Abstract
Introduction
In humanity’s ongoing fight against its common enemy of COVID-19, researchers have been relentless in finding efficient technologies to support mitigation, diagnosis, management, contact tracing, and ultimately vaccination.
Objectives
Engineers and computer scientists have deployed the potent properties of deep learning models (DLMs) in COVID-19 detection and diagnosis. However, publicly available datasets are often adulterated during collation, transmission, or storage. Meanwhile, inadequate, and corrupted data are known to impact the learnability and efficiency of DLMs.
Methods
This study focuses on enhancing previous efforts via two multimodal diagnostic systems to extract required features for COVID-19 detection using adulterated chest X-ray images. Our proposed DLM consists of a hierarchy of convolutional and pooling layers that are combined to support efficient COVID-19 detection using chest X-ray images. Additionally, a batch normalization layer is used to curtail overfitting that usually arises from the convolution and pooling (CP) layers.
Results
In addition to matching the performance of standard techniques reported in the literature, our proposed diagnostic systems attain an average accuracy of 98% in the detection of normal, COVID-19, and viral pneumonia cases using corrupted and noisy images.
Conclusions
Such robustness is crucial for real-world applications where data is usually unavailable, corrupted, or adulterated.
Keywords: Artificial intelligence, Deep learning, COVID-19, Corona virus, Chest X-ray, Image processing
1. Introduction
Recently, several computer-aided diagnosis (CAD) systems have been proposed for the diagnosis and detection of COVID-19 (Wu et al., 2020, Ozturk et al., 2020, Ardakani et al., 2020, Jain et al., 2020, Marques et al., 2020, Ucar and Korkmaz, 2019, Zabirul et al., 2020, Sedik et al., 2020, Altan and Karasu, 2020, Pathak et al., 2020, Das et al., 2020, Rahimzadeh and Attar, 2020, Oh et al., 2020, Han et al., 2020, Jamshidi et al., 2020, Hu et al., 2020, Ardakani et al., 2020, Apostolopoulos and Tzani, 2020, Civit-Masot et al., 2020). Deep learning models (DLM) are considered more robust than the traditional machine learning methods, especially when using big data. In addition, the multi-layer structure of DLMs provides tools for effective feature understanding and pattern recognition that are important, when classifying unstructured big datasets. Notwithstanding their superlative properties, there is consensus that standard DLM networks suffer from some limitations that include:
-
•
Low accuracy for multi-class classification problems.
-
•
Depleted datasets, which impede classification accuracy, especially sensitivity, and increase data overfitting.
-
•
High inter-class imbalance which leads to misclassification.
-
•
Poor performance in real-time applications.
Consequently, the study presented in this work is directed at ameliorating the above limitations via two hierarchical multi-modal DLMs for efficient feature extraction for COVID classification. In this regard, the main novel contributions of this research are:
-
•
Proposal of a novel uni-step diagnostic system for COVID detection using X-ray images.
-
•
Proposal of a novel classification system based on the dual-step diagnosis for COVID detection using X-ray images.
-
•
Presentation of a scheme that attains a creditable average accuracy of 96.3% for detection of COVID-19 and pneumonia using adulterated chest X-ray images. Our proposed scheme is tailored towards the realization of an efficient (i.e., high accuracy) yet robust (i.e. low time and cost) approach for COVID-19 detection.
2. State-of-the-art efforts for COVID-19 detection using DLMs
Even prior to COVID-19, artificial intelligence (AI) and DLMs were widely touted as the advanced solutions for many problems in image processing and pattern recognition (Alghamdi et al., 2020, Amrani et al., 2018, Hammad et al., 2018, Abou-Nassar et al., 2020), and other numerous applications (Wang et al., 2014, Sedik et al., 2020, Ahmad et al., 2021). In the ongoing fight against COVID, DLMs have been used to enhance knowledge of its etiology and pathology, which has proven useful in COVID-19 detection and diagnosis. Table 1 summarizes the state-of-the-art methods with comparison of related studies that use DLMs for COVID-19 detection.
Table 1.
