, Mich.). The identities of these isolates were confirmed by different biochemical, physiological, and serological tests according to standard methods (33). The serotyping of E. coli was done using a commercially available kit (Denka Seiken Co., Ltd., Tokyo, Japan). Monoclonal antibodies against V. cholerae O1 and O139 serogroups generated at NICED were used for serotyping the V. cholerae isolates. Representative isolates were selected at random to exclude any bias for the detection of different virulence genes by PCR, molecular typing, and antibiotic susceptibility testing.
PCR assay for virulence genes.
A multiplex PCR-based assay was used to determine the presence of the A-subunit cholera toxin gene (ctxA) and to biotype the V. cholerae isolates by targeting tcpA (encoding the major structural subunit of the toxin-coregulated pilus), which is specific for El Tor and classical isolates (19), by a method described earlier (16). All of the E. coli isolates were screened for the presence of a variety of virulence factors using a PCR assay; these factors included elt (gene encoding heat-labile toxin) and est (gene encoding heat-stable toxin) (29) for ETEC; eae (gene for enterocyte attachment and effacement) (36), bfpA (gene for bundle-forming pilli) (14), and enteropathogenic E. coli (EPEC) adherence factor (12) for EPEC; stx1 (gene encoding Shiga toxin 1) and stx2 (gene encoding Shiga toxin 2) (22) for enterohemorrhagic E. coli; and EAgg (plasmid of enteroaggregative E. coli) (26) and EAST1 (gene for enteroaggregative stable toxin) (34) for enteroaggregative E. coli. Template DNA was prepared from the whole organism by boiling in a water bath for 10 min and instantly cooling on ice. PCR amplification was done with appropriate volumes of 10× amplification buffer (500 mM KCl, 100 mM Tris HCl, 15 mM MgCl2 [pH 8.3]), 2.5 mM each deoxynucleoside triphosphate, 10 pmol of each primer, 1.25 U of rTaq DNA polymerase (Takara Shuzo, Otsu, Japan), and 5 μl of template. The reaction volume was adjusted to 25 μl using sterile triple-distilled water. Uniplex and multiplex PCRs were performed in an automated thermocycler (Perkin-Elmer) for 30 cycles using the conditions described in Table 1.
TABLE 1.
PCR primer sequences and conditions used for the detection of genes specific for diarrheagenic E. coli and V. cholerae isolates
| PCR | E. coli groupa | Target gene or encoding region | Primer sequences (5′-3′) | Amplicon size (bp) | PCR conditionsb | Reference |
|---|---|---|---|---|---|---|
| Simplex | ETEC | elt | GGCGACAGATTATACCGTGC CGGTCTCTATATTCCCTGTT | 450 | 94°C, 1.0 min; 55°C, 1.5 min; 72°C 1.5 min | 29 |
| est | ATTTTTA/CTTTCTGTATTA/GTCTT CACCCGGTACAA/GGCAGGATT | 190 | 94°C, 1.0 min; 55°C 1.5 min; 72°C, 1.5 min | 29 | ||
| EPEC | eae | AAACAGGTGAAACTGTTGCC CTCTGCAGATTAACCCTCTGC | 454 | 94°C, 1.0 min; 55°C, 1.5 min; 72°C, 1.5 min | 36 | |
| bfpA | AATGGTGCTTGCGCTTGCTGC GCCGCTTTATCCAACCTGGTA | 324 | 94°C, 1.0 min; 56°C, 1.5 min; 72°C, 1.5 min | 14 | ||
| EAF | CAGGGTAAAAGAAGATGATAA TATGGGGACCATGTATTATCA | 397 | 94°C, 1.0 min; 60°C, 1.5 min; 72°C, 1.5 min | 12 | ||
| Multiplex | EHEC | stx1 | CAACACTGGATGATCTCAG CCCCCTCAACTGCTAATA | 350 | 94°C, 1.0 min; 55°C, 1.0 min; 72°C, 1.0 min | 22 |
| stx2 | ATCAGTCGTCACTCACTGGT CTGCTGTCACAGTGACAAA | 110 | ||||
| EAgg | CTGGCGAAAGACTGTATCAT CAATGTATAGAAATCCGCTGTT | 630 | 94°C, 1.0 min; 53°C, 1.0 min; 72°C, 1.0 min | 26 | ||
| EAggEC | EAST1 | CACAGTATATCCGAAGGC CGAGTGACGGCTTTGTAG | 94 | This study | ||
| V. cholerae | ctxA | CTCAGACGGGATTTGTTAGGCACG TCTATCTCTGTAGCCCCTATTACG | 301 | 94°C, 1.0 min; 60°C, 1.5 min; 72°C, 1.5 min | ||
| tcpA (classical) | CACGATAAGAAAACCGGTCAAGAG ACCAAATGCAACGCCGAATGGAG | 617 | 16 | |||
| tcpA (El Tor) | GAAGAAGTTTGTAAAAGAAGAACAC GAAGGACCTTCTTTCACGTTG | 471 |
EHEC, enterohemorrhagic E. coli; EAggEC, enteroaggregative E. coli.
