Abstract
Forkhead box (Fox) family, an evolutionarily conserved family of transcription factors carrying the “Forkhead” motif, plays an indispensable role in human health and disease. Fox family genes are involved in cell differentiation, proliferation and apoptosis, embryonic development, aging, glucose and lipid metabolism, and immune regulation. The regulatory role of the Fox family in the context of bone metabolism and orthopedic diseases is an emerging research hotspot. In this review, we highlight the major molecular mechanisms underlying the regulatory role of Fox factors in bone metabolism, bone development, bone homeostasis, and bone diseases associated with inhibition or upregulation of Fox factors. In addition, we discuss the emerging evidence in the realm of Fox factor-based therapeutics.
Keywords: Fox family, bone metabolism, bone diseases, signaling pathways, Fox factors-based therapeutics
1 Background
Fox family, identified in 2000 (Kaestner et al., 2000), is a group of genes with “Forkhead” motif-dependent transcription factors. Recent studies have unraveled the role of Fox family genes as key sensors for bone metabolism. Members of the Fox family respond to metabolic stress in bone tissue, inflammatory signals, hypoxic/oxidative stress, and are associated with aging and autophagy.
The role of Fox in the regulation of bone metabolism was first recognized in 2001 when a defective reproductive phenotype was identified as a molecular marker of prospective rib cartilage (Sudo et al., 2001). Since then, the role of different subfamilies of Fox in bone metabolism, from FoxA to FoxS (based on the degree of homology in their forkhead domains), has been investigated. Over the years, our knowledge of the role of Fox in bone metabolism has grown exponentially along with the awareness of the key roles of Fox-regulated biological processes in bone functions (Huang et al., 2020).
Studies have demonstrated differential expression of Fox factors in osteoblasts in the setting of skeletal disease compared with normal osteoblasts; these differentially expressed factors have been shown to promote or suppress the development of osteoporosis by regulating bone metabolism (Greenblatt et al., 2010; Niedan et al., 2014; Yu et al., 2014; Guan et al., 2015; Hopkins et al., 2016; Zeng et al., 2017). Bone metabolism refers to a complex series of biological processes involving multiple signaling pathways, such as Wingless and Int-1 (Wnt)/β-catenin pathway (Huang et al., 2020), bone morphogenetic protein (BMP)/drosophila mothers against decapentaplegic (Smad) pathway (Guan et al., 2015), phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K)/Akt pathway (Hopkins et al., 2016), transforming growth factor-β (TGF-β) pathway (Niedan et al., 2014), p38/mitogen-activated protein kinase (MAPK) pathway (Zeng et al., 2017), and nuclear factor-kappa B (NF-κB) pathway (Greenblatt et al., 2010). Fox factors are involved in the regulation of bone metabolism, either directly or by acting as downstream effectors of these signaling pathways (Greenblatt et al., 2010; Niedan et al., 2014; Yu et al., 2014; Guan et al., 2015; Hopkins et al., 2016; Zeng et al., 2017). In addition to its role in osteoporosis, Fox factors are also involved in the development of osteoarthritis (Charlier et al., 2016), rheumatoid arthritis (Reedquist et al., 2006; Wasén et al., 2020), intervertebral disc degeneration (Alvarez-Garcia et al., 2018), and bone tumors (Nakamura et al., 2000; Cidre-Aranaz and Alonso, 2015; Haider et al., 2016). In particular, drugs targeting Fox factors have been reported to inhibit the progression of bone tumors (Lam and Gomes, 2014). Last but not the least, mutations in Fox factor have been implicated in inherited skeletal abnormalities (Seifi and Walter, 2018; Chen et al., 2019).
Although the close relationship between Fox factor and skeletal disease is well established, the role of Fox factors in promoting or inhibiting skeletal disease and the associated underlying mechanisms are highly controversial and perplexing. In this review, we summarize the available evidence of the functional role of the Fox family in the context of bone-associated cell metabolism and various bone diseases (Figure 1). In addition, we highlight the future research directions by identifying related novel biomarkers for cancer diagnosis and therapeutic targets.
FIGURE 1.
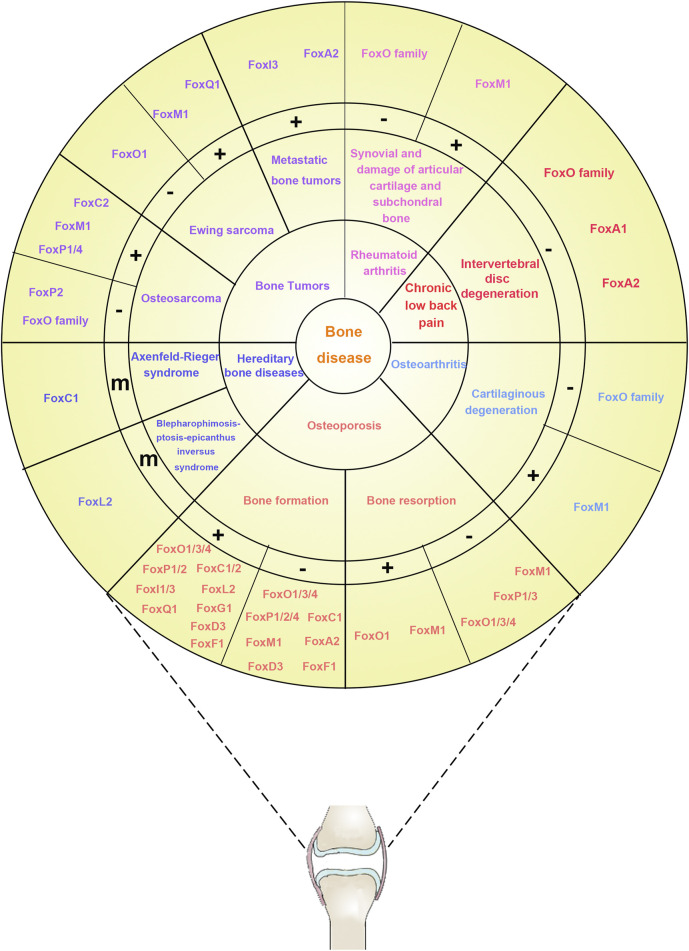
The relationship between the Fox family and bone diseases. The bone diseases related to the Fox family are divided into six types listed in the first inner ring (six types of colors, including osteoporosis, osteoarthritis, chronic low back pain, rheumatoid arthritis, bone tumors, and hereditary bone diseases). The characteristics of different diseases are listed in the second inner ring. Osteoporosis, a metabolic bone disease caused by dyshomeostasis of bone metabolism, is closely related with bone formation and bone resorption; Osteoarthritis, a chronic joint disease characterized by degenerative changes in joint cartilage, is closely related with cartilaginous degeneration; Chronic low back pain is closely related with intervertebral disc degeneration; Rheumatoid arthritis, a chronic joint disease characterized by persistent synovitis and associated damage to the articular cartilage and subchondral bone, is closely related with synovial and damage of articular cartilage and subchondral bone; Bone tumors is separated into metastatic bone tumors (a subtumor formed in bone originated from other parts of body), Ewing sarcoma (a rare and highly aggressive cancer that occurs primarily in the bones and surrounding tissues of children and adolescents), and osteosarcoma (the most common primary malignant tumor of bone, and it occurs mainly in children and adolescents); Hereditary bone diseases, caused by genetic factors, is separated into Axenfeld–Rieger syndrome and blepharophimosis-ptosis-epicanthus inversus syndrome. The correlations between the characteristics of different diseases and Fox-related genes are listed in the third inner ring. “+,” Fox-related genes enhance this symptom (or activity); “−,” Fox-related genes inhibit this symptom (or activity); “m,” mutation of Fox-related genes. The Fox-related genes are listed in the outer ring.
2 Role of Fox factors in osteoporosis
Osteoporosis is a metabolic bone disease (Bellavia et al., 2021) caused by dyshomeostasis of bone metabolism. The dynamic balance of bone metabolism depends primarily on the interaction between osteoblasts, which synthesize the bone matrix, and osteoclasts, which absorb the bone matrix. This dynamic balance is essential for preventing bone disease in the human body (Huang et al., 2020). Recent studies have indicated a key role of the Fox family in the process of bone metabolism (Figure 2). Among these, most Fox factors have been shown to be involved in osteogenic differentiation via different signal pathways (Table 1 and Table 2); however, FoxO, FoxP, and FoxM1 play their respective roles in both osteoblast differentiation and osteoclast differentiation.
FIGURE 2.
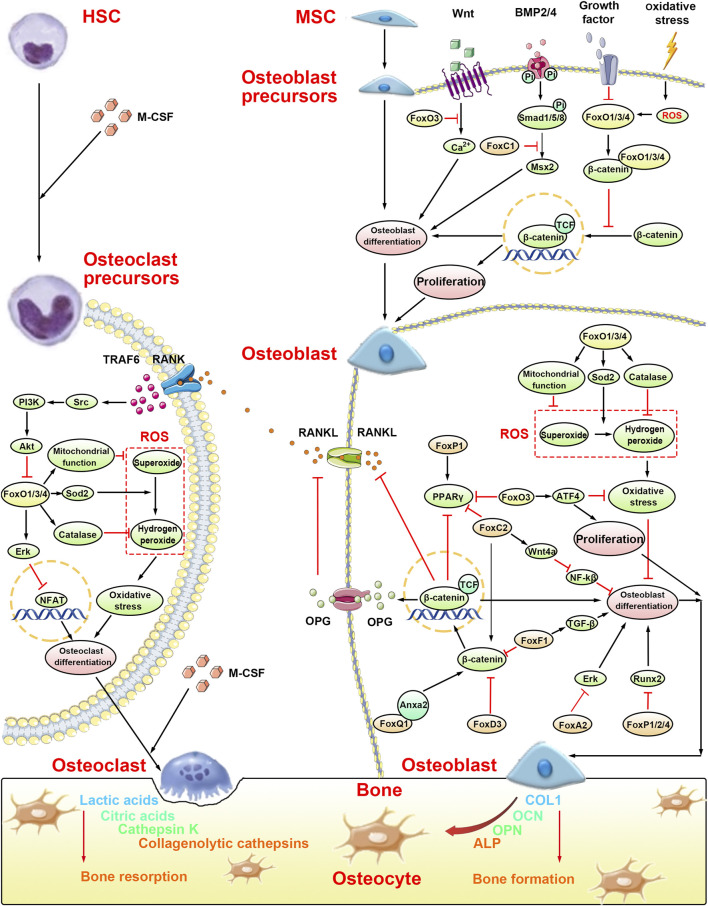
Molecular mechanisms of bone formation and bone resorption in osteoporosis by the Fox family. The occurence of osteoporosis is caused by the imbalance of bone formation, which is caused by osteoblasts, and bone resorption, which is caused by osteoclasts. Osteoblast precursors, originated from MSC, will be suppressed in osteoblast differentiation by FoxC1 and FoxO1/3/4. Meanwhile, FoxO1/3/4 can suppress the proliferation of osteoblast precursors by binding with β-catenin. In osteoblast, FoxA2, FoxC2, FoxD3, and FoxP1/2/4 suppress osteoblast differentiation. However, FoxO1/3/4 and FoxQ1 promote osteoblast differentiation. Interestingly, FoxF1 not only suppress osteoblast differentiation by the Wnt/β-catenin signaling pathway but also promote osteoblast differentiation by the TGF-β signaling pathway. Furthermore, FoxO3 can promote proliferation of osteoblast precursors by activating ATF4. Osteoclast precursors, originated from HSC, will be suppressed in osteoclast differentiation by FoxO1/3/4 through the Erk signaling pathway and ROS-dependent pathway.