Comparison of related studies that use DLMs for COVID-19 detection.
| Study | Methodology | Dataset | Weaknesses |
|---|---|---|---|
| Wu et al. (2020) | Multi-view fusion model | CT images of 495 patients |
|
| Ozturk et al. (2020) | Deep learning model (DarkCovidNet) | 125 chest X-ray images |
|
| Ardakani et al. (2020) | ResNet-101 and Xception | 1020 CT slices from 194 patients |
|
| Jain et al. (2020) | ResNet-101 | 565 chest X-ray images |
|
| Marques et al. (2020) | EfficientNet | 404 chest X-ray images |
|
| Ucar and Korkmaz (2019) | Bayes-SqueezeNet | 5949 posteroanterior chest radiography images |
|
| Islam et al. (Zabirul et al., 2020) | CNN-LSTM | 4575 X-ray images |
|
| Sedik et al. (2020) | CNN and ConvLSTM | 2880 X-ray and CT images |
|
| Altan and Karasu (2020) | EfficientNet-B0 | 7980 chest X-ray images |
|
| Pathak et al. (2020) | ResNet-50 | 852 chest CT images |
|
| Das et al. (2020) | Xception | 127 X-ray images |
|
| Rahimzadeh and Attar (2020) | Xception ResNet50V2 |
180 X-ray images |
|
| Oh et al., (2020) | attention-based deep 3D multiple instance learning | 460 chest CT images |
|
3. Proposed stepwise multimodal diagnostic systems
Our proposed DLM consists of a hierarchy of convolutional and pooling layers that are combined to support efficient COVID-19 detection using chest X-ray images. Each convolution layer is made up of a set of two-dimensional (2D) digital filters that are convolved with the input image to generate a feature map that aggregates information in the input image. Like the weight initialization in neural networks, the values of each filter are randomly set and initialized with small values. The kernels of the filters are 3 × 3 in size and the stride is 2. The filtering is efficient for the analysis of such images, with great flexibility for medical applications, especially our case (COVID-19 diagnosis) and low cost. Similarly, pooling layers consisting of Max and mean pooling operations are used to retain much information about the less important elements of a block, or pool. The Max pooling operation selects the maximum value from a window of pixels, while the mean pooling operation generates the average value of pixels in the window, which is called the kernel of the pooling layers or simply the pooling kernel.
Our proposed scheme consists of combined phases (feature extraction, feature reduction, regularization and classification). The feature extraction process is performed by a set of five convolutional layers. The feature reduction process is carried out by five Max pooling layers, which are utilized after each convolution layer. In addition, the pooling layers is inserted to execute the enumerated operations. Additionally, the regularization phase is utilized using a batch normalization layer, which is also used to curtail overfitting that usually arises from the convolution and pooling (CP) layers. We have applied deep learning techniques (Sedik, 2021) with a fully-connected layer, which converts the feature map generated from the CP group into a feature vector, and a classification layer (a dense layer with SoftMax activation function) that executes the final classification to determine the category of the input image.
Furthermore, we adopt wavelet denoising to remove the noise from the COVID-19 X-ray images. The discrete wavelet transform (DWT) has been deployed in several research fields. Wavelet transform is usually performed on signals. The assumptions used by WaveLab routines would be violated if a 1-D threshold selection scheme were used in a 2-D DWT, for example by applying a 1-D thresholding scheme in both spatial dimensions. This is because the threshold for detail coefficients would be determined from approximation and detail coefficients in some cases, and from detail coefficients only in other cases. In a 2-D DWT, the coefficients must be set in the correct order. The thresholding algorithms used in WaveLab are implemented in the case of additive white Gaussian noise (AWGN). It is desirable to apply a level-dependent threshold when the noise autocorrelation is unknown (Siam et al., 2021). The WaveLab procedures threshold each direction channel independently for each resolution. The variance varies between detail channels at different resolutions, but it is constant within each channel.
