Abstract
Molecular genotyping of strains of Trichophyton rubrum and T. mentagrophytes from patients with onychomycosis of the toes was performed to ascertain whether the fungal genotype changes over the course of time as sequential samples were obtained from patients receiving antifungal therapy and during follow-up. Sixty-six serial strains of T. rubrum and 11 strains of T. mentagrophytes were obtained from 20 patients (16 patients with T. rubrum, 4 with T. mentagrophytes) who were treated with oral antifungal therapy and observed over periods of up to 36 months. These strains were screened for genetic variation by hybridization of EcoRI-digested genomic DNAs with a probe amplified from the small-subunit (18S) ribosomal DNA and adjacent internal transcribed spacer regions. A total of five restriction fragment length polymorphism (RFLP) types were observed among 66 strains of T. rubrum. Two major RFLP types, differentiated by one band shift, represented 68% of the samples. None of the patients had a unique genotype. More than one RFLP type was often observed from a single patient (same nail) over a period of 1, 2, or 3 years, even in cases that did not appear cured at any time. Samples taken from different nails of the same patient had either the same or a different genotype. The genotypic variation did not correspond to any detectable phenotypic variation. Furthermore, no correlation was observed between the efficacy of the treatment administered and the genotype observed. While the DNA region studied distinguished among T. rubrum, T. mentagrophytes, and T. tonsurans, intraspecific RFLP variation was observed for T. rubrum and T. mentagrophytes strains. While independent multiple infection and coinhabitation of multiple strains may explain the presence of different genotypes in a nail, microevolutionary events such as rapid substrain shuffling, as seen in studies of repetitive regions in Candida species, may also produce the same result. The recovery of multiple strains during the course of sequential sampling of uncured patients further suggests that the typing system is not able to distinguish between relapse or reinfection, ongoing infection, and de novo infection.
Onychomycosis affects approximately 6 to 13% of the North American population (5, 12, 13, 14). The most significant causal organism is Trichophyton rubrum. Less commonly, T. mentagrophytes is involved (11, 13); other dermatophytes are also occasionally implicated (29). Toenail onychomycosis is notoriously difficult to treat successfully (11, 13). The newer oral antifungal agents terbinafine, itraconazole, and fluconazole have greatly improved prospects for its successful treatment in comparison to the older drugs griseofulvin and ketoconazole (11, 13). Despite this improvement, however, recent studies still show mycological cure rates ranging from roughly 40 to 80% (10, 11), indicating that successful therapy of this condition remains a challenge.
Establishing that a cure has successfully been achieved may be difficult in itself. Many patients with chronic T. rubrum onychomycosis and tinea pedis appear to be specifically genetically predisposed to this condition (34). Although cured, they may soon acquire another strain from the environment, e.g., in swimming or sporting facilities. This reacquisition of the disease (or reinfection) may be difficult to distinguish from a relapse caused by an incomplete cure of the original infecting isolate. Studies of drug efficacy, however, must make this distinction, since the complete cure that may precede environmental reacquisition of the organism represents successful therapy, whereas recrudescent disease indicates inadequate therapy. In the former type of case, the cautious patient, even if genetically predisposed to infection, may avoid reinfection through such sanitary measures as avoiding communal aquatic facilities or wearing appropriate footwear in such locations. The distinction between newly infecting and recrudescing strains, and thus between a true cure and an illusory cure, requires some method of detecting intraspecific variability in Trichophyton species. Although T. rubrum strains in some cases may appear very different from one another in culture (20), this is by no means always the case. Even in the most divergent isolates, moreover, the differences seen are seldom sufficiently dramatic to allow definite strain recognition. Furthermore, the kind of genotypic biotyping that has been developed for many medically important organisms is problematical with T. rubrum, which is highly clonal (6, 8, 30; H. T. Mitchell, W. A. Hutchins, R. C. Summerbell, and P. F. Lehmann, Abstr. 94th Gen. Meet. Am. Soc. Microbiol. 1994, abstr. F-93, p. 604, 1994). Recently, however, the detection of intraspecific variation in T. rubrum using amplification and hybridization of the ribosomal DNA (rDNA) intergenic spacer region was reported (19).
In the present study, we have used this intraspecific typing technique to analyze serial samples from patients with onychomycosis due to T. rubrum and T. mentagrophytes who were being treated with oral antifungal therapy. We wished to determine if the strain isolated at baseline remained constant during and after therapy and if an apparent relapse could have been caused by different strains. We also probed the ability of the same technique to show strain differences among T. tonsurans isolates from cases of tinea capitis.
MATERIALS AND METHODS
Clinical samples.
Sequential specimens from patients with onychomycosis caused by T. rubrum and T. mentagrophytes were collected as the patients received therapy for this indication. The nail samples were obtained from patients in Ontario and were sent to the regional Mycology Laboratory for examination. Among these isolates, there were 66 strains isolated at successive time points from 16 patients with onychomycosis caused by T. rubrum and 11 strains from 4 patients with T. mentagrophytes infection. For all of the patients included in this study, at least four positive isolations were made over a period of 3 years, from December 1996 to December 1999. Additionally, to further test the scope of the technique described by Jackson et al. (19), 10 strains of T. tonsurans from patients with tinea capitis were included in screening for intraspecific variation.
Isolation of fungal DNA.