TABLE 1.
The role of the Fox subfamilies in osteogenic differentiation.
| Fox genes | Expression | Target genes/pathway | Effects on osteogenesis | Cells/animal model | Reference |
|---|---|---|---|---|---|
| FoxO1 | Increased | Runx2, ALP, OCN | Promoted | C3H10T1/2 cells | Teixeira et al. (2010) |
| FoxO1 | Increased | miR-424/FGF2, Runx2 | Promoted | BMSCs | Li et al. (2017) |
| FoxO1/3/4 | Deleted | PPARγ, Runx2, Osx, p66shc | Inhibited | Deletion of FoxO1/3/4 mice | Ambrogini et al. (2010) |
| FoxO1/3/4 | Deleted | Wnt/β-catenin, cyclin D1 | Promoted | Bipotential progenitors of osteoblast and adipocytes | Iyer et al. (2013) |
| FoxO3a | Increased | ALP, OCN, Runx2, LRP5, LRP6 | Promoted | BMSCs | Sun et al. (2018) |
| FoxO3a | Increased | OCN, Runx2 | Inhibited | MC3T3-E1 cells | Tang et al. (2019) |
| FoxC1 | Decreased | Msx2, Runx2, ALP | Inhibited | C2C12 cells | Hopkins et al. (2016) |
| Decreased | Runx2, osterix | Promoted | MC3T3 cells | Hopkins et al. (2016) | |
| FoxC1 | Increased | Msx2, Runx2, ALP | Promoted | C2C12 cells | Mirzayans et al. (2012) |
| FoxC1 | KD | Msx2, ALP, OCN, Runx2 | Promoted | O9-1 cells | Sun et al. (2013) |
| FoxC2 | Increased | ALP, OCN, Cbfα1, Wnt/β-catenin, BSP, PPARγ2 | Promoted | BMSCs | (Lin et al., 2016) |
| FoxP1 | OE | Recombination signal-binding protein, ALP | Promoted | C3H10T1/2 cells | Li et al. (2017) |
| KO | ALP, COL1A1, PPARG, CEBPA, FABP4 | Inhibited | BMSCs | Li et al. (2017) | |
| FoxP1/2/4 | Decreased | Runx2 | Promoted | Skeletal progenitor cells | Zhao et al. (2015) |
| FoxA2 | KD | ERK, ALP, OPN, OCN, Runx2, Col1a1 | Promoted | BMMSCs | Ye et al. (2018) |
| FoxD3 | OE | OSX, TNAP, SOX9, OPN | Promoted | hESCs | Kamaldinov et al. (2018) |
| FoxD3 | Increased | Wnt/β-catenin | Inhibited | BMMSCs | Huang and Chen, (2017) |
| FoxF1 | KD | Wnt/β-catenin | Promoted | BMSCs | Shen et al. (2020) |
| FoxF1 | Increased | TGF/β-catenin | Promoted | BMSCs | (Weng et al., 2019) |
| FoxM1 | Decreased | RANKL/OPG, ALP | Promoted | PDLCs | Li et al. (2019) |
| FoxQ1 | Increased | Wnt/β-catenin, ANXA2, ALP, OCN, OPG, Runx2 | Promoted | BMSCs | Xiang et al. (2020) |
| FoxI1 | Deleted | Fgf8 | Inhibited | Foxi1 mutant embryo | Nissen et al. (2003) |
| FoxI3 | Deleted | Pax8, Fgf3 | Inhibited | Foxi3 mutant Mice | Edlund et al. (2014) |
| FoxL2 | Deleted | GH/IGF1, SOX9 | Inhibited | FoxL2−/− mice | Marongiu et al. (2015) |
| FoxG1 | Decreased | Osteocalcin, ALP | Inhibited | Osteoblasts | Uchida et al. (2018) |
Note. OE, overexpression; KD, knock down; BMSCs, bone marrow mesenchymal stem cells; BMMSCs, bone marrow‐derived mesenchymal stem cells; PDLCs, periodontal ligament cells; hESCs, human embryonic stem cells; ALP, alkaline phosphatase; Runx2, Runt-related transcription factor 2; OCL, osteocalcin; PPARγ, proliferator-activated receptor γ2; msx2, msh homeobox 2; ERK, extracellular-regulated protein kinases; RANKL, receptor activator of nuclear factor-κB ligand.
TABLE 2.
The role of the Fox subfamilies in osteoclast differentiation.
| Fox genes | Expression | Target genes/pathway | Effects on osteoclastogenesis | Cells/animal model | Reference |
|---|---|---|---|---|---|
| FoxO1 | Decreased | PI3K/AKT | Promoted | Mice with conditional loss | Bartell et al. (2014) |
| FoxO1 | OE | Myc, ERK | Inhibited | RAW264.7 cells | Tan et al. (2015) |
| FoxO1 | Decreased | NFATc1 | Promoted | Bone marrow macrophages/RAW264.7 osteoclast precursors | Wang et al. (2015) |
| FoxM1 | KD | RANKL/OPG | Promoted | RAW264.7 cells | Li et al. (2019) |
| FoxM1 | Deletion | RANKL/OPG/RANK | Inhibited | Mice AtoMs | Hasegawa et al. (2019) |
| FoxP1 | OE | INF-γ, MCP-1 | Inhibited | FoxP1-overexpressing transgenic mice | Shi et al. (2008) |
| FoxP3 | Increased | RANKL/OPG/RANK and IFNγ | Promoted | Osteoclast precursors | (Zaiss et al., 2007; Zaiss et al., 2010) |
2.1 Fox factors with dual action on bone metabolism
2.1.1 FoxO family
In mammals, FoxO family consists of four members: FoxO1, FoxO3, FoxO4, and FoxO6. Among these, FoxO1, FoxO3, and FoxO4 are expressed in the bone, while FoxO6 is confined to specific structures of the developing brain (Jacobs et al., 2003). In this review, due to the limitation of technology, some papers did not mention the specific member of the FoxO family but mentioned FoxO in general in their papers. Thus, in our paper, FoxO was represented as FoxO1/3/4. FoxO activity is mainly regulated by the PI3K/Akt pathway (Brunet et al., 1999) and posttranslational modifications (phosphorylation, acetylation, and ubiquitination) (Ambrogini et al., 2010). In addition, it can reduce the production of reactive oxygen species (ROS) by regulating antioxidant enzymes (superoxide dismutase and catalase) (Salih and Brunet, 2008) and upregulating mitochondrial function (Zorov et al., 2014) to mediate oxidative stress, thereby stimulating the differentiation of bone marrow mesenchymal stem cells (BMSCs) into osteoblasts and inhibiting their senescence (Sun et al., 2018) (Figure 2 “Osteoblast” part). Consistently, silencing of FoxO1 was found to inhibit the expression of osteogenic markers, such as Runt-related transcription factor 2 (Runx2), alkaline phosphatase (ALP), and osteocalcin (OCN), even in the presence of strong osteogenic stimulants, resulting in decreased culture calcification (Teixeira et al., 2010). Moreover, mice with conditional deletions of FoxO1, FoxO3, and FoxO4 showed profound loss of bone mass in calvaria, vertebrae, and femoral bone, which was caused by the upregulation of osteoblast apoptosis and oxidative stress (Ambrogini et al., 2010). In addition, three main FoxO isoform mice with deletion in growth plate chondrocytes showed a distinct elongation of the hypertrophic zone of the growth plate in the neonatal period; these mice showed severe skeletal deformities at older ages, indicating the importance of FoxO signaling in chondrocytes during endochondral ossification (Salih and Brunet, 2008).
However, studies have provided different insights on the function of FoxO family (Figure 2 “Osteoblast precursors” part). Increased oxidative stress was found to activate the FoxO family, which inhibited the Wnt/β-catenin signaling pathway through competing with T-cell factor transcription factor for binding with β‐catenin, and then decreased bone formation (Iyer et al., 2013). Moreover, overexpression of FoxO3 inhibited alterations in the expression of the calcium channel and decreased calcium absorption and calcium deposition, thereby preventing osteoblast differentiation (Tang et al., 2019). These studies suggest that the regulatory effect of FoxO on osteogenic differentiation is closely associated with the type of cell; for instance, in osteoblast precursors and osteoblasts, the FoxO family showed an absolutely opposite regulatory effect on osteoblast differentiation.
Similarly, contradictory findings have been reported about the regulatory effect of FoxO family on osteoclast differentiation (Figure 2 “Osteoclast precursors” part). In mice with conditional loss, receptor activator of nuclear factor-κB ligand (RANKL) was shown to induce activation of the Src pathway, subsequently activating the PI3K–Akt pathway, and downregulating FoxO1/3/4, thereby decreasing catalase, and increasing the accumulation of H2O2 and the levels of ROS (Bartell et al., 2014). This ultimately enhanced osteoclast formation, activation, and survival. Furthermore, Tan et al. found that the inhibitory effect of FoxO1 on osteoclast development was partly mediated by suppression of MYC and upregulation of extracellular-regulated protein kinases (Erk, Tan et al., 2015). However, in the study by Wang et al., FoxO1 deletion decreased osteoclastogenesis and RANKL-induced osteoclast activity in both experimental bone marrow macrophages and in RAW264.7 cells (Wang et al., 2015). These two contradictory results actually reflect the different effects of the FoxO family at different time periods on osteoclast differentiation; the long-term effect was associated with oxidative stress and aging, which enhanced osteoclast formation, and the short-term effect was associated with modulation of RANKL-stimulated osteoclast formation, which inhibited osteoclast formation.