3.1. COVID-19 dataset
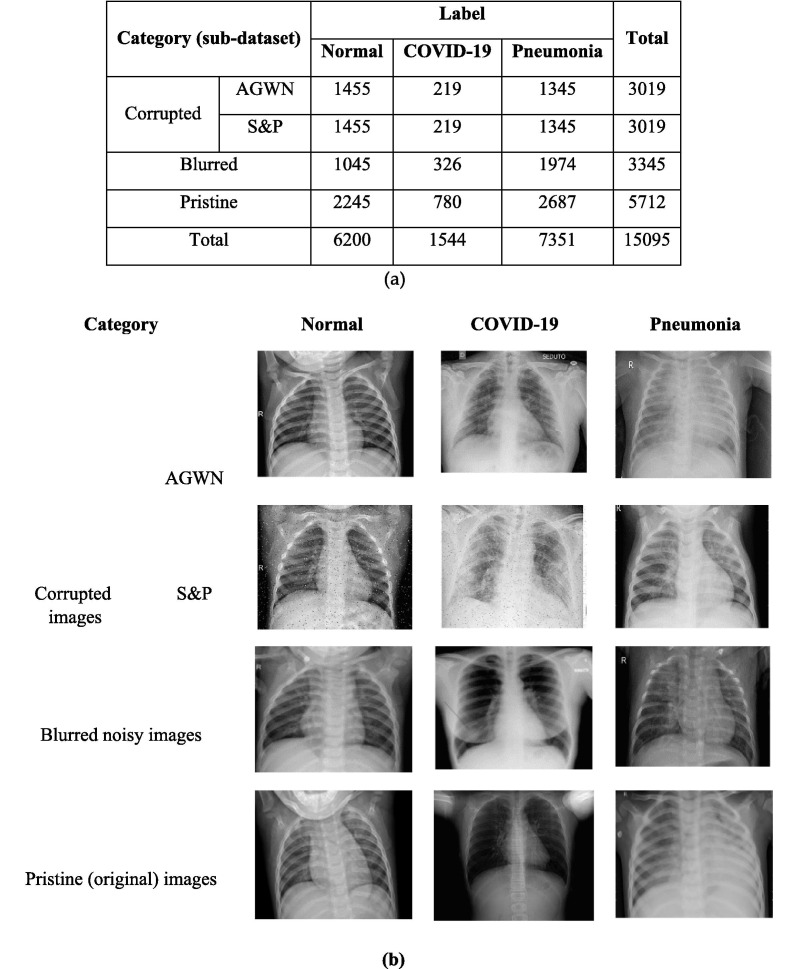
A dataset comprising 15,095 X-ray images of which 6038 are noisy images made up of 3019 images corrupted with AWGN and the remaining 3019 images corrupted with Salt and Pepper (S&P) noise has been considered. Each category of the corrupted images consists of 219 images with COVID-19 label, 1455 images with normal label and 1345 images with pneumonia label. Moreover, 3345 images are deformed by blurring. Finally, the remaining 3693 X-ray images complete the three categories of our dataset, which is summarized in Fig. 1 -a. Sample images from each category are presented in Fig. 1-b (Sedik et al., 2020).
Fig. 1.
(a) Distribution of images in dataset by their categories and labels, (b) Sample images from the COVID-19 dataset 25]
3.2. Implementation of the proposed scheme
Depending on whether the DLM could discriminate and determine the classification of the image in one or more steps, we delineate the implementation as uni- or multi-step. The frameworks are outlined in this subsection.
3.2.1. Uni-step diagnosis of COVID-19
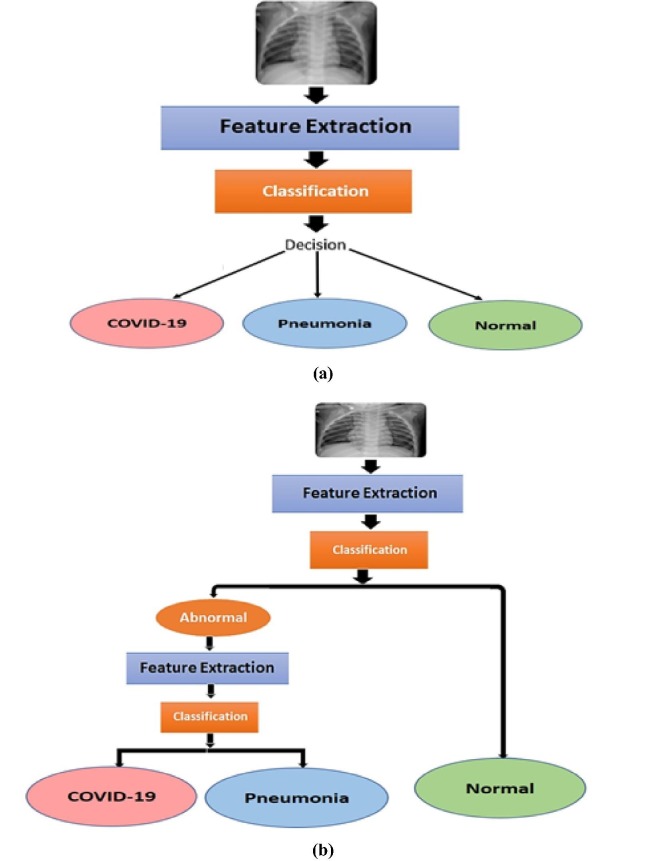
This system is a typical traditional hierarchically structured DLM, but with the binary decision activation function replaced with a SoftMax activation function. This provides better classification, since it normalizes the output of the last layer in the network to a probability distribution over predicted output classes. Fig. 2 -a illustrates the execution of the feature extraction and classification steps to discriminate the three pre-classified categories (i.e. normal, COVID-19 and pneumonia) in the dataset.