The miniprep procedure of Mochizuki et al. (26) was used to extract fungal DNA from lyophilized mycelium. In brief, lyophilized mycelium was ground with glass beads in a conical 1.5-ml microcentrifuge tube. Ground mycelium was suspended in 500 to 600 μl of cetyltrimethylammonium bromide (CTAB) extraction buffer (0.7 M NaCl, 1% CTAB, 50 mM Tris-HCl [pH 8], 250 mM EDTA, 1% 2-mercaptoethanol) and incubated at 65°C for 1 h, followed by chloroform extraction and centrifugation. One volume of dilution buffer (1% CTAB, 50 mM Tris-HCl, 10 mM EDTA) was added to the supernatant, and the crude DNA was pelleted out of solution. The crude DNA was resuspended in 1 M NaCl–Tris-EDTA (TE), and 1 volume of 7.5 M ammonium acetate was added. After the precipitate was again pelleted out, DNA was recovered by adding 1 volume of isopropanol. The resulting pellet was resuspended in 300 μl of 1 M NaCl-TE and was RNase treated with incubation at 37°C for ∼1 h followed by chloroform extraction and centrifugation. The final DNA was precipitated in 2 volumes of cold ethanol and washed with 70% ethanol. The DNA pellet was resuspended in 50 μl of TE, and 10 μl of this DNA was further used for total genomic digestion with restriction endonucleases.
Detection of RFLPs in the rDNAs of Trichophyton species.
By using the protocol for strain typing described by Jackson et al. (19), restriction fragment length polymorphisms (RFLPs) were detected by hybridization of EcoRI-digested total genomic DNAs with a probe amplified from the small-subunit (18S) rDNA and the adjacent internal transcribed spacer (ITS) region. Such EcoRI digests produce two rDNA fragments binding with the probe, including a small (approximately 3-kb) fragment which is constant in size and one which differs in size according to the number of repeat units in the intergenic nontranscribed spacer (NTS) region (13). The fragment of constant size in effect serves as an internal positive control for probe specificity.
Total genomic DNA digested with EcoRI (New England Biolabs, Mississauga, Ontario, Canada) were electrophoresed in 0.8% agarose gels and stained with ethidium bromide. Gel denaturation, neutralization, and immobilization of DNA fragments onto nylon membrane (GeneScreen; NEN LifeScience Products Inc., Boston, Mass.) by Southern transfer were performed in accordance with standard protocols.
Universal fungal primers NS5 and ITS4 (32; oligonucleotides were synthesized by Dalton Chemicals, North York, Ontario, Canada) were used to PCR amplify an ∼1,220-bp fragment of 18S rDNA plus the adjacent ITS region from a T. rubrum strain under the amplification conditions mentioned by Jackson et al. (19). A no-DNA negative control was included in every PCR mixture. The resulting amplification mixture was purified by using a QIAquick column (QIAGEN, Mississauga, Ontario, Canada). Three microliters of purified PCR mixture was radioactively labeled with [α-32P]dCTP using the Nick Translation Kit (NEN LifeScience Products Inc.). Hybridization of the probed, amplified fragment to total genomic DNA on nylon membrane was carried out at 65°C for 18 h, followed by six stringent washes (twice for 5 min each time at room temperature [RT] with 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], three times for 20 min each at 65°C with 0.5× SSC–0.1% sodium dodecyl sulfate heated to 65°C, and once for 10 min with 0.1× SSC at RT). Radioactive membranes were autoradiographed at −70°C, and the X-ray films were developed after 30 to 48 h of exposure to detect the signal.
Growth characteristics of genetically different isolates.
Isolates from the same patient showing genetic variation were also tested for morphological or growth variation on various culture media (20). Strains to be tested were inoculated simultaneously on five different growth media. These were Sabouraud agar supplemented with chloramphenicol, cycloheximide, and gentamicin (SAB-CCG), brain heart infusion (BHI) agar, bromcresol purple-milk solids-glucose agar, vitamin-free Casamino Acids agar, and Christensen's urea broth (20). Cultures were incubated at RT, and variation, if any, in growth was recorded after 7 days. Colony pigmentation, including reverse coloration and any soluble pigmentation, was also recorded.
RESULTS
Sixty-six strains were isolated from 16 patients with T. rubrum infection. Clinical observation of the patients indicated that only a few of the patients yielding sufficient serial isolates for study could be pronounced to be cured, either clinically and mycologically, during the course of the study. None of these patients developed a recurrence of disease during the follow-up period. Therefore, observations were limited to cases of apparently continuous infection rather than apparent recurrence of disease.
RFLPs in T. rubrum.
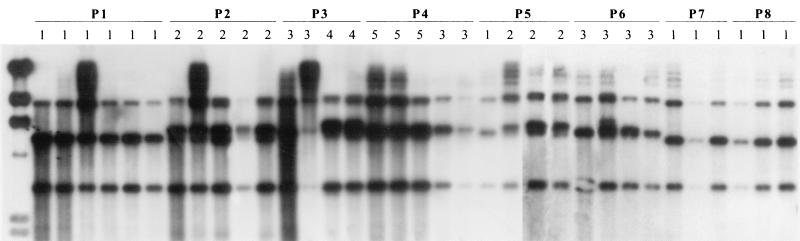
Five RFLP types (Tr-1 to Tr-5) were observed among the patients studied (Fig. 1). None of the RFLP types were unique to any individual; rather, genotypes were shared among individuals. The RFLP types observed among successive samples from all patients are shown in Table 1. RFLP type Tr-3 was the most common, representing 26 (39.4%) of 66 strains; it was followed by Tr-1 (19 [28.8%] of 66 strains), Tr-2 (10 [15.1%] of 66 strains), Tr-5 (8 [12.1%] of 66 strains), and Tr-4 (3 [4.5%] of 66 strains).