Collectively, although there is no clear consensus about the ultimate effect of FoxO on osteoblast differentiation and osteoclast differentiation, we were able to identify the importance of FoxO transcription factors involved in the regulation of bone metabolism, including through antioxidative stress and the PI3K/Akt pathway, as potential therapeutic targets for osteoporosis. In particular, it seems that the PI3K/Akt pathway may be a coupling target of FoxO on osteoblast differentiation and osteoclast differentiation.
2.1.2 FoxP family
The FoxP family consists of four members: FoxP1, FoxP2, FoxP3, and FoxP4 (Takahashi et al., 2009). Among these, FoxP1/2/4, which play a key role in the development of proper long bone in transgenic mice, are suggested to be negative regulators of Runx2 (Zhao et al., 2015) (Figure 2 “Osteoblast” part). Overexpression of FoxP1/2/4 in chondrocytes inhibits the endochondral ossification pathway and severely impairs chondrocyte hypertrophy and osteoblast differentiation (Zhao et al., 2015). In addition, FoxP1 significantly affected proliferator-activated receptor γ2 (PPARγ2) transcription, increasing lipogenic differentiation of mesenchymal progenitors at the cost of osteogenic differentiation (Wang et al., 2020) (Figure 2 “Osteoblast precursors” part). On the contrary, because of the dual role in regulating the fate switch and aging of MSCs, a study conducted by Li et al. found that FoxP1 favors bone formation over adipogenesis and may be a potential target gene for the treatment of osteoporosis (Li et al., 2017). Meanwhile, loss of Foxp2 in skeletal tissue also led to pleiotropic deficits in skull shaping and bone strengthening, indicating that Foxp2 played a key role in the process of endochondral ossification (Xu et al., 2018). These different phenomena may be due to the difference in cells, tissues, and physiological microenvironment. FoxP1/2/4 play an important role in bone formation, in particular, FoxP1; however, further studies are required to unravel the complex regulatory effect on osteogenic, chondrogenic, and lipogenic differentiation.
Interestingly, FoxP family also plays a pivotal role in osteoclast differentiation. Osteoclasts originate from the monocyte lineage, and defects in monocyte differentiation are usually accompanied by disordered osteoclastogenesis (Dai et al., 2002). Upregulation of FoxP1 has been shown to result in impaired monocyte and macrophage function. Compared with wild-type mice, reduced total tartrate-resistant acid phosphatase (TRAP)-positive cells and decreased osteolytic viability were observed in transgenic mice overexpressing human FoxP1 under induction of macrophage colony-stimulating factor and RANKL (Shi et al., 2008). FoxP3, a transcription factor expressed by T-regulatory cells, is a spectrum master regulator of Treg cell development and suppressor activity (Deng et al., 2019). FoxP3 + Treg cells inhibit RANKL-induced osteoclastogenesis through various mechanisms that may be cytokine dependent, such as IL-4, IL-10, and TGF-β, or cell dependent via cytotoxic T-lymphocyte-associated antigen-4 contact (Zaiss et al., 2007). FoxP3 overexpression in mice resulted in decreased numbers of osteoclasts, resulting in reduced bone resorption activity; the reduced osteoclast numbers were not caused by an intrinsic defect in osteoclast differentiation. Nevertheless, FoxP3-deficient bone marrow increased local and systemic bone loss (Zaiss et al., 2010). Thus, the FoxP family inhibits osteoclast differentiation.
Collectively, the available evidence suggests a key role of the FoxP family in the prevention and treatment of osteoporosis through its involvement in bone metabolism and its protective effect against cellular senescence. However, there is a paucity of studies investigating the roles of the FoxP family in bone formation. Further studies are required to unravel the specific mechanisms and effects of FoxP on osteoblastogenesis and osteoclastogenesis.
2.1.3 FoxM1
Inhibition of FoxM1 has been shown to enhance osteogenic differentiation of human periodontal ligament cells (PDLCs) (Li et al., 2019). Besides, the Wnt/β-catenin signaling pathway, an important pathway in osteogenic differentiation (Zhou et al., 2017; Huang et al., 2019), has been shown to be linked with FoxM1 in the field of oncology (Zhang et al., 2011; Gong and Huang, 2012; Chen et al., 2016). However, whether FoxM1 acts on the Wnt/β-catenin signaling pathway to regulate osteogenic differentiation has not been investigated.
There is no clear consensus on the regulatory role of FoxM1 in osteoclast differentiation. On the one hand, TRAP staining showed an increase in the number of multinucleated osteoblasts in Raw264.7 cells and upregulated RANKL/osteoprotegerin ratio in PDLCs when human PDLCs were inhibited with siRNA and specific inhibitor Siomycin A of FoxM1 cocultured with Raw264.7 cells (Li et al., 2019). On the other hand, FoxM1 deletion was found to partially inhibit synovial R3 cell osteoclastogenesis in vitro, and the bone degradation attenuated by tamoxifen-induced FoxM1 deletion was partly reversed by the overt transfer of FoxM1+/+ CX3CR1+ monocytes in vivo (Hasegawa et al., 2019). These findings suggest that deletion of FoxM1 inhibits the ability of osteoclast precursors to differentiate into osteoclasts both in vivo and in vitro.
There is obvious evidence of the involvement of FoxM1 in the regulation of bone metabolism and influencing the development of osteoporosis. However, the underlying mechanisms are not well understood due to a paucity of related studies.
1.2 Fox factors that affect osteogenic differentiation alone in bone metabolism
In addition to FoxO family, FoxP family, and FoxM1, several Fox subfamilies have been reported to be involved in the regulation of bone metabolism, which is limited to the regulation of osteogenic differentiation.
FoxC genes are pivotal in the regulation of bone development and cartilage formation (Chen et al., 2019; Yoshida et al., 2015; Xu et al., 2021). In particular, FoxC1 is an important regulator both in the initial steps of intramembranous osteogenesis (Hopkins et al., 2016) and in early and late endochondral ossification (Yoshida et al., 2015). Mice with spontaneous loss of function mutations (FoxC1ch/ch) die shortly after birth and exhibit skeletal abnormalities and defects (Hong et al., 1999). In vivo, FoxC1 exhibits different osteogenic differentiation effects on different cells (Hopkins et al., 2016). FoxC1 regulates osteogenic precursor cell differentiation and cranial bone development through its action on msh homeobox 2, a key regulator of bone formation and craniofacial skeletal development (Mirzayans et al., 2012) (Figure 2 “Osteoblast precursors” part); however, its ultimate effects are divergent (Rice et al., 2003; Mirzayans et al., 2012; Sun et al., 2013). Besides, FoxC2 acts on the Wnt signaling pathway to promote bone formation. On the one hand, FoxC2 directly activates the classical Wnt/β-catenin signaling pathway, increasing the expression of osteogenic markers, such as Runx2, COL1A1, OCN, and osteopontin, and inhibiting the expression of PPARγ2 (You et al., 2013; You et al., 2014; Zhou et al., 2019a). On the other hand, FoxC2 binds to the Wnt4 promoter and stimulates the nonclassical Wnt signaling pathway by activating the p38/MAPK pathway (Chang et al., 2007) and inhibiting NF-κB to promote differentiation and bone formation in BMSCs (Yu et al., 2014) (Figure 2 “Osteoblast” part).
FoxI1/3 and FoxL2 genes are also important regulators of bone development (Ohyama and Groves, 2004; Uda et al., 2004). Both FoxI1 deletion in zebrafish (Nissen et al., 2003) and FoxI3 mutants in mice (Edlund et al., 2014) cause severe structural defects of the facial skeleton, such as malformation and absence of the external ear and jaws (Edlund et al., 2015). FoxL2−/− mice, which died in large numbers soon after birth (Uda et al., 2004), showed abnormal cranial, vertebral, and pelvic development, with bone loss and impaired cartilage formation (Marongiu et al., 2015).
In addition, knockdown of FoxA2 (FoxA2-KD) promotes osteogenic differentiation of BMSCs partially activating the ERK pathway (Ye et al., 2018) and overexpression of FoxQ1 promotes osteogenic differentiation of BMSCs through the Wnt/β-catenin pathway by binding with annexin a267 (Figure 2 “Osteoblast” part). FoxG1 knockout (KO) osteoblasts exhibit lower mRNA expressions of Runx2, Osterix, and ALP; however, the underlying mechanism is still unknown (Kimira et al., 2017; Uchida et al., 2018).
There is inconsistency in the reported effects of FoxD3 and FoxF1 on osteoblast differentiation (Figure 2 “Osteoblast” part). Upregulation of FoxF1 during MOTS-c-induced osteogenesis activates the TGF-β pathway, thereby promoting fracture healing (Zeng et al., 2017). However, in another study, FoxF1 knockdown significantly increased osteogenic-specific gene expression and mineralization, which was associated with partial activation of the Wnt/β-catenin pathway (Shen et al., 2020). In ovariectomized (OVX) mice, knockdown of FoxF1 with siRNA significantly reduced OVX-induced bone loss by enhancing bone formation, suggesting that FoxF1 may be a marker factor for bone formation and a therapeutic target for postmenopausal osteoporosis (Shen et al., 2020). The mRNA and protein expressions of FoxD3 were upregulated in bone marrow‐derived mesenchymal stem cells treated with IL-1β. Huang et al. pointed out that FoxD3 may mediate transcriptional activation of miR-496 triggered by IL-1β, thereby repressing the Wnt/β-catenin signaling pathway and reducing osteoblast differentiation (Huang and Chen, 2017). Interestingly, a study by Kamaldinov et al. showed that overexpressed FoxD3 may enhance osteogenesis in human embryonic stem cells via the endochondral ossification pathway (Kamaldinov et al., 2018).