Fig. 2.
Layout of proposed (a) uni-step diagnostic system, and (b) dual-step diagnostic system.
3.2.2. Multi-step diagnosis of COVID-19
Unlike the uni-step diagnostic system, the multi-step diagnosis uses multiple steps to discriminate different labels of the pre-classified dataset. In the first step of our implementation, the dataset is delineated into normal and abnormal categories. Subsequently, in the second step, the features in the abnormal category are extracted for the purpose of classification of such images COVID-19 or pneumonia. Fig. 2-b presents outlines of the dual-step diagnosis procedure.
4. Simulation framework and results
Inspecting Fig. 2-b, we note that relative to the uni-step diagnosis (i.e., in Fig. 2-a), the dual-step diagnosis requires two feature extraction steps for the delineation and classification procedures.
Using the dataset described earlier and the two implementation scenarios outlined in the previous section, we present a simulation-based evaluation of our diagnosis systems.
4.1. Simulation-based evaluation of the proposed DLMs
Our experiments are designed to assess the impact of the proposed systems on the accuracy of COVID-19 detection. We present results based on the multimodal (i.e., uni- and dual-step) diagnosis scenarios presented in Section 3.
Each diagnosis scenario is implemented on chest X-ray images from the publicly available COVID-19 dataset that was explained in Section 3. Our simulation was implemented on a computing workstation (Laptop) equipped with NVIDIA GPU (4 GB), Intel CPU (Core i7) and, 16 GB RAM.
In addition to the composition explained in Fig. 1, the dataset is partitioned with 70:30 ratio for training and testing. Additionally, the training sub-dataset is fragmented into batches, each of size 20 with a number of epochs set at 50. The results reported in the remainder of this section are based on the use of CNN- and ConvLSTM-based DLMs.
4.2. Results of Uni-step diagnostic system
The uni-step COVID diagnosis system depends on the framework outlined in Fig. 2 to classify images in terms of the pre-assigned labels of the dataset, i.e., normal, COVID, and pneumonia. Results for the two (i.e. CNN and ConvLSTM) DLMs used are presented in the sequel.
4.2.1. CNN-based DLM for uni-step diagnosis
A summary of the detection accuracy and testing time for all sub-datasets is presented in Table 2 , where the bold entries highlight the recorded detection accuracy, true positive rates, and testing time for COVID-19, pneumonia, and normal cases of the dataset.
Table 2.
Performance of the proposed CNN method on different cases to classify three classes.
| Category |
Original images |
AWGN corrupted images |
Blurred images |
S&P noisy images |
Denoised images |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | |
| COVID-19 | 0.96 | 0.01 | 0.03 | 0.81 | 0.02 | 0.17 | 0.94 | 0.02 | 0.04 | 0.94 | 0.01 | 0.05 | 0.97 | 0 | 0.03 |
| Normal | 0 | 1 | 0 | 0 | 0.99 | 0.01 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Pneumonia | 0 | 0.03 | 0.97 | 0 | 0.04 | 0.96 | 0 | 0.03 | 0.97 | 0 | 0 | 1 | 0.04 | 0 | 0.96 |
| Accuracy | 98.2 % | 96.78 % | 98 % | 97.5 % | 98.2 % | ||||||||||
| Testing time | 2.19 Sec. | 2.29 Sec. | 2.5 Sec. | 2.7 Sec. | 2.6 Sec. | ||||||||||
As presented in the table, the average detection accuracies for COVID-19 are 97.4%, 98.2%, and 98.2% for the corrupted, pristine and denoised image categories of our dataset. Similarly, the average test times are 2.49 s, 2.19 s, and 2.60 s for corrupted, pristine, and denoised image categories, respectively. Further discussions on these results are presented later in Section 5.
In the meantime, results of the second DLM, i.e. ConvLSTM-based DLM for the dual-step diagnostic system are presented in the sequel.
4.2.2. ConvLSTM-based DLM for Dual-step diagnosis
Table 3 presents a summary of the detection accuracies and testing times for the proposed ConvLSTM-based DLM in classifying the different sub-datasets reported. We note that the average detection accuracies for all the sub-datasets are 91.8%, 96.8% and 96.8% for the corrupted, pristine, and denoised image categories of the dataset, respectively. Similarly, average testing times of 8.1 s, 8.6 s, and 8.6 s are reported for the corrupted, pristine and denoised image categories of the dataset, respectively.
Table 3.