FIG. 1.
Southern hybridizations of EcoRI-digested genomic DNAs of strains from eight onychomycosis patients (P1 to P8) with T. rubrum infection. A PCR-amplified fragment (∼1,220 bp) of 18S rDNA plus the adjacent ITS region from a T. rubrum strain was used as a probe. The numbers above the lanes represent the RFLP types found among T. rubrum strains. Five distinct RFLP types (1 to 5) are shown. Lanes marked by solid lines represent strains obtained from the same patient during the course of therapy. Example lanes are not in chronological order. Types 2 and 4 have double bands in the 4.3- to 6.5-kb region. The leftmost lane contains a molecular weight marker (HindIII-digested lambda DNA).
TABLE 1.
RFLP patterns and clinical and mycological status of T. rubrum and T. mentagrophytes strains from 20 onychomycosis patients
| Patient and time | No. of samples | Toenail sampleda | Microscopyb | % of area involvedc | RFLP typed | |
|---|---|---|---|---|---|---|
| Patients with T. rubrum infection | ||||||
| Patient 1 | ||||||
| Aug. 97e | R3 | Pos. | 100 | Tr-1 | ||
| Sept. 97e | Pos. | 60 | Tr-1 | |||
| Nov. 97 | Pos. | 40 | Tr-1 | |||
| Dec. 97 | Pos. | 20 | Tr-1 | |||
| Jan. 98 | Pos. | 5 | Tr-1 | |||
| June 98e | 6 | Neg. | 0 | Tr-1 | ||
| Patient 2 | ||||||
| May 97e | R1 | Pos. | 40 | Tr-2 | ||
| Jun. 97e | Pos. | 30 | Tr-2 | |||
| Sept. 97 | Neg. | 20 | Tr-2 | |||
| Nov. 97e | Pos. | 20 | Tr-2 | |||
| Feb. 98 | 5 | Pos. | 15 | Tr-2 | ||
| Patient 3 | ||||||
| Nov. 97e | R1 | Pos. | 70 | Tr-3 | ||
| Dec. 97e | L1 | Pos. | 70 | Tr-4 | ||
| Jan. 98e | R1 | Pos. | 60 | Tr-4 | ||
| Mar. 98 | 4 | Pos. | 50 | Tr-3 | ||
| Patient 4 | ||||||
| Dec. 96e | L1 | Pos. | 50 | Tr-5 | ||
| Sept. 97e | Pos. | 20 | Tr-3 | |||
| Nov. 97e | Neg. | 7.5 | Tr-3 | |||
| Dec. 97 | Pos. | 7.5 | Tr-5 | |||
| Mar. 98e | 5 | Neg. | 7.5 | Tr-5 | ||
| Patient 5 | ||||||
| Dec. 96 | R1 | Pos. | 80 | Tr-1 | ||
| July 97e | L1 | Pos. | 60 | Tr-2 | ||
| Aug. 97 | Pos. | 50 | Tr-1 | |||
| Nov. 97 | Pos. | 10 | Tr-2 | |||
| Feb. 98 | 5 | Pos. | 0 | Tr-2 | ||
| Patient 6 | ||||||
| Sept. 97e | R1 | Pos. | 70 | Tr-3 | ||
| Oct. 97e | L1 | Neg. | 70 | Tr-3 | ||
| Dec. 97 | Pos. | 50 | Tr-3 | |||
| Jan. 98 | Pos. | 50 | Tr-3 | |||
| Dec. 98 | Pos. | 10 | Tr-3 | |||
| Apr. 99 | 6 | Neg. | 7.5 | Tr-5 | ||
| Patient 7 | ||||||
| Nov. 97e | R1 | Pos. | 70 | Tr-1 | ||
| Feb. 98 | Pos. | 60 | Tr-1 | |||
| Apr. 98e | Pos. | 55 | Tr-1 | |||
| May 98e | Pos. | 55 | Tr-1 | |||
| Dec. 98 | 5 | Pos. | 10 | Tr-1 | ||
| Patient 8 | ||||||
| July 97e | L1 | Pos. | 70 | Tr-1 | ||
| Aug. 97 | Pos. | 60 | Tr-1 | |||
| Sept. 97 | Pos. | 50 | Tr-1 | |||
| Jan. 98 | Pos. | 50 | Tr-1 | |||
| Apr. 98e | Pos. | 50 | Tr-1 | |||
| July 98 | 6 | Neg. | 7.5 | Tr-1 |
| Patient 9 | |||||
| Jan. 98e | L1 | Pos. | 95 | Tr-4 | |
| Feb. 98e | Pos. | 95 | Tr-3 | ||
| Mar. 98e | Pos. | 90 | Tr-3 | ||
| Apr. 98 | 4 | Pos. | 80 | Tr-3 | |
| Patient 10 | |||||
| July 97e | R1 | Pos. | 40 | Tr-3 | |
| Aug. 97e | Pos. | 40 | Tr-3 | ||
| Oct. 97 | 3 | Pos. | 20 | Tr-3 | |
| Patient 11 | |||||
| May 97e | R1 | Pos. | 30 | Tr-3 | |
| Sept. 97e | Pos. | 80 | Tr-3 | ||
| Jan. 98 | 3 | Pos. | 70 | Tr-3 | |
| Patient 12 | |||||
| Aug. 97e | L3 | Pos. | 80 | Tr-5 | |
| Sept. 97e | Pos. | 60 | Tr-5 | ||
| Oct. 97 | 3 | Pos. | 20 | Tr-5 | |
| Patient 13 | |||||
| July 97e | R1 | Pos. | 90 | Tr-3 | |
| Dec. 97 | 2 | Neg. | 40 | Tr-3 | |
| Patient 14 | |||||
| Jan. 97 | L1 | Pos. | 100 | Tr-3 | |
| Apr. 97e | 2 | R1 | Pos. | 80 | Tr-3 |
| Patient 15 | |||||
| Mar. 97 | L1 | Pos. | 100 | Tr-2 | |
| Apr. 97e | R1 | Pos. | 100 | Tr-2 | |
| Dec. 97 | 3 | Neg. | 0 | Tr-5 | |
| Patient 16 | |||||
| May 98e | R1 | Pos. | 70 | Tr-3 | |
| June 98e | Pos. | 60 | Tr-3 | ||
| July 98e | 4 | Pos. | 60 | Tr-3 | |
| Aug 98 | Neg. | 50 | Tr-3 | ||
| Patients with T. mentagrophytes infection | |||||
| Patient 1 | |||||
| June 97e | L1 | Pos. | 85 | Tm-1 | |
| Aug. 97 | Pos. | 50 | Tm-1 | ||