Thirty cycles consisting of denaturation, annealing, and extension.
Detection of CFAs.
All of the ETEC isolates were tested for colonization factor antigens (CFAs) by the procedure detailed by Qadri et al. (24). Briefly, ETEC isolates were inoculated on CFA agar plates with and without bile salts and incubated at 37°C overnight. The colonies from CFA agar plates were tested for the expression of CFA/I, CS1 to CS7, CS12 (PCFO159), CS14 (PCFO166), CS17, and CFA/III (CS8) by monoclonal antibody-based dot blot assay (24) with appropriate reference strains.
Antimicrobial susceptibility.
Antimicrobial susceptibility testing was done using standard methods (3). E. coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as quality control strains. Representative isolates of V. cholerae were tested for susceptibility using commercially available discs (HiMedia, Mumbai, India) of ampicillin (10 μg), chloramphenicol (30 μg), co-trimoxazole (25 μg), ciprofloxacin (5 μg), furazolidone (100 μg), norfloxacin (10 μg) gentamicin (10 μg), neomycin (30 μg), streptomycin (10 μg), and tetracycline (30 μg). All of the E. coli isolates included in this study were examined for susceptibility to cephalothin (30 μg), amikacin (30 μg), ceftazidime (10 μg), kanamycin (30 μg), ceftriaxone (30 μg), and nalidixic acid (30 μg) in addition to the antibiotics used for V. cholerae isolates, with the exception of furazolidone. Characterization of isolates as susceptible, intermediately resistant, or resistant was based on the size of the inhibition zones according to the manufacturer's instructions, which matched the interpretative criteria recommended by the NCCLS (21). In addition, we analyzed three representative quinolone-resistant ETEC isolates for norfloxacin and ciprofloxacin MICs using the E-test method (AB Biodisk, Solnea, Sweden).
Ribotyping of V. cholerae.
The 7.5-kb BamHI fragment of plasmid pKK3535 containing 16S and 23S rRNA genes of E. coli was used as the rRNA probe (6). The modified method of Murray and Thompson (20) was used for genomic DNA extraction. For ribotyping, the transfer of digested DNA from gels to Hybond N+ membranes (Amersham International PLC, Buckinghamshire, England) and hybridization with rRNA probes were performed as described previously (1), using the ECL nucleic acid detection system (Amersham). The membranes were washed, exposed to Biomax film (Eastman Kodak Co., Rochester, N.Y.), and developed according to the manufacturer's instruction.
PFGE.
Pulsed-field gel electrophoresis (PFGE) of V. cholerae and E. coli isolates was performed by preparing agarose plugs as described previously (17, 35). NotI (Takara)-digested inserts of V. cholerae and XbaI (Takara)-digested inserts of E. coli were applied to a contour-clamped homogenous electric field in a CHEF Mapper system (Bio-Rad, Richmond, Calif.) using 1% PFGE-grade agarose in 0.5× Tris-borate-EDTA (44.5 mM Tris-HCl, 44.5 mM boric acid, 1.0 mM EDTA [pH 8.0]) for 40 h 24 min at 14°C. Run conditions were generated by the autoalgorithm mode of the CHEF Mapper PFGE system using a size range of 20 to 300 kb for V. cholerae and 20 to 350 kb for E. coli isolates. After electrophoresis, the gels were stained in distilled water containing 1.0 μg of ethidium bromide per ml for 30 min, destained in distilled water for 15 min, and photographed under UV light using the Gel Doc 2000 documentation system (Bio-Rad). A DNA size standard (ladder; New England Biolabs, Beverly, Mass.) was used as the molecular size standard.