Performance analysis for the proposed uni-step diagnostic system using ConvLSTM DLM on different sub-datasets.
| Category |
Original images |
AWGN corrupted images |
Blurred images |
S&P noisy images |
Denoised images |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | |
| COVID-19 | 0.94 | 0.03 | 0.03 | 0.89 | 0.04 | 0.07 | 0.87 | 0 | 0.13 | 0.89 | 0.02 | 0.09 | 0.92 | 0.01 | 0.07 |
| Normal | 0.01 | 0.99 | 0 | 0 | 0.99 | 0.01 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Pneumonia | 0.01 | 0 | 0.99 | 0.15 | 0 | 0.85 | 0.03 | 0 | 0.97 | 0.2 | 0.02 | 0.79 | 0.04 | 0 | 0.96 |
| Accuracy | 96.8 % | 90.9 % | 94.6 % | 89.9 % | 96.8 % | ||||||||||
| Testing time | 8.6 Sec. | 7.3 Sec. | 8.9 Sec. | 8.1 Sec. | 8.6 Sec. | ||||||||||
As in Table 2, the bold entries in Table 4 highlight the recorded detection accuracies, true positive rates, and testing times for the COVID-19, pneumonia, and normal classifications of the dataset using the ConvLSTM-based DLM.
Table 4.
Performance analysis (in %) for the discrimination step of the proposed dual-step diagnostic system using the CNN DLM resulting in delineation of images as Normal or Abnormal.
| Category | Sensitivity | Specificity | TP | TN | Accuracy | F1 Score | MCC |
|---|---|---|---|---|---|---|---|
| Original images | 100 | 98.1 | 98.7 | 100 | 99.2 | 99.4 | 98.4 |
| AWGN images | 95.5 | 98.4 | 98.6 | 94.7 | 96.8 | 97.1 | 93.6 |
| Blurred images | 97.5 | 98.2 | 98.4 | 97.2 | 97.8 | 97.9 | 95.7 |
| S&P noisy images | 93.9 | 98.9 | 98.9 | 93.6 | 96.3 | 96.4 | 92.7 |
| Denoised images | 97.8 | 94.5 | 96.1 | 96.9 | 96.4 | 96.9 | 92.7 |
4.3. Results of dual-step diagnosis
As outlined in Section 3, the first stage of the dual-step diagnosis system entails discrimination of the dataset in terms of normal and abnormal categories. Following that, images in the abnormal category are further classified as either COVID or pneumonia images.
Outcomes of the implementation of the system using the proposed CNN- and ConvLSTM-based DLMs are presented in the remainder of this section.
4.3.1. Discrimination phase of the dual-step diagnostic system
Table 5A, Table 5B presents summaries of the Sensitivity (T), Specificity (S), positive predictive value (PPV) or simply True Positive (TP), negative predictive value (NPV) or simply True Negative (TN) accuracy (A), F1 Score (F) and Matthew’s correlation coefficient (MCC) for both (i.e. CNN and ConvLSTM) DLMs in the discrimination stage of the dual-step diagnosis systems. The parameters reported in the tables are defined in the binary quality metrics equation (Metrics, 2022).
Table 5A.
Performance analysis (in %) for discrimination step of the proposed dual-step diagnostic system using the ConvLSTM DLM resulting in delineation of images as Normal or Abnormal.
| Category | Sensitivity | Specificity | TP | TN | Accuracy | F1 Score | MCC | Testing Time (s) |
|---|---|---|---|---|---|---|---|---|
| Original images | 98.9 | 99.1 | 98.9 | 99.1 | 99.0 | 98.9 | 98.0 | 5.9 |
| AWGN images | 96.5 | 97.3 | 96.9 | 96.8 | 96.9 | 96.7 | 93.8 | 6.1 |
| Blurred images | 96.4 | 99.7 | 99.8 | 94.4 | 97.7 | 98.1 | 95.2 | 5.7 |
| S&P noisy images | 99.8 | 93.2 | 94.3 | 99.7 | 96.7 | 96.9 | 93.8 | 5.8 |
| Denoised images | 99.8 | 93.4 | 93.5 | 99.8 | 96.5 | 96.5 | 93.2 | 6.2 |
Table 5B.
Performance analysis (in %) for classification step of the proposed dual-step diagnostic system using the CNN-based DLM.
| Category | Sensitivity | Specificity | TP | TN | Accuracy | F1 Score | MCC |
|---|---|---|---|---|---|---|---|
| Original images | 100 | 97.0 | 97.0 | 100 | 98.6 | 98.5 | 97.1 |
| AWGN images | 98.9 | 89.2 | 88.0 | 99.0 | 93.5 | 93.1 | 87.5 |
| Blurred images | 100 | 96.2 | 96.0 | 100 | 98.0 | 97.9 | 96.1 |
| S&P noisy images | 100 | 92.6 | 92.0 | 100 | 96.0 | 95.8 | 92.3 |
| Denoised images | 96.0 | 98.0 | 98.0 | 96.0 | 97.0 | 97.0 | 94.0 |
Following the discrimination of the dataset as normal and abnormal categories, a feature extraction process is used to further classify images in the abnormal category as COVID-19 or pneumonia.