The above evidence demonstrates that Fox factor plays a key role in osteoporosis pathology by acting on both osteoblast differentiation and osteoclast differentiation or by influencing osteoblast differentiation alone to regulate bone metabolism.
3 Role of Fox factors in osteoarthritis
Osteoarthritis is a chronic joint disease characterized by degenerative changes in joint cartilage. Aging and inflammation are the main risk factors for osteoarthritis (Duan et al., 2020). Studies have indicated the involvement of the FoxO family and FoxM1 in the development of osteoarthritis. In an in vitro experiment, upregulation of FoxO increased the expressions of autophagic genes (Map1lc3b, Atg4b, Becn1, Gabarapl1, and Bnip3), which could prevent aging, protecting cartilage by mediating apoptosis, and elimination of ROS (Charlier et al., 2016). Moreover, by upregulating mitochondrial function and reducing intracellular ROS (Bartell et al., 2014), and reducing the production of inflammatory factors (chemerin) and cartilage-degrading enzymes (Akasaki et al., 2014), FoxO1 delayed chondrocyte senescence and reduced chondrocyte apoptosis, respectively. Matsuzaki et al. found that FoxO1 acts synergistically with TGF-β to activate recombinant proteoglycan 4 expression, which is essential for maintaining the integrity of the superficial cartilage region. FoxO triple KO mice (AcanCreERT-TKO) exhibited complete cartilage defects, more severe synovial inflammation, and subchondral bone changes after administration of tamoxifen for 5 months (Matsuzaki et al., 2018). FoxM1 impairs chondrocyte viability and accelerates the development of osteoarthritis. In a lipopolysaccharide-induced osteoarthritis model, FoxM1 was shown to bind with signal transducer and activator of transcription3 (STAT3) in the nucleus, leading to its upregulation and phosphorylation, impairing chondrocyte viability. Knockdown/silencing of FoxM1 inhibited the production of inflammatory factors and NF-κB activation, enhancing cell viability in an osteoarthritis model (Zeng et al., 2019; Zeng et al., 2021). Moreover, miR-877-5p was shown to improve chondrocyte function by inhibiting FoxM1 in both in vivo and in vitro experiments (Zhu et al., 2020a).
4 Role of Fox factors in rheumatoid arthritis
Rheumatoid arthritis is a chronic joint disease characterized by persistent synovitis and associated damage to the articular cartilage and subchondral bone. Smoking is a major environmental risk factor for rheumatoid arthritis (Scott et al., 2010). Activation of the PI3K–Akt axis by differentially expressed miRNAs in smokers leads to rheumatoid arthritis, in part through FoxO inactivation (Reedquist et al., 2006; Wasén et al., 2020). However, rapid downregulation of FoxO1 in rheumatoid arthritis fibroblast-like synoviocytes in response to IL-1β or PDGF stimulation is independent of Akt and results from accelerated c-Jun N-terminal kinase (JNK)-mediated degradation of FoxO1 mRNA (Grabiec et al., 2015). Moreover, autophagy protects chondrocytes from glucocorticoid-induced apoptosis through upregulation of the ROS/Akt/FoxO3 signaling pathway (Shen et al., 2015). In addition to the FoxO family, FoxM1, which is related to damage to subchondral bone, may also play a role in the pathogenesis of rheumatoid arthritis (Hasegawa et al., 2019). High energy is required for osteoclastogenesis under arthritic conditions, and FoxM1 directly drives mitochondrial biogenesis (De Luca et al., 2015), promoting differentiation of AtoMs into osteoclasts. In vivo, FoxM1 inhibition alleviated not only articular bone destruction but also joint inflammation (Hasegawa et al., 2019). Thus, thiostrepton, a direct inhibitor of FoxM1, which inhibits FoxM1 binding to genomic target sites (Hegde et al., 2011), may be a new approach to rheumatoid arthritis treatment.
5 Role of Fox factors in intervertebral disc homeostasis and intervertebral disc degeneration
Intervertebral disc (IVD) is a fibrocartilaginous tissue that lies between two vertebrae and functions as a shock absorber. It includes the jelly-like nucleus pulposus, the surrounding fibrocartilaginous annulus fibrosus, and the cartilaginous endplate anchoring the IVD to the corpus vertebrae (Kamali et al., 2021). Intervertebral disc degeneration (IDD), the major cause of chronic low back pain (Maher et al., 2017), was recently shown to be closely related with the Fox family. FoxO is required for intervertebral disc homeostasis during aging, and its deficiency promotes disc degeneration (Alvarez-Garcia et al., 2018). In a study by Xia et al., upregulation of FoxO3 promoted proliferation and inhibited apoptosis of nucleus pulposus cells in IDD (Xia et al., 2021). Moreover, FoxO3 was shown to retard IDD by antioxidative stress (Zhou et al., 2019b). Furthermore, FoxA1−/−, FoxA2c/c, and ShhcreERT2 double mutant animals showed severely deformed nucleus pulposus, increase in cell death in the tail, decreased hedgehog signaling, defects in the notochord sheath, and aberrant dorsal–ventral patterning of the neural tube (Maier et al., 2013). Recently, Zhou et al. revealed that FoxA2 regulates the type II collagen-induced nucleus pulposus-like differentiation of adipose-derived stem cells via activation of the Shh signaling pathway (Zhou et al., 2018).
6 Role of fox factors in bone tumors
6.1 Osteosarcoma
Osteosarcoma (OS) is the most common primary malignant tumor of bone, and it occurs mainly in children and adolescents (Kansara et al., 2014). Recently, researchers have conducted in-depth studies on the mechanisms of FoxO, FoxM1, and FoxC2 in the development of OS and the drugs that target these Fox factors. First, FoxO induces G1 cell cycle arrest, apoptosis, and DNA repair (Nakamura et al., 2000), and is considered to be a tumor suppressor (Niedan et al., 2014). FoxO1 expression was absent or low in OS cells, and upregulation of FoxO1 expression induced OS cell cycle arrest and apoptosis, and reduced the number of colonies (Guan et al., 2015). For instance, E2F transcription factor 1 induces the expression of FoxO1 and interacts with it to activate the target gene apoptotic protease-activating factor-1, promoting apoptosis in U2OS cells (Shats et al., 2013). Therefore, a number of drugs targeting FoxO are already being used in studies for OS treatment (Herman et al., 2020). Grifolin inhibits the PI3K/Akt/FoxO1 pathway in human OS cells suppressing their proliferation and inducing apoptosis (Jin et al., 2007). Brazilin was shown to increase the expression of autophagy-related genes and promote death of human OS cells (MG-63 cells) by interfering with the steady-state phosphorylation of the Ser7 site of FoxO3a by Ca2+ (Kang et al., 2018).
Second, FoxM1 is associated with tumor cell proliferation, migration, invasion, and angiogenesis, suggesting that FoxM1 is an oncogenic factor (Halasi and Gartel, 2013). FoxM1 is highly expressed in human OS tissues and cell lines, and downregulation of FoxM1 expression was found to inhibit the viability, migration, and invasive growth of OS cells (Zhu et al., 2020b). Recent studies have shown that avasimibe (Wang et al., 2019a), diallyl disulfide (Li et al., 2018), thiostrepton (Cai et al., 2020), and some miRNAs [including miR-134 (Li et al., 2018), miR-370 (Duan et al., 2015), miR-216b (Wang et al., 2019b), and miR-197 (Sun et al., 2020)] inhibit the proliferation and invasive growth of OS cells by directly or indirectly downregulating FoxM1 expression.
Third, FoxC2, a transcription factor involved in epithelial–mesenchymal transition (EMT), is defined as a carcinogenic factor (Kalluri, 2009). Silencing of FoxC2 expression attenuated anchored nondependent growth of OS cells and reduced the invasive ability, which may be related to downregulation of C-X-C motif chemokine receptor 4 (Gozo et al., 2016). FoxC2 can regulate chemoresistance in OS; for instance, siRNA transfection-mediated knockdown of FoxC2 increased the sensitivity of two OS cell lines to doxorubicin (Zhang et al., 2017).
Lastl, the FoxP family was found to be closely related with OS (Gascoyne et al., 2015; Yin et al., 2017; Li et al., 2021). In the study by Li et al., FoxP1 was shown to promote proliferation, tumor sphere formation, migration and invasion, and inhibit anoikis by FOXP1 overexpression and knockdown in OS cell lines (Li et al., 2021). In 143B OS cells with minimal endogenous expression, FOXP2 induced by growth arrest is required for upregulation of p21WAF1/CIP1 107. Upregulation of miR-491-5p suppressed proliferation of human OS cells and induced apoptosis by targeting FoxP4 (Yin et al., 2017).
6.2 Ewing sarcoma
Ewing sarcoma (ES) is a rare and highly aggressive cancer that occurs primarily in the bones and surrounding tissues of children and adolescents. It is the second most common primary malignant bone tumor in children and adolescents (Balamuth and Womer, 2010; Dowless et al., 2018). The pathogenetic mechanism of ES is still not well understood. Studies have focused on the downstream target genes of the oncogenic transcription factor EWS/FLI1 (Balamuth and Womer, 2010). FoxO1, FoxM1, and FoxQ1, members of the Fox transcription factor family were shown to be three potent targets downstream of EWS/FLI1 (Cidre-Aranaz and Alonso, 2015).
FoxO1, as previously mentioned, is a tumor-suppressor factor with low expression in ES caused by the repressive effect of EWS/FLI1 binding to the FoxO1 promoter (Yang et al., 2010). Besides, negative regulation of FoxO1 activity and nuclear localization, caused by CDK2 (a negative regulator of EWS–FLI1-induced FoxO1 transcriptional activity) and PI3K/Akt-mediated FoxO1 phosphorylation, accelerated proliferation and promoted soft agar colony formation in two Ewing sarcoma cell lines (A673sh and SK-N-MC) (Niedan et al., 2014). Thus, methylseleninic acid, a drug reported to induce elevated expression of FoxO1 in ES cells, apoptosis of ES cells, and significant reduction of tumor growth in an orthotopic mouse xenotransplantation model, may be a potential target drug for Ewing sarcoma (Cidre-Aranaz and Alonso, 2015).