RESULTS
Examination of the 103 isolates of V. cholerae revealed that 72 (70%) were V. cholerae O1 Ogawa serotype, El Tor biotype, while 31 (30%) were identified as belonging to the V. cholerae O139 serogroup. All of the 25 representative V. cholerae isolates, including 17 serogroup O1 and 8 serogroup O139 isolates, were positive in multiplex PCR for ctxA and tcpA of the El Tor variant. Six different serotypes of E. coli were seen (Table 2), with the O1 serotype being dominant (41.6%), followed by O146 (16.6%); 16.6% of the isolates were untypeable. In the PCR assay, 18 (75%) of the E. coli isolates harbored the elt gene, of which 9 (50%) belonged to serotype O1 (Table 2). None of the E. coli isolates tested harbored the est, stx1, or stx2 gene, and all were negative in the EAgg PCR assay. None of the ETEC isolates possessed any of the 12 commonly prevalent CFAs that were examined in this study.
TABLE 2.
Serotyping, PCR, and CFA results for the E. coli isolates from the Ahmedabad outbreak
| Strain | Serotype | PCR resultsa
|
CFA | ||||
|---|---|---|---|---|---|---|---|
| elt | eae | bfpA | EAFb | EAST1 | |||
| E-2 | O146 | + | − | − | − | − | − |
| E-3 | O1 | + | − | − | − | − | − |
| E-4 | O1 | + | − | − | − | + | − |
| E-5 | ONTc | + | − | − | − | − | − |
| E-6 | O8 | + | − | − | − | − | − |
| E-7 | O146 | + | − | − | − | − | − |
| E-8 | ONT | + | − | − | − | − | − |
| E-9 | O146 | + | − | − | − | − | − |
| E-10 | O1 | + | − | − | − | − | − |
| E-11 | O158 | − | + | + | + | − | NDd |
| E-13 | O1 | + | − | − | − | − | − |
| E-14 | O1 | + | − | − | − | − | − |
| E-15 | O1 | + | − | − | − | − | − |
| E-16 | O1 | + | − | − | − | − | − |
| E-17 | O146 | + | − | − | − | − | − |
| AV-155 | O114 | − | − | − | − | − | ND |
| AV-169 | O1 | − | − | − | − | − | ND |
| AV-170 | O8 | − | − | − | − | − | ND |
| AV-185 | O1 | + | − | − | − | − | − |
| AV-188 | O114 | − | + | − | − | − | ND |
| AV-189 | O25 | + | − | − | − | − | − |
| AV-193 | ONT | + | − | − | − | − | − |
| AV-195 | O1 | + | − | − | − | + | − |
| A-89 | ONT | − | − | − | − | − | ND |
None of the E. coli isolates yielded positive results with est, stx1, and stx2.
EAF, EPEC adherence factor.
ONT, not typeable.
ND, not done.
Antibiotic susceptibility results for 23 O1 isolates and 9 O139 isolates revealed that all of these isolates were resistant to ampicillin, furazolidone, and nalidixic acid (Table 3). In addition, V. cholerae O1 isolates were resistant to co-trimoxazole and streptomycin, and 21.7% of them were resistant to chloramphenicol, whereas V. cholerae O139 isolates were susceptible to these antibiotics. The majority of the E. coli isolates showed high resistance to several antibiotics, including members of the quinolone group of antimicrobial drugs (Table 3). MICs for three ETEC isolates (E2, E14, and E15) were found to be ≥32 μg/ml for ciprofloxacin and ≥256 μg/ml for norfloxacin.
TABLE 3.