Discussions of the results reported in this section as well as a brief performance analysis are presented in the next section.
5. Result discussion
The results presented in Table 3, Table 4 reveal better performance for the CNN-based DLM compared to the ConvLSTM-based DLM in the uni-step diagnostic system, where average accuracies of 98.2% and 96.8% are reported for COVID-19 and pneumonia detection, respectively.
Furthermore, we note that in terms of the four categories in the adulterated sub-dataset (i.e., original, AGWN corrupted, blurred, and S&P corrupted X-ray images) that were considered, the uni-step diagnostic system yields average increases of 1.4%, 5.9%, 3.4% and 7.6% in detection accuracy for the normal, COVID, and pneumonia classifications of the dataset. A similar trend is observed in terms of the testing time, where an average gain of 6 s is reported for the CNN-based DLM over the ConvLSTM-based DLM.
Despite the encouraging performance, a major shortcoming of the uni-step diagnostic system is the low incidence of misclassification, which leads to lower accuracy. A preprocessing stage with appropriate filters to remove noise would help ameliorate this weakness.
In terms of the dual-step diagnostic system, whereas, as reported in Table 5A, Table 5B, the ConvLSTM-based DLM performs better in the discrimination phase to delineate normal and abnormal image categories for three (i.e. the two corrupted image categories (AGWN and S&P)) and denoised images) out of the five sub-datasets reported. The CNN-based DLM offers marginal improvements of 0.2% and 0.1% in accuracy for the pristine (i.e., unadulterated) and blurred X-ray image categories of the dataset, respectively.
However, in terms of the classification phase of the dual-step diagnostic system to detect COVID-19 and pneumonia, the ConvLSTM-based DLM performs better than the CNN-based DLM in all categories except for the blurred X-ray images, where it records an accuracy of 98% compared to 96.5% reported for the ConvLSTM-based DLM. Similar trends are observed in terms of the other binary quality metrics reported in Table 6 .
Table 6.
Summary of COVID-19 detection accuracy for proposed diagnostic systems.
| Category |
Detection accuracy (in %) |
||
|---|---|---|---|
| Uni step | Dual step | ||
| Pristine images | 98.2 | 99.5 | |
| Adulterated | AGWN | 96.8 | 96.0 |
| S&P | 98.0 | 96.5 | |
| Blurred | 97.5 | 96.5 | |
| Average | 97.6 | 96.3 | |
| Denoised images | 98.2 | 98.0 | |
We finish our discussion on the results reported with a performance analysis that compares the proposed DLM diagnostic systems with other state-of-the-art methods reported in the literature as shown in Table7 . On this note, we recall that in the introductory parts of this study, some advanced DLM approaches deployed for COVID-19 detection (Jain et al., 2020, Marques et al., 2020, Ucar and Korkmaz, 2019, Zabirul et al., 2020, Sedik et al., 2020, Altan and Karasu, 2020, Pathak et al., 2020, Das et al., 2020) as well as some of their perceived weakness were enumerated. To assess the performance of our proposed diagnostic system, we reflect on these reported approaches (Jain et al., 2020, Marques et al., 2020, Ucar and Korkmaz, 2019, Zabirul et al., 2020, Sedik et al., 2020, Altan and Karasu, 2020, Pathak et al., 2020, Das et al., 2020) by presenting a comparison of their results with those reported in this study.
Table 7.