| Sept. 97 | 3 | Pos. | 50 | Tm-1 | |
| Patient 2 | |||||
| Sept. 97e | L1 | Pos. | 20 | Tm-1 | |
| Nov. 97 | Pos. | 20 | Tm-1 | ||
| Dec. 97 | 3 | Pos. | 20 | Tm-1 | |
| Patient 3 | |||||
| Aug. 97e | L1 | Pos. | 60 | Tm-2 | |
| Sept. 97 | Pos. | 55 | Tm-2 | ||
| Nov. 97 | 3 | Pos. | 55 | Tm-2 | |
| Patient 4 | |||||
| July 97e | L1 | Pos. | 90 | Tm-1 | |
| Aug. 97 | 2 | Pos. | 80 | Tm-1 |
Infected toenails were collected from either the right (R) or the left (L) toes.
Result of microscopic examination of the clinical material; Pos. represents the presence of fungal filaments, and Neg. represents no fungal filaments in the specimen under the microscope. All specimens were culture positive for T. mentagrophytes.
The percentage of the nail infected was estimated visually.
RFLP pattern observed by hybridization of the PCR-amplified ITS region used as a probe against EcoRI-digested total genomic DNA. Tr, T. rubrum RFLP type; Tm, T. mentagrophytes RFLP type.
Time period when the patient was under treatment. The two oral antifungals used for treatment were itraconazole and terbinafine. Time periods not marked represent follow-up examinations.
In 10 of 16 patients, the same RFLP type was observed in all isolations. In each of the six remaining patients, more than one RFLP type was observed in successive samples taken over a 1-to 3-year period (Table 1; Fig. 1). In four of six cases, RFLP type Tr-3 was observed on at least one occasion. Only two strains were recovered from the same nail sampled at different times.
No specific correlation between a particular genotype and a particular type of nail (e.g., the hallux) was observed. Strains from different nails of the same patient either had the same (patient 14) or a mixture of the same and different (patients 3, 5, 6, and 15) genotypes (Table 1). No particular genotype seemed to predominate or emerge in connection with patients undergoing antifungal treatment.
Morphological variation among genetically different serial samples.
Fifteen strains from five patients showing different RFLP types were inoculated on five diagnostic and growth-testing media. Growth characteristics of these strains, as recorded on day 7 after inoculation, are presented in Table 2. Growth on SAB-CCG and Casamino Acids agar ranged from granular to cottony (with aerial mycelium). Three strains, all with different RFLPs, occasionally produced dark, soluble pigment suggestive of melanin on BHI agar. Pigmentation of the colony reverse varied from yellow to red. Overall, no correlation of morphological variation with the genotype was observed.
TABLE 2.
Morphological variation among T. rubrum strains from patients showing genetic differences
| Patient and time | RFLP pattern | Mycelium type on SAB-CCG | Presence of melanin on BHIa | Mycelium type on Casamino Acids plain agar | Pigment on BCPMSGb | Presence of urease on Christensen's urea brothc |
|---|---|---|---|---|---|---|
| Patient 4 | ||||||
| Dec. 96 | Tr-5 | Cottony | − | Cottony | Yellow-red | − |
| Sept. 97 | Tr-3 | Cottony | − | Cottony | Yellow | ± |
| Nov. 97 | Tr-3 | Cottony | − | Cottony | Yellow | − |
| Dec. 97 | Tr-5 | Cottony | − | Cottony | Red | − |
| Patient 5 | ||||||
| Dec. 96 | Tr-1 | Cottony | − | Cottony | Red | − |
| Aug. 97 | Tr-1 | Cottony | + | Granular | Red-yellow | − |
| Nov. 97 | Tr-2 | Cottony | + | Cottony | Red | − |
| Patient 6 | ||||||
| Dec. 97 | Tr-3 | Cottony | − | Cottony | Red | − |
| Jan. 98 | Tr-3 | Cottony | − | Cottony | Red | − |
| Apr. 99 | Tr-5 | Cottony | − | Cottony | Red | − |
| Patient 9 | ||||||
| Jan. 98 | Tr-4 | Cottony | + | Cottony | Yellow | ± |
| Mar. 98 | Tr-3 | Cottony | ± | Cottony | Yellow | ± |
| Apr. 98 | Tr-3 | Granular | − | Cottony | Red | − |
| Patient 15 | ||||||
| Mar. 97 | Tr-2 | Cottony | − | Cottony | Yellow | − |
| Dec. 97 | Tr-5 | Cottony | − | Cottony | Red-purple | − |
−, no melanin; +, melanin present; ±, melanin sometimes present.