Antimicrobial resistance of the Ahmedabad outbreak isolates of V. cholerae and E. coli
| Antimicrobial | % Resistant strains (no. of strains screened)
|
||
|---|---|---|---|
| V. cholerae O1 (23) | V. cholerae O139 (9) | E. coli (24) | |
| Amikacin | NDa | ND | 4.16 |
| Ampicillin | 100 | 100 | 95.8 |
| Ceftazidime | ND | ND | 54.2 |
| Ceftriaxone | ND | ND | 66.7 |
| Cephalothin | ND | ND | 100 |
| Chloramphenicol | 21.7 | 0 | 66.7 |
| Ciprofloxacin | 0 | 0 | 79.2 |
| Co-trimoxazole | 100 | 0 | 91.7 |
| Furazolidone | 100 | 100 | ND |
| Gentamycin | 0 | 0 | 50 |
| Kanamycin | ND | ND | 83.3 |
| Nalidixic acid | 100 | 100 | 95.8 |
| Neomycin | 65.2 | 55.2 | 91.6 |
| Norfloxacin | 0 | 0 | 79.2 |
| Streptomycin | 95 | 0 | 95.8 |
| Tetracycline | 0 | 0 | 91.7 |
ND, not done.
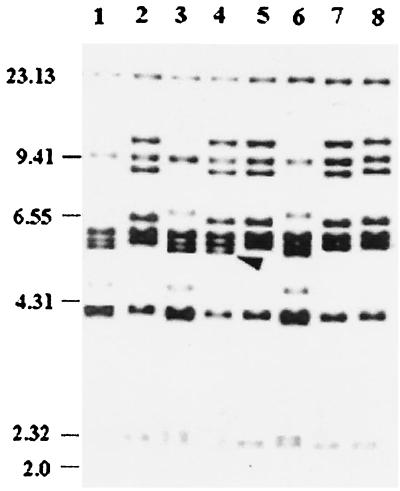
Recent findings have shown that species other than V. cholerae might act as an extraordinary reservoir for both CTXφ and VPIφ and might play an important role in the emergence of new toxigenic strains (5, 10). In view of this, we tested the E. coli isolates for the ctxA and tcpA genes, which are specifically found in V. cholerae, to detect any lateral gene transfer event. The multiplex PCR showed that none of the E. coli isolates harbored ctxA or tcpA. Ribotyping of eight representative isolates of O1 Ogawa revealed that seven of them (Fig. 1, lanes 2, 5, 7, and 8 [only representative isolates are shown]) showed the previously documented RIII type (27), while one isolate AHO94 (Fig. 1, lane 4) showed a pattern slightly different from the RIII type, which is the currently prevailing type (27), by the presence of an additional band at approximately 5.6 kb. The ribotype patterns of five representative V. cholerae O139 isolates (Fig. 1, lanes 1, 3, and 6 [only three isolates are shown]) were identical to the most commonly found BII ribotype pattern, which is the ribotype currently prevailing among V. cholerae O139 isolates in Calcutta and Bangladesh (11).
FIG. 1.
Ribotypes of the representative V. cholerae isolates using BglI. Lanes: 1, AHO92 (O139); 2, AHO86 (O1, El Tor, Ogawa); 3, AHO78 (O139); 4, AHO94 (O1, El Tor, Ogawa); 5, AHO66 (O1, El Tor, Ogawa); 6, AHO82 (O139); 7, AHO74 (O1 El Tor Ogawa); 8, AHO80 (O1, El Tor Ogawa). Positions of λ-HindIII molecular size markers (in kilobases) are indicated by bars. The arrow indicates an extra band in the 5.66-kb region.
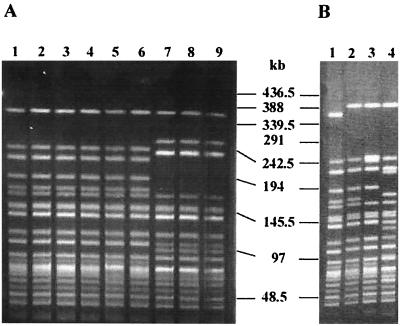
PFGE of all six representative O1 Ogawa isolates (Fig. 2A, lanes 1 to 6) exhibited identical patterns which were comparable to the H pattern of the new clone of O1 reported in Calcutta (35) (Fig. 2B, lane 1). PFGE analysis of the three V. cholerae O139 isolates (Fig. 2A, lanes 7 to 9) showed identical patterns which differed from the PFGE patterns of O139 isolates reported during 1992 to 1997 in Calcutta (2).
FIG. 2.