Performance analysis alongside similar DLM techniques (N.R = Not reported)
| Method | Remarks |
Performance |
||||
|---|---|---|---|---|---|---|
| Accuracy (%) | Specificity (%) | Sensitivity (%) | Testing time (s) | |||
| (Jain et al., 2020) | 2 class | 76.0 | 81.1 | 61.5 | N.R | |
| (Marques et al., 2020) | 2 class | 98.1 | N.R | N.R | N.R | |
| 3 class | 87.0 | N.R | N.R | N.R | ||
| (Ucar and Korkmaz, 2019) | ResNet | 99.5 | 100 | 99.0 | N.R | |
| Xception | 99.0 | 98.0 | 100 | N.R | ||
| (Zabirul et al., 2020) | 3 class | 98.9 | 98.9 | 98.6 | N.R | |
| (Sedik et al., 2020) | 2 class | 99.6 | N.R | N.R | N.R | |
| 3 class | 98.7 | N.R | N.R | N.R | ||
| (Altan and Karasu, 2020) | 3 class | 98.3 | 99.1 | 99.2 | N.R | |
| (Pathak et al., 2020) | 3 class | 99.4 | 99.3 | 98.7 | 113.0 | |
| (Das et al., 2020) | 2 class | CNN | 99.0 | 100 | 98.6 | 2.6 |
| ConvLSTM | 96.0 | 100 | 99.0 | 5.2 | ||
| Proposed | 2 class | CNN | 99.2 | 100 | 100 | 2.5 |
| ConvLSTM | 98.0 | 97.0 | 99.0 | 7.3 | ||
| 3 class | CNN | 98.2 | 99.2 | 97.2 | 5.7 | |
| ConvLSTM | 96.8 | 99.6 | 94 | 4.3 | ||
From the table, we can deduce that the proposed system matches all the reported methods in terms of detection accuracy (A), Sensitivity (T), and Specificity (S). Furthermore, a comparison with (Das et al., 2020) indicates that the proposed system outperforms it in terms of detection accuracy (A) for the two DLMs. i.e., CNN– and ConvLSTM-based DLMs, where our proposed diagnostic system offers a 3% improvement in COVID-19 detection accuracy.
Considering the precarious circumstances that we find ourselves involved in; this improvement is significant in assisting clinicians to make an accurate diagnosis of COVID-19; thus, contributing towards the defeat of the common enemy.
6. Conclusions
This study presented two simple yet robust deep learning diagnostic models for the detection of COVID-19 using corrupted and noisy images. This is crucial because real and publicly available datasets are adulterated at different stages of collation, transmission, and storage. The proposed CNN-based model achieved 99.2% and 98.2% classification accuracies for two and three classes, respectively. Similarly, an accuracy of 99% was reported for the ConvLSTM-based model in two classes and 96.8% for multi-class classification. Overall, in addition to matching the performance of similar state-of-the-art deep learning models, our system attains a creditable average accuracy of 96.3% for detection of COVID-19 and pneumonia using adulterated chest X-ray images. This robustness of our proposed system enhances efforts to support clinicians for accurate diagnosis of COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by Texas A&M University-San Antonio, TX, USA.
Footnotes
Peer review under responsibility of King Saud University.
References
- Wu Xiangjun, Hui Hui, Niu Meng, Li Liang, Wang Li, He Bingxi, et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur. J. Radiol. 2020;2020:109041. doi: 10.1016/j.ejrad.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;103792 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardakani Ali Abbasian, Kanafi Alireza Rajabzadeh, Rajendra Acharya U., Khadem Nazanin, Mohammadi Afshin. Computers in Biology and Medicine. 2020. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks; p. 103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain G., Mittal D., Thakur D., Mittal M.K. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybern. Biomed. Eng. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques Gonçalo, Agarwal Deevyankar, de la Torre Isabel, Díez. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar Ferhat, Korkmaz Deniz. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based Diagnostic of the Coronavirus Disease 2019 (COVID-19) from X-Ray Images. Med. Hypotheses. 2019;2020 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabirul Islam, Md, Islam Md Milon, Asraf Amanullah. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inf. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedik, Ahmed, Abdullah M. Iliyasu, Abd El-Rahiem, Mohammed E. Abdel Samea, Asmaa Abdel-Raheem, Mohamed Hammad, Jialiang Peng, Abd El-Samie, E. Fathi, and Ahmed A. Abd El-Latif. “Deploying machine and deep learning models for efficient data-augmented detection of covid-19 infections.” Viruses 12, no. 7 (2020): 769. [DOI] [PMC free article] [PubMed]
- Altan Aytaç, Karasu Seçkin. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos, Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, Yadunath, Prashant Kumar Shukla, Akhilesh Tiwari, Shalini Stalin, Saurabh Singh, and Piyush Kumar Shukla. “Deep Transfer Learning based Classification Model for COVID-19 Disease.” IRBM (2020). [DOI] [PMC free article] [PubMed]
- Das N Narayan, Kumar Naresh, Kaur Manjit, Kumar Vijay, Singh Dilbag. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. Irbm. 2020 doi: 10.1016/j.irbm.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimzadeh, Mohammad, and Abolfazl Attar. “A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2.” Informatics in Medicine Unlocked (2020): 100360. [DOI] [PMC free article] [PubMed]
- Oh Yujin, Park Sangjoon, Ye Jong Chul. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- Han Zhongyi, Wei Benzheng, Hong Yanfei, Li Tianyang, Cong Jinyu, Zhu Xue, Wei Haifeng, Zhang Wei. Accurate Screening of COVID-19 using Attention Based Deep 3D Multiple Instance Learning. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- Jamshidi, Mohammad, Ali Lalbakhsh, Jakub Talla, Zdeněk Peroutka, Farimah Hadjilooei, Pedram Lalbakhsh, Morteza Jamshidi et al. “Artificial intelligence and COVID-19: deep learning approaches for diagnosis and treatment.” IEEE Access 8 (2020): 109581-109595. [DOI] [PMC free article] [PubMed]
- Hu, Shaoping, Yuan Gao, Zhangming Niu, Yinghui Jiang, Lao Li, Xianglu Xiao, Minhao Wang et al. “Weakly supervised deep learning for covid-19 infection detection and classification from ct images.” IEEE Access 8 (2020): 118869-118883.