BCPMSG, bromeresol purple-milk solids-glucose agar; pigment on colony reverse was recorded.
−, urease negative (orange); +, urease positive (red); ±, a change in color was noticed.
Strain typing of T. mentagrophytes and T. tonsurans.
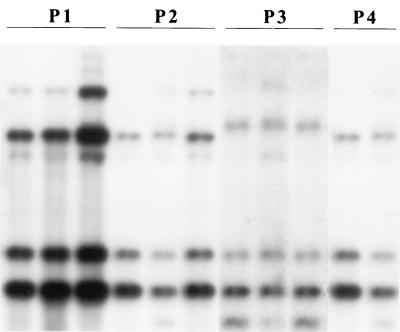
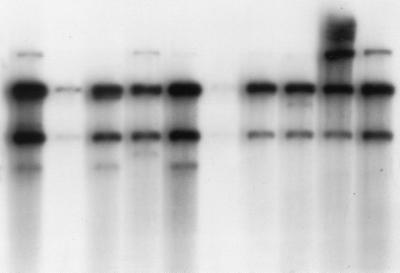
Two RFLP types were observed among 11 serial isolates from four patients with T. mentagrophytes infection. RFLP types Tm-1 and Tm-2 differed by only one band shift (Fig. 2). Based on the RFLP pattern, however, T. mentagrophytes strains could be readily distinguished from both T. rubrum and T. tonsurans. Similarly, 10 strains of T. tonsurans taken from different patients had the same RFLP type, Tt-1, which was different from that of T. mentagrophytes and T. rubrum (Fig. 3).
FIG. 2.
Southern hybridizations of EcoRI-digested genomic DNAs of strains from four onychomycosis patients (P1 to P4) with T. mentagrophytes infection. A PCR-amplified fragment (∼1,220 bp) of 18S rDNA plus the adjacent ITS region from a T. rubrum strain was used as a probe. Two RFLP types are shown. Lanes marked by a solid line represent strains obtained from the same patient during the course of therapy.
FIG. 3.
Southern hybridizations of EcoRI-digested genomic DNAs of T. tonsurans strains. A PCR-amplified fragment (∼1,220 bp) of 18S rDNA plus the adjacent ITS region from a T. rubrum strain was used as a probe. Ten different tinea capitis patients show the same RFLP type.
DISCUSSION
In this study, we have focused on the molecular genotyping of T. rubrum and T. mentagrophytes strains from patients with toe onychomycosis to determine whether the strain (genotype) of the species changes during the course of therapy and follow-up. The original purpose of the study was to distinguish between recrudescence and postcure reinfection; however, it was found retrospectively that none of the patients presenting with sufficient consistency to be included in the study had at any time appeared to be cured and then reinfected. The study therefore focused purely on its control dimension, namely, the question of the genetic stability and homogeneity of isolates from established infections undergoing treatment. Such a control study, in any case, is an absolute prerequisite for meaningful interpretation of any longitudinal studies of continuous or successive infections. It should be noted that there was no detectable bias in the selection of the patients in our study toward matters correlated with predicted resistance to therapy: hundreds of dermatophyte cultures were saved from all available patients. Retrospective analysis, however, indicated that patients who appeared to be cured after initial therapy did not return with new dermatophytoses over the 3-year period of our study. A protocol involving recall of all or randomly selected, treated patients for a 3-year follow-up, as recommended by De Cuyper and Hindryckx (4) for pharmaceutical clinical trials, was not possible in this study, which was based entirely on self-motivated patient visits and was not funded for additional consultations.
Our results confirm the findings of Jackson et al. (19) that there is substantial variability in the rDNA repeat region of T. rubrum. These authors indicated that the molecular polymorphisms are due to a variation in the copy number of a repetitive element present in the NTS regions of the rDNA cistrons. Given the now well-demonstrated uniformity of the coding and ITS regions of the T. rubrum ribosomal gene (7, 30), the variation seen in the present study is presumed to reflect the same NTS copy number polymorphism. The predominant RFLP types, Tr-1 and Tr-3, seen in the present study appear to correspond to types A and B, reported as the most common types seen by Jackson et al. (19) in their multipatient survey. Recent studies by Gräser et al. (6) have further highlighted the uniformity in the T. rubrum complex by comparing the morphological and physiological features with the results of sequencing of the ITS region of the ribosomal operon, PCR fingerprinting, and amplified fragment length polymorphism analysis.