PFGE profiles generated with NotI-digested genomic DNAs of V. cholerae O1 El Tor and O139 isolates. (A) Lanes: 1, AHO66 (O1, Ogawa); 2, AHO72 (O1, Ogawa); 3, AHO74 (O1, Ogawa); 4, AHO80 (O1, Ogawa); 5, AHO94 (O1, Ogawa); 6, AHO86 (O1, Ogawa); 7, AHO78 (O139); 8, AHO82 (O139); 9, AHO92 (O139). (B) Lanes: 1, CO366 (O1, Ogawa), pattern H; 2, CO370 (O1, Ogawa), pattern I; 3, CO388 (O1, Ogawa), pattern J; 4, CO392 (O1, Ogawa), pattern K. The various patterns referred to are those reported by Yamasaki et al. (35). Positions of the bacteriophage λ ladder molecular size markers are indicated by bars.
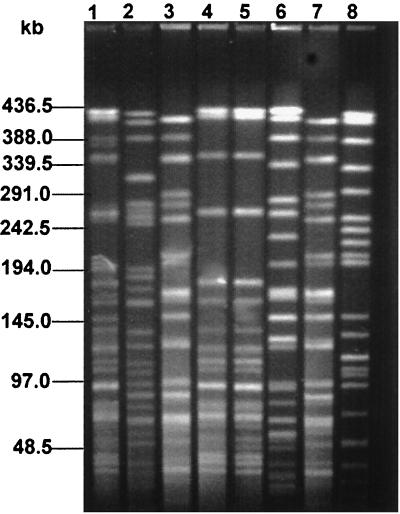
PFGE analysis was done on eight representative E. coli isolates. Of five of these isolates belonging to E. coli serotype O1 (Fig. 3, lanes 1 to 5), two isolates, E14 and AV185 (Fig. 3, lanes 4 and 5), exhibited identical patterns, while the remaining three (Fig. 3, lanes 1 to 3) were different from each other despite belonging to the same serotype. Two isolates of E. coli serotype O146 (Fig. 3, lanes 6 and 7) showed different PFGE profiles. Surprisingly, the PFGE pattern of isolate E10, belonging to the O1 serotype (Fig. 3, lane 3), was identical to that of isolate E2, belonging to the O146 serotype (Fig. 3, lane 7). The PFGE pattern of one representative E. coli O-untypeable isolate (Fig. 3, lane 8) was different from those of E. coli isolates belonging to either the O1 or O146 serotype.
FIG. 3.
PFGE profiles of the representative E. coli isolates using XbaI. Lanes: 1, E3 (O1); 2, E4 (O1); 3, E10 (O1); 4, E14 (O1); 5, AV185 (O1); 6, E9 (O146); 7, E2 (O146); 8, E5 (O nontypeable). Positions of the bacteriophage λ ladder molecular size markers are indicated by bars.
DISCUSSION
The outbreak of acute diarrhea in Ahmedabad was unusual in that two major enterotoxic enteropathogens, namely, V. cholerae and ETEC were involved. To our knowledge, this is the first report describing the involvement of more than one enteric pathogen in an outbreak setting in India. Involvement of more than one pathogen during outbreaks has been reported elsewhere and was usually attributed to gross contamination of food or drinking water (9, 18). However in this study, the concurrent incidence of two different enteric pathogens, V. cholerae and ETEC, in a single patient was not encountered.
This outbreak was predominantly due to V. cholerae O1, Ogawa serotype, biotype El Tor, the serotype and biotype currently prevailing in India. Uniquely, however, V. cholerae serogroup O139 and multiple serotypes of ETEC were also involved. When the 18 ETEC isolates were analyzed in detail, we were unable to detect any of the 12 commonly occurring CFAs (24). The probable reasons for this result might be either the loss of CFAs due to repeated subculture in vitro or the prevalence of a hitherto-unrecognized CFA different from the 12 CFAs assayed in this study. V. cholerae O1 Ogawa isolates had antibiotic resistance patterns similar to those of the prevailing O1 Ogawa strains in the rest of the country (13). The other significant observation was that all of the isolates of V. cholerae O139 examined were resistant to nalidixic acid, and such a high percentage of resistance to nalidixic acid has not been previously reported for this serogroup (13). Interestingly, all of the ETEC and other E. coli isolates were resistant to almost all of the antimicrobial drugs tested (Table 3) and showed alarmingly high levels of resistance to ciprofloxacin, norfloxacin, and nalidixic acid. As far as E. coli-mediated diarrhea is concerned, a prevalence of ETEC strains resistant to fluoroquinolones has rarely been reported (31).