- Ardakani Ali Abbasian, Rajendra Acharya U., Habibollahi Sina, Mohammadi Afshin. COVIDiag: a clinical CAD system to diagnose COVID-19 pneumonia based on CT findings. Eur. Radiol. 2020:1–10. doi: 10.1007/s00330-020-07087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos, Ioannis D., and Tzani A. Mpesiana. “Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks.” Physical and Engineering Sciences in Medicine (2020): 1. [DOI] [PMC free article] [PubMed]
- Civit-Masot Javier, Luna-Perejón Francisco, Morales Manuel Domínguez, Civit Anton. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Applied Sciences. 2020;10(13):4640. [Google Scholar]
- Alghamdi Ahmed, Hammad Mohamed, Ugail Hassan, Abdel-Raheem Asmaa, Muhammad Khan, Khalifa Hany S., Abd Ahmed A., El-Latif. Detection of myocardial infarction based on novel deep transfer learning methods for urban healthcare in smart cities. Multimedia Tools and Applications. 2020:1–22. [Google Scholar]
- Amrani Moussa, Hammad Mohamed, Jiang Feng, Wang Kuanquan, Amrani Amel. Very deep feature extraction and fusion for arrhythmias detection. Neural Comput. Appl. 2018;30(7):2047–2057. [Google Scholar]
- Hammad Mohamed, Maher Asmaa, Wang Kuanquan, Jiang Feng, Amrani Moussa. Detection of abnormal heart conditions based on characteristics of ECG signals. Measurement. 2018;125:634–644. [Google Scholar]
- Abou-Nassar Eman M., Iliyasu Abdullah M., El-Kafrawy Passent M., Song Oh-Young, Bashir Ali Kashif, Abd El-Latif Ahmed A. DITrust chain: towards blockchain-based trust models for sustainable healthcare IoT systems. IEEE Access. 2020;8:111223–111238. [Google Scholar]
- Wang Ning, Li Qiong, Abd El-Latif Ahmed A., Zhang Tiejun, Niu Xiamu. Toward accurate localization and high recognition performance for noisy iris images. Multimedia Tools and Applications. 2014;71(3):1411–1430. [Google Scholar]
- Sedik, Ahmed; El-Hag, Noha A.; El-Banby, Ghada M.; Khalaf, Ashraf A. M.; Abd El-Rahiem, Basma; Abd El-Samie, Fathi; Abd El-Latif, Ahmed; Hammad, Mohamed (2020), “Noisy COVID-19 X-ray Dataset”, Mendeley Data, V3, doi: 10.17632/fjg5cbzffh.3.
- Ahmad Rasheed, Alsmadi Izzat, Alhamdani Wasim, Tawalbeh Lo'ai. A Comprehensive Deep Learning Benchmark for IoT IDS. Computers & Security. 2021;102588 [Google Scholar]
- Sedik A., et al. Deep Learning Modalities for Biometric Alteration Detection in 5G Networks-Based Secure Smart Cities. IEEE Access. 2021;9:94780–94788. doi: 10.1109/ACCESS.2021.3088341. [DOI] [Google Scholar]
- Siam A.I., Sedik A., El-Shafai W., Elazm A.A., El-Bahnasawy N.A., El Banby G.M., Abd El-Samie F.E. Biosignal classification for human identification based on convolutional neural networks. Int. J. Commun Syst. 2021;34(7) [Google Scholar]
- Evaluation Metrics for Binary Classificaiotn On line: Last accessed January 4th, 2022 https://neptune.ai/blog/evaluation-metrics-binary-classification.