Techniques for the detection of intraspecific variation have long been sought in order to allow population genetic studies in Trichophyton species, especially in the often greatly predominant T. rubrum. Anthropophilic (human-specific) dermatophyte species, however, are products of evolutionarily recent adaptive radiation events (16, 31) and also have an ecological niche strongly favoring clonality over sexual recombination (31), resulting in species with an unusually high degree of genetic uniformity (7, 30). Previous attempts to find genetic differences in T. rubrum by using normally sensitive techniques such as RFLP typing (17, 25, 36; Mitchell et al., Abstr. Gen. Meet. Am. Soc. Microbiol.), arbitrarily primed PCR (22), amplified fragment length polymorphisms (8), and ITS sequencing have had little or no success. The technique used in the present study was sufficiently sensitive to distinguish a high proportion of the strains seen from one another, as was previously found by Jackson et al (19), and appeared at first to have considerable promise for epidemiological study. Although intraspecies strain variability was observed by using this technique, this typing system may not be very useful in studies of relapse and reinfection for the reasons discussed below.
Interestingly, however, it became clear over the course of our study that many of the patients and individual nails studied varied in their resident T. rubrum ribosomal RFLP type. Although some patient nails remained stable with regard to T. rubrum genotype over at least a 1-year period, sufficient variability was seen in individual, uncured nails to ensure that the presence of a different genotype on a nail could not automatically be interpreted as infection by a novel strain. It is not impossible that individual dermatophytosis patients may be infected independently by multiple strains of the same organism, especially considering recent evidence that patients with T. rubrum foot infection may have a genetic predisposition making them vulnerable to this organism (35). The results of the present study, however, also suggest another natural pattern that is increasingly seen in fungal genetic studies. There is evidence in numerous studies with other fungi, especially Candida species, that an incompletely understood process typically causes rapid genotypic change in at least some repetitive gene loci (3, 18, 23, 24, 27). This process, generically referred to as reorganization of repetitive regions (23), may be based on deletions and duplications of repeat segments, with these events deriving from unequal mitotic crossing over taking place among the different copies of multicopy repetitive genes within single nuclei (27). Jackson et al. (19) have noted that the sequence length increment distinguishing their four major successively larger T. rubrum NTS molecular size types was 100 to 150 bp, a length similar to the well-characterized 172-bp alt element of the Candida albicans RPS family of repeat sequences (2).
Lockhart et al. (24) described one chronic C. albicans vaginitis patient in detail who carried two coderived clonal “substrains” that were distinguished by minor microevolutionary differences elucidated with two repetitive fingerprinting probes, the hypervariable C1 subfragment of the Ca3 probe, and the unrelated CARE2 probe. As judged by the degrees of interrelatedness seen in strains isolated from single patients, as opposed to diverse patient populations, the patient's two genotypes clearly had a very recent common ancestor, possibly native to the patient herself. Analysis of strains isolated over a 2-year period indicated that there were successive sweeps to pathogenic prominence wherein each genotype caused some, but not all, of the recurrent episodes of vaginitis. A correlated coincidence of minor variations in the two independent probes allowed the investigators to rule out the possibility that each change to a new prevalent strain was a newly arising genetic switch and, instead, pointed to successive ecological niche occupation by two stable (over the study period) genotypes. This alteration in niche predominance was referred to by the investigators as substrain shuffling. The pattern seen in this patient is highly suggestive of the pattern seen in our T. rubrum patients bearing more than one genotype. It appears unlikely that microevolution is so rapid in these patients that their isolates frequently switch genetic type within a period of less than 3 years. On the other hand, cohabitation not just in the nails but on the adjacent foot skin (35) by relatively recently derived, genetically distinct substrains may well yield the result that each new dermatologic mycology examination tends to isolate the substrain that is pathogenically predominant in the affected nail at the time.
Whether massive sampling of such a patient's nails and foot skin at one time could readily yield multiple genetic types at once has yet to be demonstrated; our results predict that, given a patient with a history of yielding more than one genotype in successive studies, the simultaneous co-occurrence of these genotypes should be detectable if sought with sufficient diligence, a study that remains to be done. In fact, however, the degree of ongoing microevolution in the T. rubrum NTS may be even greater than such a scenario would indicate. Jackson et al. (19) proposed that a small number of strains they obtained with multiple fragment sizes in ribosomal probing “may be the result of heterogeneities in the number of repeat units within different copies of the rDNA cistrons of individual strains.” Although other possible interpretations were also mentioned, the possibility that single strains may contain multiple ribosomal types suggests the possibility of a very labile genetic region subject to unusually rapid microevolution. Very rapid, spontaneous microevolution of repetitive regions in an experimental population of C. albicans has recently been demonstrated in drug-free control cultures in a time-limited, directed drug resistance evolution study (3). It should be noted that diploidy, one of the alternative suggestions proffered by Jackson et al. (19), is highly unlikely, since T. rubrum is known via the Stockdale test to show the characteristic partial mating response of the ancestral minus mating type (28, 33) and appears to be a typical single mating type, haploid, asexual, pathogenic lineage analogous to species seen in many other fungal and oomycetous groups (1, 31).
It should be pointed out that, in contrast to the situation with C. albicans repetitive elements, there is currently no means by which to assign a time scale to microevolution in the T. rubrum NTS region, and two genotypes coexisting in the same nail, even though their genetic differences may represent the most minimal degree of microevolution, may have independent evolutionary histories going back years, decades, or millennia. The study of the population genetics of this organism are at the most rudimentary stage, and many more repetitive and variable loci require exploration before a clear picture emerges. Similarly, further studies are needed to determine if there is any correlation between genotypes and drug responses in vivo or in vitro in T. rubrum.