FoxM1, an oncogenic factor, is highly expressed in Ewing sarcoma and cell lines. Reduction of FoxM1 expression impairs the ability of Ewing cell lines to grow in an anchorage-independent manner (Christensen et al., 2013). Based on this, some studies have shown that thiazole antibiotics and proteasome inhibitors represented by thiostrepton (Gartel, 2011) and Siomycin A (Bhat et al., 2009) may have a role in the treatment of Ewing sarcoma by inhibiting FoxM1 expression.
High expression of FoxQ1 was detected in human ES lines KH and EWS. FoxQ1 may be associated with enhanced activation of downstream target genes through interaction with EWS–FLI1 at the N-terminal end of EWS, thereby promoting ES proliferation (Shimizu et al., 2018).
6.3 Metastatic bone tumors
During EMT, elevated FoxI3 expression correlates with the dedifferentiated state and motility of cells (Ye et al., 2015). Recent studies have demonstrated high expression of FoxI3 in bone metastases from prostate and breast cancers, suggesting that FoxI3 may promote bone metastasis and tumor growth and infiltration in the bone (Haider et al., 2016; Mukherjee et al., 2018). In addition, knockdown of FoxA2 inhibited bone metastasis of prostate cancer. Osteolytic lesions and tumor incidence were significantly lower in the tibia of FoxA2-suppressed mice compared with the control group (Connelly et al., 2020), which may be related to decreased expression of parathyroid hormone-related protein, a major factor mediating cancer-induced osteoclast production (Martin and Johnson, 2019), encoded by the PTHLH gene (Connelly et al., 2020).
7 Role of Fox factors in hereditary bone diseases
FoxC1 mutation is associated with Axenfeld–Rieger syndrome, an autosomal dominant disorder characterized by major skeletal abnormalities, such as mild craniofacial deformities, including forehead protrusion, limb hypertrophy, anterior sphenoid, and maxillary hypoplasia (Seifi and Walter, 2018; Chen et al., 2019). Heterozygous mutation in FoxL2 is associated with blepharophimosis-ptosis-epicanthus inversus syndrome, an autosomal dominant disorder that manifests primarily as eyelid and mild craniofacial defects (Jin et al., 2007).
8 Conclusion and perspectives
The role of Fox factors in skeletal development and skeletal dynamic homeostasis has attracted much academic attention. The role of Fox factors as effectors in many signaling pathways and the associated regulatory mechanisms, including their actions on downstream targets, is an emerging research hotspot. Uncontrolled expression of Fox factor can lead to a variety of bone diseases, such as osteoporosis, osteoarthritis, rheumatoid arthritis, and bone tumors. Regulation of Fox expression has important clinical implications for the prevention and treatment of these bone diseases. In the context of progressive population aging and increase in the number of elderly patients with bone diseases (such as fractures, osteoporosis, and rheumatoid arthritis), development of new and more effective treatments is a key imperative. The role of the Fox gene family represents an entirely new area of research in bone metabolism that is expected to address this challenge.
Although Fox factors have been identified to play a pivotal role in the maintenance of bone homeostasis, much of the contemporary research is limited to cellular and animal studies. The use of drugs targeting Fox factors in the treatment of bone tumors has been attempted; however, there are few reports of clinical efficacy. Apart from this, development of targeted drugs for Fox factors in the clinical treatment of other skeletal diseases is still at a theoretical stage. Therefore, clarifying the prospects for clinical application of the Fox factors and development of Fox-targeted drugs represent key areas of future research.
Acknowledgments
We thank all clients from the Clinical Research Center of the Second Affiliated Hospital, Zhejiang University, including Xing Zhang, Jiayu Yang, Am Liu, Chunc Li et al. We also appreciate the general help of Xiaoh Y, Lingl Zhang, and Sm Feng from the Orthopedic Research Center of the Second Affiliated Hospital, Zhejiang University. We thank Medjaden Inc. for the scientific editing of this manuscript.
Author Contributions
DX, WL, and ZP designed this review. JX, KW, and ZZ made the figures and tables. JX, DX, WL, and ZP wrote the paper. JX and ZP revised the paper.
funding
The National Natural Science Foundation of China (Grant No. 81874007 and 82172404) funds were received in support of this work. The authors declare no relevant financial activities outside the submitted work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- Fox:
forkhead box
- SIRT:
sirtuin
- WNT:
wingless and int-1
- BMP:
bone morphogenetic protein
- Smad:
drosophila mothers against decapentaplegic
- PI3K:
phosphatidylinositol 4,5-bisphosphate 3-kinase
- TGF-β:
transforming growth factor-β
- MAPK:
mitogen-activated protein kinase
- NF-κB:
nuclear factor-kappa B
- ROS:
reactive oxygen species
- BMSCs:
bone marrow mesenchymal stem cells
- Runx2:
Runt-related transcription factor 2
- TCF:
T-cell factor
- RANKL:
receptor activator of nuclear factor-κB ligand
- Erk:
extracellular-regulated protein kinases
- PPARγ2:
proliferator-activated receptor γ2
- TRAP:
tartrate-resistant acid phosphatase
- M-CSF:
macrophage colony-stimulating factor
- Tregs:
T-regulatory cells
- IL:
interleukin
- CTLA-4:
cytotoxic T lymphocyte-associated antigen-4
- OPG:
osteoprotegerin
- PDLCs:
periodontal ligament cells
- Msx2:
msh homeobox two
- COL1A1:
collagen type I a 1
- OCN:
osteocalcin
- OPN:
osteopontin
- ANXA2:
annexin a2
- Osx:
osterix
- ALP:
alkaline phosphatase
- OVX:
ovariectomized
- hESCs:
human embryonic stem cells
- Prg4:
recombinant proteoglycan four
- OE:
overexpression
- KD:
knockdown
- KO:
knock out
- LPS:
lipopolysaccharide
- STAT3:
signal transducer and activator of transcription3
- FLS:
fibroblast-like synoviocytes
- JNK:
c-Jun N-terminal kinase
- IDD:
intervertebral disc degeneration
- OS:
osteosarcoma
- E2F1:
E2F transcription factor 1
- APAF1:
apoptotic protease activating factor-1
- CXCR4:
C-X-C motif chemokine receptor four
- ES:
Ewing sarcoma
- MSA:
methylseleninic acid
- PTHLH:
parathyroid hormone-related protein
- ARS:
Axenfeld–Rieger syndrome
- BPES:
blepharophimosis-ptosis-epicanthus inversus syndrome
References
- Akasaki Y., Alvarez-Garcia O., Saito M., Caramés B., Iwamoto Y., Lotz M. K. (2014). FoxO Transcription Factors Support Oxidative Stress Resistance in Human Chondrocytes. Arthritis Rheumatol. 66, 3349–3358. 10.1002/art.38868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia O., Matsuzaki T., Olmer M., Miyata K., Mokuda S., Sakai D., et al. (2018). FOXO Are Required for Intervertebral Disk Homeostasis during Aging and Their Deficiency Promotes Disk Degeneration. Aging Cell 17, e12800. 10.1111/acel.12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogini E., Almeida M., Martin-Millan M., Paik J. H., Depinho R. A., Han L., et al. (2010). FoxO-mediated Defense against Oxidative Stress in Osteoblasts Is Indispensable for Skeletal Homeostasis in Mice. Cell Metab 11, 136–146. 10.1016/j.cmet.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamuth N. J., Womer R. B. (2010). Ewing's Sarcoma. Lancet Oncol. 11, 184–192. 10.1016/S1470-2045(09)70286-4 [DOI] [PubMed] [Google Scholar]
- Bartell S. M., Kim H. N., Ambrogini E., Han L., Iyer S., Serra Ucer S., et al. (2014). FoxO Proteins Restrain Osteoclastogenesis and Bone Resorption by Attenuating H2O2 Accumulation. Nat. Commun. 5, 3773. 10.1038/ncomms4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia D., Dimarco E., Costa V., Carina V., De Luca A., Raimondi L., et al. (2021). Flavonoids in Bone Erosive Diseases: Perspectives in Osteoporosis Treatment. Trends Endocrinol. Metab. 32, 76–94. 10.1016/j.tem.2020.11.007 [DOI] [PubMed] [Google Scholar]
- Bhat U. G., Halasi M., Gartel A. L. (2009). Thiazole Antibiotics Target FoxM1 and Induce Apoptosis in Human Cancer Cells. PloS one 4, e5592. 10.1371/journal.pone.0005592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., et al. (1999). Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell 96, 857–868. 10.1016/s0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- Cai X., Xiao W., Shen J., Lian H., Lu Y., Liu X., et al. (2020). Thiostrepton and miR-216b Synergistically Promote Osteosarcoma Cell Cytotoxicity and Apoptosis by Targeting FoxM1. Oncol. Lett. 20, 391. 10.3892/ol.2020.12254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Sonoyama W., Wang Z., Jin Q., Zhang C., Krebsbach P. H., et al. (2007). Noncanonical Wnt-4 Signaling Enhances Bone Regeneration of Mesenchymal Stem Cells in Craniofacial Defects through Activation of P38 MAPK. J. Biol. Chem. 282, 30938–30948. 10.1074/jbc.M702391200 [DOI] [PubMed] [Google Scholar]