PCR results indicated that all V. cholerae O1 and O139 isolates tested harbored ctxA and that 75% of the ETEC isolates harbored elt only (Table 2). Ribotyping of the 13 representative isolates of V. cholerae was done using the BglI restriction endonuclease, which is known to produce good discriminatory patterns for V. cholerae (23). Ribotyping of representative isolates of V. cholerae O1 Ogawa (Fig. 2) showed that 87.5% had an identical ribotype, which was similar to the reported ribotype of the new clone of O1 (27). Ribotyping analysis of representative isolates of V. cholerae O139 (Fig. 2) indicated that all of the isolates were identical to the BII clone (11), which is the prevailing ribotype in many parts of India. Overall, based on the ribotyping results, it appears that the Ahmedabad outbreak was caused by the prevailing clones of V. cholerae O1 and O139 found in Calcutta and rest of the country.
In the PFGE analysis, all of the V. cholerae O1 isolates exhibited the H pattern of the new clone of O1 (35). PFGE analysis of three representative O139 isolates (Fig. 2) clearly showed that the pattern was very different from that of the prevailing O139 clone in Calcutta (2). PFGE of E. coli isolates revealed very interesting results. Two E. coli isolates, E14 and AV185, belonging to serotype O1 had identical patterns (Fig. 3) although their antibiograms were very different: E14 was resistant to gentamicin, norfloxacin, and ciprofloxacin, while AV185 was sensitive to all of these drugs.
Generally, the clonal diversity among E. coli is high even though the strains are phenotypically identical but genetically dissimilar. The existence of such genetic heterogeneity among E. coli strains belonging to the same serotype has been recorded previously (7, 32). Surprisingly, two E. coli isolates (E10 and E2) were identical in both PFGE and antibiotic susceptibility testing although they belonged to serotypes O1 and O146, respectively. Such a phenomenon has been observed among pandemic Vibrio parahaemolyticus isolates (8). The outbreak reported in the present study was due to the contamination of drinking water with sewage. What is intriguing in this outbreak is why instead of having V. cholerae infection some patients were infected with ETEC, even though the same population was exposed to the common source of infection. It could be possible that such patients had protective levels of antibody due to previous exposures to toxigenic V. cholerae, and thus ETEC prevailed in these individuals. More detailed analysis of patients in such concomitant outbreaks would provide a wealth of information which would be useful from the perspective of development of vaccines for enteric infections.
ACKNOWLEDGMENTS
We acknowledge F. Qadri, International Centre for Diarrhoeal Disease Research, Bangladesh, Dhaka, and Ann-Mari Svennerholm, Göteborg University, Göteborg, Sweden, for examining the ETEC strains for colonization factors and Sheth V. S. General Hospital, Ahemdabad, India, for providing the epidemiological data.
This work was supported in part by the Council of Scientific and Industrial Research Projects no. 27 (0103)/EMR-II and no. 37 (1019)/99/EMRII and by the Japan International Co-operation Agency (JICA/NICED project 054-1061-E-O).