Although T. mentagrophytes patients were less frequently seen in our study population than T. rubrum patients, sufficient repeat cultures were obtained to show that the technique of Jackson et al. (19) elucidates some genetic differences in this species as well. T. mentagrophytes sensu lato has long been known to be a species complex, and it is presumed, but has not been demonstrated, that our isolates belong to the purely genetically distinguished species recently redefined under the name T. interdigitale Priestley by Gräser et al. (9). The anthropophilic forms of this species are known to have a high degree of genetic uniformity, approaching that seen in T. rubrum (26). Therefore, further exploration of this technique within this species is warranted. The complete uniformity of T. tonsurans isolates preliminarily investigated over the course of our study may simply suggest that this technique is not applicable to that species. On the other hand, most T. tonsurans infections in Ontario, our study area, derive from a recent outbreak (15), and the prevalence of a uniform NTS type in our pilot study may simply reflect the predominance of a single outbreak strain. T. tonsurans, in general, is a typically highly genetically homogeneous anthropophilic dermatophyte species (21).
ACKNOWLEDGMENTS
We thank Saleh Albreish, Ursula Bunn, and Maria Witkowska for provision of documented cultures. We also thank Linda M. Kohn, Department of Biology, University of Toronto at Mississauga, Mississauga, Ontario, Canada, for providing laboratory space in which to conduct the radioactive hybridization work for detection of RFLPs.
REFERENCES
- 1.Brasier C M. The dynamics of fungal speciation. In: Rayner A D M, Brasier C M, Moore D, editors. Evolutionary biology of the fungi. Cambridge, England: Cambridge University Press; 1987. pp. 231–260. [Google Scholar]
- 2.Chibana H, Iwaguchi S-I, Homma M, Chindamporn A, Nakagawa Y, Tanaka K. Diversity of tandemly repeated sequences due to short periodic repetitions in the chromosomes of Candida albicans. J Bacteriol. 1994;176:3851–3858. doi: 10.1128/jb.176.13.3851-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowen L E, Sanglard D, Calabrese D, Sirjusingh C, Anderson J B, Kohn L M. Evolution of drug resistance in experimental populations of Candida albicans. J Bacteriol. 2000;182:1515–1522. doi: 10.1128/jb.182.6.1515-1522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Cuyper C, Hindryckx P H. Long-term outcomes in the treatment of toenail onychomycosis. Br J Dermatol. 1999;141:15–20. doi: 10.1046/j.1365-2133.1999.00009.x. [DOI] [PubMed] [Google Scholar]
- 5.Elewski B E, Charif M A. Prevalence of onychomycosis in patients attending a dermatology clinic in northeastern Ohio for other conditions. Arch Dermatol. 1997;133:1172–1173. [PubMed] [Google Scholar]
- 6.Gräser Y, Kuijpers A F A, Presber W, de Hoog G S. Molecular taxonomy of the Trichophyton rubrum complex. J Clin Microbiol. 2000;38:3329–3336. doi: 10.1128/jcm.38.9.3329-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gräser Y, Fari M L, Presber W, Sterry W, Tietz H J. Identification of common dermatophytes (Trichophyton, Microsporum, Epidermophyton) using polymerase chain reactions. Br J Dermatol. 1998;138:576–582. doi: 10.1046/j.1365-2133.1998.02165.x. [DOI] [PubMed] [Google Scholar]
- 8.Gräser Y, Kühnisch J, Presber W. Molecular markers reveal exclusively clonal reproduction in Trichophyton rubrum. J Clin Microbiol. 1999;37:3713–3717. doi: 10.1128/jcm.37.11.3713-3717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gräser Y, Kuijpers A F A, Presber W, de Hoog G S. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med Mycol. 1999;37:315–330. doi: 10.1046/j.1365-280x.1999.00234.x. [DOI] [PubMed] [Google Scholar]
- 10.Gupta A K, Shear N H. The new oral antifungal agents for onychomycosis of the toenails. J Eur Acad Dermatol Venereol. 1999;13:1–13. [PubMed] [Google Scholar]
- 11.Gupta A K, Shear N H. A risk-benefit assessment of the newer oral antifungal agents used to treat onychomycosis. Drug Safety. 2000;22:33–52. doi: 10.2165/00002018-200022010-00004. [DOI] [PubMed] [Google Scholar]
- 12.Gupta A K, Jain H C, Lynde C W, Macdonald P, Cooper E, Summerbell R C. Prevalence and epidemiology of onychomycosis in patients visiting physicians' offices: a multicenter Canadian survey of 15,000 patients. J Am Acad Dermatol. 2000;43:244–248. doi: 10.1067/mjd.2000.104794. [DOI] [PubMed] [Google Scholar]
- 13.Gupta A K, Jain H C, Lynde C W, Watteel G N, Summerbell R C. Prevalence and epidemiology of unsuspected onychomycosis in patients visiting dermatologists' offices in Ontario, Canada—a multicenter survey of 2001 patients. Int J Dermatol. 1997;36:783–787. doi: 10.1046/j.1365-4362.1997.00349.x. [DOI] [PubMed] [Google Scholar]
- 14.Gupta A K, Sauder D N, Shear N H. Antifungal agents: an overview. Part I. J Am Acad Dermatol. 1994;30:677–698. [PubMed] [Google Scholar]