- Charlier E., Relic B., Deroyer C., Malaise O., Neuville S., Collée J., et al. (2016). Insights on Molecular Mechanisms of Chondrocytes Death in Osteoarthritis. Int. J. Mol. Sci. 17, 2146. 10.3390/ijms17122146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wei H., Li J., Liang X., Dai S., Jiang L., et al. (2019). Structural Basis for DNA Recognition by FOXC2. Nucleic Acids Res. 47, 3752–3764. 10.1093/nar/gkz077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li Y., Xue J., Gong A., Yu G., Zhou A., et al. (2016). Wnt-induced Deubiquitination FoxM1 Ensures Nucleus β-catenin Transactivation. EMBO J. 35, 668–684. 10.15252/embj.201592810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen L., Joo J., Lee S., Wai D., Triche T. J., May W. A. (2013). FOXM1 Is an Oncogenic Mediator in Ewing Sarcoma. PloS one 8, e54556. 10.1371/journal.pone.0054556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cidre-Aranaz F., Alonso J. (2015). EWS/FLI1 Target Genes and Therapeutic Opportunities in Ewing Sarcoma. Front. Oncol. 5, 162. 10.3389/fonc.2015.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly Z. M., Jin R., Zhang J., Yang S., Cheng S., Shi M., et al. (2020). FOXA2 Promotes Prostate Cancer Growth in the Bone. Am. J. Transl Res. 12, 5619–5629. [PMC free article] [PubMed] [Google Scholar]
- Dai X. M., Ryan G. R., Hapel A. J., Dominguez M. G., Russell R. G., Kapp S., et al. (2002). Targeted Disruption of the Mouse colony-stimulating Factor 1 Receptor Gene Results in Osteopetrosis, Mononuclear Phagocyte Deficiency, Increased Primitive Progenitor Cell Frequencies, and Reproductive Defects. Blood 99, 111–120. 10.1182/blood.v99.1.111 [DOI] [PubMed] [Google Scholar]
- De Luca A., Fiorillo M., Peiris-Pagès M., Ozsvari B., Smith D. L., Sanchez-Alvarez R., et al. (2015). Mitochondrial Biogenesis Is Required for the anchorage-independent Survival and Propagation of Stem-like Cancer Cells. Oncotarget 6, 14777–14795. 10.18632/oncotarget.4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G., Song X., Fujimoto S., Piccirillo C. A., Nagai Y., Greene M. I. (2019). Foxp3 Post-translational Modifications and Treg Suppressive Activity. Front. Immunol. 10, 2486. 10.3389/fimmu.2019.02486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowless M., Lowery C. D., Shackleford T., Renschler M., Stephens J., Flack R., et al. (2018). Abemaciclib Is Active in Preclinical Models of Ewing Sarcoma via Multipronged Regulation of Cell Cycle, DNA Methylation, and Interferon Pathway Signaling. Clin. Cancer Res. 24, 6028–6039. 10.1158/1078-0432.CCR-18-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan N., Hu X., Yang X., Cheng H., Zhang W. (2015). MicroRNA-370 Directly Targets FOXM1 to Inhibit Cell Growth and Metastasis in Osteosarcoma Cells. Int. J. Clin. Exp. Pathol. 8, 10250–10260. [PMC free article] [PubMed] [Google Scholar]
- Duan R., Xie H., Liu Z. Z. (2020). The Role of Autophagy in Osteoarthritis. Front Cel Dev Biol 8, 608388. 10.3389/fcell.2020.608388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund R. K., Birol O., Groves A. K. (2015). The Role of Foxi Family Transcription Factors in the Development of the Ear and Jaw. Curr. Top. Dev. Biol. 111, 461–495. 10.1016/bs.ctdb.2014.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund R. K., Ohyama T., Kantarci H., Riley B. B., Groves A. K. (2014). Foxi Transcription Factors Promote Pharyngeal Arch Development by Regulating Formation of FGF Signaling Centers. Dev. Biol. 390, 1–13. 10.1016/j.ydbio.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel A. L. (2011). Thiostrepton, Proteasome Inhibitors and FOXM1. Cell Cycle 10, 4341–4342. 10.4161/cc.10.24.18544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne D. M., Spearman H., Lyne L., Puliyadi R., Perez-Alcantara M., Coulton L., et al. (2015). The Forkhead Transcription Factor FOXP2 Is Required for Regulation of p21WAF1/CIP1 in 143B Osteosarcoma Cell Growth Arrest. PloS one 10, e0128513. 10.1371/journal.pone.0128513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong A., Huang S. (2012). FoxM1 and Wnt/β-Catenin Signaling in Glioma Stem Cells. Cancer Res. 72, 5658–5662. 10.1158/0008-5472.CAN-12-0953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozo M. C., Jia D., Aspuria P. J., Cheon D. J., Miura N., Walts A. E., et al. (2016). FOXC2 Augments Tumor Propagation and Metastasis in Osteosarcoma. Oncotarget 7, 68792–68802. 10.18632/oncotarget.11990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabiec A. M., Angiolilli C., Hartkamp L. M., van Baarsen L. G., Tak P. P., Reedquist K. A. (2015). JNK-dependent Downregulation of FoxO1 Is Required to Promote the Survival of Fibroblast-like Synoviocytes in Rheumatoid Arthritis. Ann. Rheum. Dis. 74, 1763–1771. 10.1136/annrheumdis-2013-203610 [DOI] [PubMed] [Google Scholar]
- Greenblatt M. B., Shim J. H., Zou W., Sitara D., Schweitzer M., Hu D., et al. (2010). The P38 MAPK Pathway Is Essential for Skeletogenesis and Bone Homeostasis in Mice. J. Clin. Invest. 120, 2457–2473. 10.1172/JCI42285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H., Tan P., Xie L., Mi B., Fang Z., Li J., et al. (2015). FOXO1 Inhibits Osteosarcoma Oncogenesis via Wnt/β-Catenin Pathway Suppression. Oncogenesis 4, e166. 10.1038/oncsis.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider M., Zhang X., Coleman I., Ericson N., True L. D., Lam H. M., et al. (2016). Epithelial Mesenchymal-like Transition Occurs in a Subset of Cells in Castration Resistant Prostate Cancer Bone Metastases. Clin. Exp. Metastasis 33, 239–248. 10.1007/s10585-015-9773-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasi M., Gartel A. L. (2013). FOX(M1) News-Iit Is Cancer. Mol. Cancer Ther. 12, 245–254. 10.1158/1535-7163.MCT-12-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T., Kikuta J., Sudo T., Matsuura Y., Matsui T., Simmons S., et al. (2019). Identification of a Novel Arthritis-Associated Osteoclast Precursor Macrophage Regulated by FoxM1. Nat. Immunol. 20, 1631–1643. 10.1038/s41590-019-0526-7 [DOI] [PubMed] [Google Scholar]
- Hegde N. S., Sanders D. A., Rodriguez R., Balasubramanian S. (2011). The Transcription Factor FOXM1 Is a Cellular Target of the Natural Product Thiostrepton. Nat. Chem. 3, 725–731. 10.1038/nchem.1114 [DOI] [PubMed] [Google Scholar]
- Herman L., Todeschini A., Veitia R. (2020). Forkhead Transcription Factors in Health and Disease. Trends Genetics 37 (5), 460–475. 10.1016/j.tig.2020.11.003 [DOI] [PubMed] [Google Scholar]
- Hong H. K., Lass J. H., Chakravarti A. (1999). Pleiotropic Skeletal and Ocular Phenotypes of the Mouse Mutation Congenital Hydrocephalus (ch/Mf1) Arise from a Winged helix/forkhead Transcriptionfactor Gene. Hum. Mol. Genet. 8, 625–637. 10.1093/hmg/8.4.625 [DOI] [PubMed] [Google Scholar]
- Hopkins A., Mirzayans F., Berry F. (2016). Foxc1 Expression in Early Osteogenic Differentiation Is Regulated by BMP4-SMAD Activity. J. Cel Biochem 117, 1707–1717. 10.1002/jcb.25464 [DOI] [PubMed] [Google Scholar]
- Huang J., Shen G., Ren H., Zhang Z., Yu X., Zhao W., et al. (2020). Role of Forkhead Box Gene Family in Bone Metabolism. J. Cel Physiol 235, 1986–1994. 10.1002/jcp.29178 [DOI] [PubMed] [Google Scholar]
- Huang J., Chen L. (2017). IL-1β Inhibits Osteogenesis of Human Bone Marrow-Derived Mesenchymal Stem Cells by Activating FoxD3/microRNA-496 to Repress Wnt Signaling. New York, N.Y.: Genesis, 55. [DOI] [PubMed] [Google Scholar]
- Huang P., Yan R., Zhang X., Wang L., Ke X., Qu Y. (2019). Activating Wnt/β-Catenin Signaling Pathway for Disease Therapy: Challenges and Opportunities. Pharmacol. Ther. 196, 79–90. 10.1016/j.pharmthera.2018.11.008 [DOI] [PubMed] [Google Scholar]
- Iyer S., Ambrogini E., Bartell S. M., Han L., Roberson P. K., de Cabo R., et al. (2013). FOXOs Attenuate Bone Formation by Suppressing Wnt Signaling. J. Clin. Invest. 123, 3409–3419. 10.1172/JCI68049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs F. M., van der Heide L. P., Wijchers P. J., Burbach J. P., Hoekman M. F., Smidt M. P. (2003). FoxO6, a Novel Member of the FoxO Class of Transcription Factors with Distinct Shuttling Dynamics. J. Biol. Chem. 278, 35959–35967. 10.1074/jbc.M302804200 [DOI] [PubMed] [Google Scholar]
- Jin S., Pang R. P., Shen J. N., Huang G., Wang J., Zhou J. G. (2007). Grifolin Induces Apoptosis via Inhibition of PI3K/AKT Signalling Pathway in Human Osteosarcoma Cells. Apoptosis 12, 1317–1326. 10.1007/s10495-007-0062-z [DOI] [PubMed] [Google Scholar]
- Kaestner K. H., Knochel W., Martinez D. E. (2000). Unified Nomenclature for the Winged helix/forkhead Transcription Factors. Genes Dev. 14, 142–146. 10.1101/gad.14.2.142 [DOI] [PubMed] [Google Scholar]
- Kalluri R. (2009). EMT: when Epithelial Cells Decide to Become Mesenchymal-like Cells. J. Clin. Invest. 119, 1417–1419. 10.1172/JCI39675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaldinov T., Erndt-Marino J., Diaz-Rodriguez P., Chen H., Gharat T., Munoz-Pinto D., et al. (2018). Tuning Forkhead Box D3 Overexpression to Promote Specific Osteogenic Differentiation of Human Embryonic Stem Cells while Reducing Pluripotency in a Three-Dimensional Culture System. J. Tissue Eng. Regen. Med. 12, 2256–2265. 10.1002/term.2757 [DOI] [PubMed] [Google Scholar]