REFERENCES
- 1.Basu A, Mukhopadhaya A K, Sharma C, Jyoti J, Gupta N, Ghosh A, Bhattacharya S K, Takeda Y, Faruque A S G, Albert M J, Nair G B. Heterogeneity in the organization of the CTX genetic element in strains of Vibrio cholerae O139 Bengal isolated from Calcutta, India and Dhaka, Bangladesh and its possible link to the dissimilar incidence of O139 cholera in two locales. Microb Pathog. 1998;24:175–183. doi: 10.1006/mpat.1997.0186. [DOI] [PubMed] [Google Scholar]
- 2.Basu A, Garg P, Datta S, Chakraborty S, Bhattacharya T, Khan A, Ramamurthy T, Bhattacharya S K, Yamasaki S, Takeda Y, Nair G B. Vibrio cholerae O139 in Calcutta, 1992–1998. Incidence, antibiograms, and genotypes. Emerg Infect Dis. 2000;6:139–147. doi: 10.3201/eid0602.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer A W, Kirby W M M, Sherris J C, Truck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 4.Birmingham M E, Lee L A, Ndayimirije N, Nkurikiye S, Hersh B S, Wells J G, Deming M S. Epidemic cholera in Brundi: patterns of transmission in the Great Rift Valley lame region. Lancet. 1997;349:981–985. doi: 10.1016/S0140-6736(96)08478-4. [DOI] [PubMed] [Google Scholar]
- 5.Boyd E F, Moyer K E, Shi L, Waldor M K. Infectious CTXφ and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun. 2000;68:1507–1513. doi: 10.1128/iai.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius J, Ullrich A, Raber M A, Garg A, Dull J J, Gutell R R, Noler H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of Escherichia coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 7.Caugant D A, Levin B R, Orskov I, Orskov F, Eden C S, Selander R K. Genetic diversity in relation to serotype in Escherichia coli. Infect Immun. 1985;49:407–413. doi: 10.1128/iai.49.2.407-413.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury N R, Chakraborty S, Ramamurthy T, Nishibuchi M, Yamasaki S, Takeda Y, Nair G B. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg Infect Dis. 2000;6:631–636. doi: 10.3201/eid0606.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Divizia M, Gabrieli R, Donia D, Ruscia V, Degener A M, Pana P. Concomitant poliovirus infection during an outbreak of hepatitis A. J Infect. 1999;39:227–230. doi: 10.1016/s0163-4453(99)90054-3. [DOI] [PubMed] [Google Scholar]
- 10.Faruque S M, Rahman M M, Asadulghani, Nasirul Islam K M, Mekalanos J J. Lysogenic conversion of environmental Vibrio mimicus strains by CTXφ. Infect Immun. 1999;67:5723–5729. doi: 10.1128/iai.67.11.5723-5729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque S M, Saha M N, Asadulghani, Bag P K, Bhadra R K, Bhattacharya S K, Sack R B, Takeda Y, Nair G B. Genomic diversity among Vibrio cholerae O139 strains isolated in Bangladesh and India between 1992 and 1998. FEMS Microbiol Lett. 2000;184:279–284. doi: 10.1111/j.1574-6968.2000.tb09027.x. [DOI] [PubMed] [Google Scholar]
- 12.Franke J, Franke S, Schmidt H, Schwarzkopf A, Wieler L H, Baljer G, Beutin L, Karch H. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J Clin Microbiol. 1994;32:2460–2463. doi: 10.1128/jcm.32.10.2460-2463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garg P, Chakraborty S, Basu I, Datta S, Rajendran K, Bhattacharya T, Yamasaki S, Bhattacharya S K, Takeda Y, Nair G B, Ramamurthy T. Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992 to 1997 in Calcutta, India. Epidemiol Infect. 2000;124:393–399. doi: 10.1017/s0950268899003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunzburg S T, Tornieporth N G, Riley L W. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol. 1995;33:1375–1377. doi: 10.1128/jcm.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Institute of Medicine. New vaccine development: establishing priorities. II. Washington, D.C.: Academy Press; 1986. pp. 178–184. [Google Scholar]
- 16.Keasler S P, Hall R H. Detecting and biotyping V. cholerae O1 with multiplex polymerase reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 17.Kurazono H, Okuda J, Takeda Y, Nair G B, Albert M J, Sack R B, Chongsa-nguan M, Chaichumpa W. Vibrio cholerae O139 Bengal isolated from India, Bangladesh and Thailand are clonal as determined by pulsed-field gel electrophoresis. J Infect. 1994;29:109–110. doi: 10.1016/s0163-4453(94)95357-0. [DOI] [PubMed] [Google Scholar]