- 15.Gupta A K, Summerbell R C. Increased incidence of Trichophyton tonsurans tinea capitis in Ontario, Canada, between 1985 and 1996. Med Mycol. 1998;36:55–60. [PubMed] [Google Scholar]
- 16.Harmsen D, Schwinn A, Weig M, Brcker E-B, Heeseman J. Phylogeny and dating of some pathogenic keratinophilic fungi using small subunit ribosomal RNA. J Med Vet Mycol. 1995;33:299–303. doi: 10.1080/02681219580000611. [DOI] [PubMed] [Google Scholar]
- 17.Howell S A, Barnard R J, Humphreys F. Application of molecular typing methods to dermatophyte species that cause skin and nail infections. J Med Microbiol. 1999;48:33–40. doi: 10.1099/00222615-48-1-33. [DOI] [PubMed] [Google Scholar]
- 18.Iwaguchi S-I, Homma M, Chibana H, Tanaka K. Isolation and characterization of a repeated sequence (RPS1) of Candida albicans. J Gen Microbiol. 1992;138:1893–1900. doi: 10.1099/00221287-138-9-1893. [DOI] [PubMed] [Google Scholar]
- 19.Jackson J J, Barton R C, Evans E G. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J Clin Microbiol. 1999;37:931–936. doi: 10.1128/jcm.37.4.931-936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kane J, Summerbell R C, Sigler L, Krajden S, Land G. Laboratory handbook of dermatophytes, a clinical guide and laboratory manual of dermatophytes and other filamentous fungi from skin, hair and nails. Belmont, Calif: Star Publishing Company; 1997. [Google Scholar]
- 21.Kim J A, Takizawa K, Fukushima K, Nishimura K, Miyaji M. Identification and genetic homogeneity of Trichophyton tonsurans isolated from several regions by random amplified polymorphic DNA. Mycopathologia. 1999;145:1–6. doi: 10.1023/a:1007008401122. [DOI] [PubMed] [Google Scholar]
- 22.Liu D, Coloe S, Pederson J, Baird R. Use of arbitrarily primed polymerase chain reaction to differentiate Trichophyton dermatophytes. FEMS Microbiol Lett. 1996;136:147–150. doi: 10.1111/j.1574-6968.1996.tb08040.x. [DOI] [PubMed] [Google Scholar]
- 23.Lockhart S R, Fritch J J, Meier A S, Schröppel K, Srikantha T, Galask R, Soll D R. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995;33:1501–1509. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart S R, Reed B D, Pierson C L, Soll D R. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, Cl, and CARE2. J Clin Microbiol. 1996;34:767–777. doi: 10.1128/jcm.34.4.767-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mochizuki T, Uehara M, Menon T, Ranganathan S. Minipreparation of total cellular DNA is useful as an alternative molecular marker of mitochondrial DNA for the identification of Trichophyton mentagrophytes and T. rubrum. Mycoses. 1996;39:31–35. doi: 10.1111/j.1439-0507.1996.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 26.Mochizuki T, Watanabe S, Uehara M. Genetic homogeneity of Trichophyton mentagrophytes var. interdigitale isolated from geographically distant regions. J Med Vet Mycol. 1996;34:139–143. [PubMed] [Google Scholar]
- 27.Pujol C, Joly S, Nolan B, Srikantha T, Soll D R. Microevolutionary changes in Candida albicans identified by the complex Ca3 fingerprinting probe involve insertions and deletions of the full-length repetitive sequence RPS at specific genomic sites. Microbiology. 1999;145:2635–2646. doi: 10.1099/00221287-145-10-2635. [DOI] [PubMed] [Google Scholar]
- 28.Stockdale P M. Sexual stimulation between Arthroderma simii Stockd., Mackenzie and Austwick and related species. Sabouraudia. 1968;6:176–181. [PubMed] [Google Scholar]
- 29.Summerbell R C, Kane J, Krajden S. Onychomycosis, tinea pedis and tinea manuum caused by non-dermatophytic fungi. Mycoses. 1989;32:609–619. doi: 10.1111/j.1439-0507.1989.tb02192.x. [DOI] [PubMed] [Google Scholar]
- 30.Summerbell R C, Haugland R A, Li A, Gupta A K. rRNA gene internal transcribed spacer 1 and 2 sequences of asexual, anthropophilic dermatophytes related to Trichophyton rubrum. J Clin Microbiol. 1999;37:4005–4111. doi: 10.1128/jcm.37.12.4005-4011.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Summerbell R C, Li A, Haugland R. What constitutes a functional species in the asexual dermatophytes? Microbiol Cult Coll. 1997;13:29–37. [Google Scholar]
- 32.White T J, Bruns T, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, editor. PCR protocols: a guide to methods and applications. London, United Kingdom: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 33.Young C N. Pseudo-cleistothecia in Trichophyton rubrum. Sabouraudia. 1968;6:160–162. [PubMed] [Google Scholar]
- 34.Zaias N, Rebell G. Chronic dermatophytosis syndrome due to Trichophyton rubrum. Int J Dermatol. 1996;35:614–617. doi: 10.1111/j.1365-4362.1996.tb03682.x. [DOI] [PubMed] [Google Scholar]
- 35.Zaias N, Tosti A, Rebell G, Morelli R, Bardazzi F, Bieley H, Zaiac M, Glick B, Paley B, Allevato M, Baran R. Autosomal dominant pattern of distal subungual onychomycosis caused by Trichophyton rubrum. J Am Acad Dermatol. 1996;34:302–304. doi: 10.1016/s0190-9622(96)80142-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhong Z, Li R, Li D, Wang D. Typing of common dermatophytes by random amplification of polymorphic DNA. Jpn J Med Mycol. 1997;38:239–246. [Google Scholar]