- Kamali A., Ziadlou R., Lang G., Pfannkuche J., Cui S., Li Z., et al. (2021). Small Molecule-Based Treatment Approaches for Intervertebral Disc Degeneration: Current Options and Future Directions. Theranostics 11, 27–47. 10.7150/thno.48987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., He P., Wang H., Ye Y., Li X., Xie P., et al. (2018). Brazilin Induces FOXO3A-dependent Autophagic Cell Death by Disturbing Calcium Homeostasis in Osteosarcoma Cells. Cancer Chemother. Pharmacol. 82, 479–491. 10.1007/s00280-018-3633-5 [DOI] [PubMed] [Google Scholar]
- Kansara M., Teng M. W., Smyth M. J., Thomas D. M. (2014). Translational Biology of Osteosarcoma. Nat. Rev. Cancer 14, 722–735. 10.1038/nrc3838 [DOI] [PubMed] [Google Scholar]
- Kimira Y., Odaira H., Nomura K., Taniuchi Y., Inoue N., Nakatani S., et al. (2017). Collagen-derived Dipeptide Prolyl-Hydroxyproline Promotes Osteogenic Differentiation through Foxg1. Cell Mol Biol Lett. 22, 27. 10.1186/s11658-017-0060-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. W., Gomes A. R. (2014). Forkhead Box Transcription Factors in Cancer Initiation, Progression and Chemotherapeutic Drug Response. Front. Oncol. 4, 305. 10.3389/fonc.2014.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Han X., Yang S., Wang Y., Dong Y., Tang T. (2021). FOXP1 Drives Osteosarcoma Development by Repressing P21 and RB Transcription Downstream of P53. Oncogene 40, 2785–2802. 10.1038/s41388-021-01742-4 [DOI] [PubMed] [Google Scholar]
- Li H., Liu P., Xu S., Li Y., Dekker J. D., Li B., et al. (2017). FOXP1 Controls Mesenchymal Stem Cell Commitment and Senescence during Skeletal Aging. J. Clin. Invest. 127, 1241–1253. 10.1172/JCI89511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhang J., Liu D., Liu Y., Zhou Y. (2019). Force-induced Decline of FOXM1 in Human Periodontal Ligament Cells Contributes to Osteoclast Differentiation. Angle Orthod. 89, 804–811. 10.2319/072418-536.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Z., Li J., Sang X. (2018). Diallyl Disulfide Suppresses FOXM1-Mediated Proliferation and Invasion in Osteosarcoma by Upregulating miR-134. J. Cell. Biochem. 120, 7286. 10.1002/jcb.28003 [DOI] [PubMed] [Google Scholar]
- Maher C., Underwood M., Buchbinder R. (2017). Non-specific Low Back Pain. Lancet 389, 736–747. 10.1016/S0140-6736(16)30970-9 [DOI] [PubMed] [Google Scholar]
- Maier J. A., Lo Y., Harfe B. D. (2013). Foxa1 and Foxa2 Are Required for Formation of the Intervertebral Discs. PloS one 8, e55528. 10.1371/journal.pone.0055528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marongiu M., Marcia L., Pelosi E., Lovicu M., Deiana M., Zhang Y., et al. (2015). FOXL2 Modulates Cartilage, Skeletal Development and IGF1-dependent Growth in Mice. BMC Dev. Biol. 15, 27. 10.1186/s12861-015-0072-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T., Johnson R. (2019). Multiple Actions of Parathyroid Hormone-Related Protein in Breast Cancer Bone Metastasis. Br. J. Pharmacol. 178 (9), 1923–1935. 10.1111/bph.14709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T., Alvarez-Garcia O., Mokuda S., Nagira K., Olmer M., Gamini R., et al. (2018). FoxO Transcription Factors Modulate Autophagy and Proteoglycan 4 in Cartilage Homeostasis and Osteoarthritis. Sci. Transl Med. 10, eaan0746. 10.1126/scitranslmed.aan0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans F., Lavy R., Penner-Chea J., Berry F. B. (2012). Initiation of Early Osteoblast Differentiation Events through the Direct Transcriptional Regulation of Msx2 by FOXC1. PloS one 7, e49095. 10.1371/journal.pone.0049095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Hollern D. P., Williams O. G., Rayburn T. S., Byrd W. A., Yates C., et al. (2018). A Review of FOXI3 Regulation of Development and Possible Roles in Cancer Progression and Metastasis. Front. Cel Dev Biol 6, 69. 10.3389/fcell.2018.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Ramaswamy S., Vazquez F., Signoretti S., Loda M., Sellers W. R. (2000). Forkhead Transcription Factors Are Critical Effectors of Cell Death and Cell Cycle Arrest Downstream of PTEN. Mol. Cel Biol. 20, 8969–8982. 10.1128/mcb.20.23.8969-8982.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedan S., Kauer M., Aryee D. N., Kofler R., Schwentner R., Meier A., et al. (2014). Suppression of FOXO1 Is Responsible for a Growth Regulatory Repressive Transcriptional Sub-signature of EWS-FLI1 in Ewing Sarcoma. Oncogene 33, 3927–3938. 10.1038/onc.2013.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen R. M., Yan J., Amsterdam A., Hopkins N., Burgess S. M. (2003). Zebrafish Foxi One Modulates Cellular Responses to Fgf Signaling Required for the Integrity of Ear and Jaw Patterning. Development 130, 2543–2554. 10.1242/dev.00455 [DOI] [PubMed] [Google Scholar]
- Ohyama T., Groves A. K. (2004). Expression of Mouse Foxi Class Genes in Early Craniofacial Development. Developmental Dyn. : official Publ. Am. Assoc. Anatomists 231, 640–646. 10.1002/dvdy.20160 [DOI] [PubMed] [Google Scholar]
- Reedquist K. A., Ludikhuize J., Tak P. P. (2006). Phosphoinositide 3-kinase Signalling and FoxO Transcription Factors in Rheumatoid Arthritis. Biochem. Soc. Trans. 34, 727–730. 10.1042/BST0340727 [DOI] [PubMed] [Google Scholar]
- Rice R., Rice D. P., Olsen B. R., Thesleff I. (2003). Progression of Calvarial Bone Development Requires Foxc1 Regulation of Msx2 and Alx4. Dev. Biol. 262, 75–87. 10.1016/s0012-1606(03)00355-5 [DOI] [PubMed] [Google Scholar]
- Salih D. A., Brunet A. (2008). FoxO Transcription Factors in the Maintenance of Cellular Homeostasis during Aging. Curr. Opin. Cel Biol. 20, 126–136. 10.1016/j.ceb.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. L., Wolfe F., Huizinga T. W. (2010). Rheumatoid Arthritis. Lancet 376, 1094–1108. 10.1016/S0140-6736(10)60826-4 [DOI] [PubMed] [Google Scholar]
- Seifi M., Walter M. A. (2018). Axenfeld-Rieger Syndrome. Clin. Genet. 93, 1123–1130. 10.1111/cge.13148 [DOI] [PubMed] [Google Scholar]
- Shats I., Gatza M. L., Liu B., Angus S. P., You L., Nevins J. R. (2013). FOXO Transcription Factors Control E2F1 Transcriptional Specificity and Apoptotic Function. Cancer Res. 73, 6056–6067. 10.1158/0008-5472.CAN-13-0453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Cai G. Q., Peng J. P., Chen X. D. (2015). Autophagy Protects Chondrocytes from Glucocorticoids-Induced Apoptosis via ROS/Akt/FOXO3 Signaling. Osteoarthritis Cartilage 23, 2279–2287. 10.1016/j.joca.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Shen G., Ren H., Shang Q., Zhao W., Zhang Z., Yu X., et al. (2020). Foxf1 Knockdown Promotes BMSC Osteogenesis in Part by Activating the Wnt/β-Catenin Signalling Pathway and Prevents Ovariectomy-Induced Bone Loss. EBioMedicine 52, 102626. 10.1016/j.ebiom.2020.102626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Sakuma M., Mooroka T., Liscoe A., Gao H., Croce K. J., et al. (2008). Down-regulation of the Forkhead Transcription Factor Foxp1 Is Required for Monocyte Differentiation and Macrophage Function. Blood 112, 4699–4711. 10.1182/blood-2008-01-137018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R., Tanaka M., Tsutsumi S., Aburatani H., Yamazaki Y., Homme M., et al. (2018). EWS-FLI1 Regulates a Transcriptional Program in Cooperation with Foxq1 in Mouse Ewing Sarcoma. Cancer Sci. 109, 2907–2918. 10.1111/cas.13710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo H., Takahashi Y., Tonegawa A., Arase Y., Aoyama H., Mizutani-Koseki Y., et al. (2001). Inductive Signals from the Somatopleure Mediated by Bone Morphogenetic Proteins Are Essential for the Formation of the Sternal Component of Avian Ribs. Dev. Biol. 232, 284–300. 10.1006/dbio.2001.0198 [DOI] [PubMed] [Google Scholar]
- Sun C., Dai J., Ma S., Pan Y., Liu F., Wang Y. (2020). MicroRNA-197 Inhibits the Progression of Osteosarcoma through Targeting FOXM1. Minerva Endocrinol. 45, 153–156. 10.23736/S0391-1977.19.03108-0 [DOI] [PubMed] [Google Scholar]
- Sun J., Ishii M., Ting M. C., Maxson R. (2013). Foxc1 Controls the Growth of the Murine Frontal Bone Rudiment by Direct Regulation of a Bmp Response Threshold of Msx2. Development 140, 1034–1044. 10.1242/dev.085225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Qiao W., Zhou B., Hu Z., Yan Q., Wu J., et al. (2018). Overexpression of Sirt1 in Mesenchymal Stem Cells Protects against Bone Loss in Mice by FOXO3a Deacetylation and Oxidative Stress Inhibition. Metabolism 88, 61–71. 10.1016/j.metabol.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Takahashi K., Liu F. C. (2009). FOXP Genes, Neural Development, Speech and Language Disorders. Adv. Exp. Med. Biol. 665, 117–129. 10.1007/978-1-4419-1599-3_9 [DOI] [PubMed] [Google Scholar]