- 18.Maurer A M, Sturchler D. A water borne outbreak of small round structured virus. Campylobacter and Shigella co-infections in La Neuveville, Switzerland, 1998. Epidemiol Infect. 2000;125:325–332. doi: 10.1017/s0950268899004495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhyay A K, Garg S, Mitra R, Basu A, Rajendran K, Dutta D, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Temporal shifts in traits of V. cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J Clin Microbiol. 1996;34:2537–2543. doi: 10.1128/jcm.34.10.2537-2543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2–A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 22.Pal A, Ghosh S, Ramamurthy T, Yamasaki S, Tsukamoto T, Bhattacharya S K, Nair G B, Takeda Y. Shiga toxin producing Escherichia coli from healthy cattle in a semi-urban community in Calcutta, India. Indian J Med Res. 1999;110:83–85. [PubMed] [Google Scholar]
- 23.Popovic T, Bopp C, Olsvik O, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qadri F, Das S K, Faruque A S G, Fuchs G J, Albert M J, Sack R, Svennerholm A. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J Clin Microbiol. 2000;38:27–31. doi: 10.1128/jcm.38.1.27-31.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radhakutty G, Sirkar B K, Mondal S K, Mukhopadhyay A K, Mitra R K, Basu A, Ichhpujani R L, Nair G B, Bhattacharya S K. Investigation of the outbreak of cholera in Alleppey and Palghat districts, South India. Indian J Med Res. 1998;106:455–457. [PubMed] [Google Scholar]
- 26.Schmidt H, Knop C, Franke S, Aleksic S, Heesmann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–705. doi: 10.1128/jcm.33.3.701-705.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma C, Nair G B, Mukhopadhyay A K, Bhattacharya S K, Ghosh R K, Ghosh A. Molecular characterization of Vibrio cholerae O1 biotype El Tor strains isolated between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of the El Tor biotype. J Infect Dis. 1997;175:1134–1141. doi: 10.1086/516453. [DOI] [PubMed] [Google Scholar]
- 28.Siddique A K, Salan A, Islam M S, Akram K, Majumdar R N, Zaman K, Franczak N, Kaston S. Why treatment centres failed to prevent cholera deaths among Rwandan refugees in Goma, Zaire. Lancet. 1995;345:359–361. doi: 10.1016/s0140-6736(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 29.Stacy-Phipps S, Mecca J M, Weiss J B. Multiplex PCR assay and simple preparation method for stool specimens to detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33:1054–1059. doi: 10.1128/jcm.33.5.1054-1059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swerdlow D L, Mintz E D, Tejada E, Rodriguez E, Ocampo C, Espefo L, Greene K D, Saldana W, Seminario L, Tauxe R V, Wells J V, Bean N H, Ries A A, Pollack M, Vertiz B, Blake P A. Water borne transmission of epidemic cholera in Trufillo, Peru: lessons for a continent at risk. Lancet. 1992;340:28–32. doi: 10.1016/0140-6736(92)92432-f. [DOI] [PubMed] [Google Scholar]
- 31.Vila J, Vargas M, Ruiz J, Corachan M, Anta M T J D, Gascon J. Quinolone resistance in Escherichia coli causing diarrhea in travelers to India in comparison with other geographical areas. Antimicrob Agents Chemother. 2000;44:1731–1733. doi: 10.1128/aac.44.6.1731-1733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodward J M, Connaughton I D, Fahy V A, Lymbery A J, Hampson D J. Clonal analysis of Escherichia coli of serogroup O9, O20, and O101 isolated from Australian pigs with neonatal diarrhea. J Clin Microbiol. 1993;31:1185–1188. doi: 10.1128/jcm.31.5.1185-1188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. Manual for laboratory investigations of acute enteric infections. CDD/83.3. Geneva, Switzerland: World Health Organization; 1987. [Google Scholar]
- 34.Yamamoto T, Nakazawa M. Detection and sequence of the enteroaggregative Escherichia coli heat-stable enterotoxin gene in enterotoxigenic Escherichia coli strains isolated from piglets and calves with diarrhea. J Clin Microbiol. 1997;35:223–227. doi: 10.1128/jcm.35.1.223-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamasaki S, Nair G B, Bhattacharya S K, Yamamoto S, Kurazono H, Takeda Y. Cryptic appearance of a new clone of Vibrio cholerae O1 biotype El Tor in Calcutta, India. Microbiol Immunol. 1997;41:1–6. doi: 10.1111/j.1348-0421.1997.tb01165.x. [DOI] [PubMed] [Google Scholar]
- 36.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]