- Tan P., Guan H., Xie L., Mi B., Fang Z., Li J., et al. (2015). FOXO1 Inhibits Osteoclastogenesis Partially by Antagnozing MYC. Sci. Rep. 5, 16835. 10.1038/srep16835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K. C., Pan W., Doschak M. R., Alexander R. T. (2019). Increased FoxO3a Expression Prevents Osteoblast Differentiation and Matrix Calcification. Bone Rep. 10, 100206. 10.1016/j.bonr.2019.100206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira C. C., Liu Y., Thant L. M., Pang J., Palmer G., Alikhani M. (2010). Foxo1, a Novel Regulator of Osteoblast Differentiation and Skeletogenesis. J. Biol. Chem. 285, 31055–31065. 10.1074/jbc.M109.079962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y., Irie K., Fukuhara D., Kataoka K., Hattori T., Ono M., et al. (2018). Commensal Microbiota Enhance Both Osteoclast and Osteoblast Activities. Molecules 23, 1517. 10.3390/molecules23071517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda M., Ottolenghi C., Crisponi L., Garcia J. E., Deiana M., Kimber W., et al. (2004). Foxl2 Disruption Causes Mouse Ovarian Failure by Pervasive Blockage of Follicle Development. Hum. Mol. Genet. 13, 1171–1181. 10.1093/hmg/ddh124 [DOI] [PubMed] [Google Scholar]
- Wang F. S., Chen Y. S., Ko J. Y., Kuo C. W., Ke H. J., Hsieh C. K., et al. (2020). Bromodomain Protein BRD4 Accelerates Glucocorticoid Dysregulation of Bone Mass and Marrow Adiposis by Modulating H3K9 and Foxp1. Cells 9, 1500. 10.3390/cells9061500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu Y., Yu G. (2019). Avasimibe Inhibits Tumor Growth by Targeting FoxM1-Akr1c1 in Osteosarcoma. Onco Targets Ther. 12, 815–823. 10.2147/OTT.S165647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Guo Z., Yu H., Fan L. (2019). MiR-216b Inhibits Osteosarcoma Cell Proliferation, Migration, and Invasion by Targeting Forkhead Box M1. J. Cel Biochem 120, 5435–5443. 10.1002/jcb.27822 [DOI] [PubMed] [Google Scholar]
- Wang Y., Dong G., Jeon H. H., Elazizi M., La L. B., Hameedaldeen A., et al. (2015). FOXO1 Mediates RANKL-Induced Osteoclast Formation and Activity. J. Immunol. 194, 2878–2887. 10.4049/jimmunol.1402211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasén C., Ospelt C., Camponeschi A., Erlandsson M. C., Andersson K. M. E., Silfverswärd S. T., et al. (2020). Nicotine Changes the microRNA Profile to Regulate the FOXO Memory Program of CD8+ T Cells in Rheumatoid Arthritis. Front. Immunol. 11, 1474. 10.3389/fimmu.2020.01474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Gao X., Li F., Shao L., Sun Y. (2021). Down-Regulation of microRNA-30d Alleviates Intervertebral Disc Degeneration through the Promotion of FOXO3 and Suppression of CXCL10. Calcif Tissue Int. 108, 252–264. 10.1007/s00223-020-00760-w [DOI] [PubMed] [Google Scholar]
- Xiang L., Zheng J., Zhang M., Ai T., Cai B. (2020). FOXQ1 Promotes the Osteogenic Differentiation of Bone Mesenchymal Stem Cells via Wnt/β-Catenin Signaling by Binding with ANXA2. Stem Cel Res Ther 11, 403. 10.1186/s13287-020-01928-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Yu H. V., Tseng K. C., Flath M., Fabian P., Segil N., et al. (2021). Foxc1 Establishes Enhancer Accessibility for Craniofacial Cartilage Differentiation. Elife 10. 10.7554/elife.63595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Liu P., Chen Y., Chen Y., Zhang W., Zhao H., et al. (2018). Foxp2 Regulates Anatomical Features that May Be Relevant for Vocal Behaviors and Bipedal Locomotion. Proc. Natl. Acad. Sci. U S A. 115, 8799–8804. 10.1073/pnas.1721820115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Hu H. M., Zielinska-Kwiatkowska A., Chansky H. A. (2010). FOXO1 Is a Direct Target of EWS-Fli1 Oncogenic Fusion Protein in Ewing's Sarcoma Cells. Biochem. Biophys. Res. Commun. 402, 129–134. 10.1016/j.bbrc.2010.09.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Chen M., Chen E., Li W., Wang S., Ding Q., et al. (2018). Knockdown of FOXA2 Enhances the Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells Partly via Activation of the ERK Signalling Pathway. Cell Death Dis. 9, 836. 10.1038/s41419-018-0857-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Tam W. L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E., et al. (2015). Distinct EMT Programs Control normal Mammary Stem Cells and Tumour-Initiating Cells. Nature 525, 256–260. 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Ding H., He E., Chen J., Li M. (2017). Up-regulation of microRNA-491-5p Suppresses Cell Proliferation and Promotes Apoptosis by Targeting FOXP4 in Human Osteosarcoma. Cell Prolif 50, e12308. 10.1111/cpr.12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Hata K., Takashima R., Ono K., Nakamura E., Takahata Y., et al. (2015). The Transcription Factor Foxc1 Is Necessary for Ihh-Gli2-Regulated Endochondral Ossification. Nat. Commun. 6, 6653. 10.1038/ncomms7653 [DOI] [PubMed] [Google Scholar]
- You W., Fan L., Duan D., Tian L., Dang X., Wang C., et al. (2014). Foxc2 Over-expression in Bone Marrow Mesenchymal Stem Cells Stimulates Osteogenic Differentiation and Inhibits Adipogenic Differentiation. Mol. Cel Biochem 386, 125–134. 10.1007/s11010-013-1851-z [DOI] [PubMed] [Google Scholar]
- You W., Gao H., Fan L., Duan D., Wang C., Wang K. (2013). Foxc2 Regulates Osteogenesis and Angiogenesis of Bone Marrow Mesenchymal Stem Cells. BMC Musculoskelet. Disord. 14, 199. 10.1186/1471-2474-14-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Chang J., Liu Y., Li J., Kevork K., Al-Hezaimi K., et al. (2014). Wnt4 Signaling Prevents Skeletal Aging and Inflammation by Inhibiting Nuclear Factor-Κb. Nat. Med. 20, 1009–1017. 10.1038/nm.3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss M. M., Axmann R., Zwerina J., Polzer K., Gückel E., Skapenko A., et al. (2007). Treg Cells Suppress Osteoclast Formation: a New Link between the Immune System and Bone. Arthritis Rheum. 56, 4104–4112. 10.1002/art.23138 [DOI] [PubMed] [Google Scholar]
- Zaiss M. M., Frey B., Hess A., Zwerina J., Luther J., Nimmerjahn F., et al. (2010). Regulatory T Cells Protect from Local and Systemic Bone Destruction in Arthritis. J. Immunol. 184, 7238–7246. Baltimore, Md. : 1950. 10.4049/jimmunol.0903841 [DOI] [PubMed] [Google Scholar]
- Zeng H. C., Bae Y., Dawson B. C., Chen Y., Bertin T., Munivez E., et al. (2017). MicroRNA miR-23a Cluster Promotes Osteocyte Differentiation by Regulating TGF-β Signalling in Osteoblasts. Nat. Commun. 8, 15000. 10.1038/ncomms15000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Lu X., Lin J., Ron Z., Fang J., Liu Z., et al. (2021). FOXM1 Activates JAK1/STAT3 Pathway in Human Osteoarthritis Cartilage Cell Inflammatory Reaction. Exp. Biol. Med. (Maywood) 246, 644–653. 10.1177/1535370220974933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R. M., Lu X. H., Lin J., Hu J., Rong Z. J., Xu W. C., et al. (2019). Knockdown of FOXM1 Attenuates Inflammatory Response in Human Osteoarthritis Chondrocytes. Int. Immunopharmacol 68, 74–80. 10.1016/j.intimp.2018.12.057 [DOI] [PubMed] [Google Scholar]
- Zhang C. L., Zhu K. P., Ma X. L. (2017). Antisense lncRNA FOXC2-AS1 Promotes Doxorubicin Resistance in Osteosarcoma by Increasing the Expression of FOXC2. Cancer Lett. 396, 66–75. 10.1016/j.canlet.2017.03.018 [DOI] [PubMed] [Google Scholar]
- Zhang N., Wei P., Gong A., Chiu W. T., Lee H. T., Colman H., et al. (2011). FoxM1 Promotes β-catenin Nuclear Localization and Controls Wnt Target-Gene Expression and Glioma Tumorigenesis. Cancer Cell 20, 427–442. 10.1016/j.ccr.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhou W., Yao Z., Wan Y., Cao J., Zhang L., et al. (2015). Foxp1/2/4 Regulate Endochondral Ossification as a Suppresser Complex. Dev. Biol. 398, 242–254. 10.1016/j.ydbio.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Li Y., Di R., Yang Y., Meng S., Song F., et al. (2019). H19 and Foxc2 Synergistically Promotes Osteogenic Differentiation of BMSCs via Wnt-β-Catenin Pathway. J. Cel Physiol 234, 13799–13806. 10.1002/jcp.28060 [DOI] [PubMed] [Google Scholar]
- Zhou T. Y., Wu Y. G., Zhang Y. Z., Bao Y. W., Zhao Y. (2019). SIRT3 Retards Intervertebral Disc Degeneration by Anti-oxidative Stress by Activating the SIRT3/FOXO3/SOD2 Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 9180–9188. 10.26355/eurrev_201911_19408 [DOI] [PubMed] [Google Scholar]
- Zhou X., Ma C., Hu B., Tao Y., Wang J., Huang X., et al. (2018). FoxA2 Regulates the Type II Collagen-Induced Nucleus Pulposus-like Differentiation of Adipose-Derived Stem Cells by Activation of the Shh Signaling Pathway. FASEB J. 1, fj201800373R. official publication of the Federation of American Societies for Experimental Biology. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wang T., Hamilton J. L., Chen D. (2017). Wnt/β-catenin Signaling in Osteoarthritis and in Other Forms of Arthritis. Curr. Rheumatol. Rep. 19, 53. 10.1007/s11926-017-0679-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Deng Y., Gao H., Huang K., Nie Z. (2020). miR-877-5p Alleviates Chondrocyte Dysfunction in Osteoarthritis Models via Repressing FOXM1. J. Gene Med. 22, e3246. 10.1002/jgm.3246 [DOI] [PubMed] [Google Scholar]
- Zhu X., Lu K., Cao L., Hu Y., Yin Y., Cai Y. (2020). FoxM1 Is Upregulated in Osteosarcoma and Inhibition of FoxM1 Decreases Osteosarcoma Cell Proliferation, Migration, and Invasion. Cancer Manag. Res. 12, 9857–9867. 10.2147/CMAR.S270825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorov D. B., Juhaszova M., Sollott S. J. (2014). Mitochondrial Reactive Oxygen Species (ROS) and ROS-Induced ROS Release. Physiol. Rev. 94, 909–950. 10.1152/physrev.00026.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]