Abstract
With the increase in international traffic, the risk of introducing rare but severe infectious diseases like Ebola hemorrhagic fever is increasing all over the world. However, the system for the diagnosis of Ebola virus infection is available in a limited number of countries. In the present study, we developed an Ebola virus antigen-detection enzyme-linked immunosorbent assay (ELISA) system using a novel monoclonal antibody (MAb) to the nucleoprotein (NP). This antibody recognized an epitope defined by a 26-amino-acid stretch near the C terminus of NP. In a sandwich ELISA system with the MAb, as little as 30 ng of purified recombinant NP (rNP) was detected. Although this MAb was prepared by immunization with rNP of subtype Zaire, it also reacted to the corresponding region of NP derived from the Reston and Sudan subtypes. These results suggest that our ELISA system should work with three of four Ebola subtypes. Furthermore, our ELISA system detected the NP in subtype Reston-infected monkey specimens, while the background level in noninfected specimens was very low, suggesting the usefulness of the ELISA for laboratory diagnosis with clinical specimens.
Ebola virus infection causes one of the most severe hemorrhagic fevers and has a high fatality rate (20). Although the region of endemicity of Ebola virus is limited, the risk of infection of humans and animals in other parts of the world is increasing with the increase in international traffic and transactions. Since Ebola virus causes secondary human-to-human infections among medical personnel and family members (2, 20), it is important to diagnose the infection at the early stage of an outbreak and to alert society.
On the basis of genetic divergence, four subtypes of Ebola viruses have been defined: subtypes Zaire, Sudan, Côte d'Ivoire, and Reston (3, 5, 14). The first three subtypes cause severe clinical symptoms in both humans and monkeys, while subtype Reston has caused disease only in monkeys (4, 10, 11). Ebola virus infection has an acute onset, and frequently, no antibody production is observed at the onset of clinical symptoms (1, 7). On the other hand, the virus load in patients' blood and tissues such as liver is extremely high (7). Therefore, quick and accurate primary screening for Ebola virus infection can be achieved by detection of the viral antigens rather than by detection of specific antibodies (14).
An antigen-detection system for Ebola virus infection was reported and successfully applied in the field (6). However, the information on that enzyme-linked immunosorbent assay (ELISA) is quite limited. For example, the monoclonal antibodies (MAbs) used in that system have not been reported even in terms of their molecular specificities. Moreover, the supply of that ELISA system is rather limited. For these reasons, we decided to establish another system for the detection of Ebola viral antigen. Toward this goal, we first established MAbs to a recombinant nucleoprotein (rNP) of Ebola virus subtype Zaire.
NP is one of the major viral structural components and consists of 739 amino acid (aa) residues. It is predicted that the hydrophobic N terminus of this protein may be involved in genomic RNA binding, while the hydrophilic and extremely acidic C terminus may be involved in the binding of other viral proteins, analogous to paramyxovirus (13, 17). We chose this molecule for the target of antigen detection because of the abundance of NP in Ebola virus particles and the availability of cDNA and sequence information. Here, we report on the successful development of an antigen-capture sandwich ELISA system with a novel NP-specific MAb which recognizes 26 aa residues on the C terminus of NP.
MATERIALS AND METHODS
Cell culture.
Hybridomas and their parental cell line, P3/Ag568, were maintained in RPMI 1640 (Gibco BRL, Rockville, Md.) supplemented with 10% fetal bovine serum, nonessential amino acids (Gibco BRL), and antibiotics (streptomycin and penicillin; Gibco BRL). Hypoxanthine-aminopterin-thymidine supplement (Gibco BRL) was added to the medium during the selection of hybridomas, as recommended by the supplier. Tn5 insect cells were maintained in TC100 (Gibco BRL) supplemented with 10% fetal bovine serum, 2% tryptose phosphate broth (Difco, Detroit, Mich.), and kanamycin.
Clinical specimens.
Tissues and sera from cynomolgus monkeys (Macaca fascicularis) naturally infected with Ebola virus subtype Reston in the Philippines were used as clinical specimens. These specimens had been kept either at −80°C or in liquid nitrogen since an outbreak in 1996 (12). The status of infection with subtype Reston in these animals was established previously (12). Liver and spleen tissues (approximately 10% [wt/vol]) were homogenized in 0.05% Tween 20, 1% Triton X-100, and 5% nonfat milk in phosphate-buffered saline (PBS). After centrifugation, the supernatants were used as the starting material. Sera were inactivated by addition of 1% Triton X-100 and were diluted in 5% nonfat milk. RNA from infected tissues was extracted with RNAzol B (TEL-TEST, Inc., Friendswood, Tex.) according to the instructions of the manufacturer.
rNP.
rNP of Ebola virus subtype Zaire was expressed by a baculovirus expression system with a histidine tag at the N terminus. Briefly, the entire open reading frame of the cDNA fragment derived from a subtype Zaire strain (provided by C. J. Peters) (17) was amplified by PCR with primers EBO(Z)NP/F and EBO(Z)NP/R (Table 1) and inserted into plasmid pQE31 (Qiagen, Hilden, Germany) which adds a histidine tag in frame at the N terminus of NP. The cDNA with the histidine tag was then cloned into the transfer vector, pAcYM1 (9), as a SalI-EcoRI fragment after a fill-up reaction with the Klenow enzyme. Recombinant baculoviruses were generated by cotransfection of the transfer plasmid and virus DNA. The rNP was purified from the Tn5 cell lysate infected with the recombinant baculovirus with Ni-agarose (Qiagen). The protein concentration was determined by using protein assay kits (Bio-Rad, Hercules, Calif.). Polypeptides that represent different parts of the NP were expressed as fusion proteins with a glutathione S-transferase (GST) tag in Escherichia coli with the pGEX2T vector (Amersham Pharmacia, Little Chalfont, United Kingdom) after PCR amplification (18). The primers used in the study are summarized in Table 1. To express the 26-aa peptides of the Sudan and Reston subtypes, primers SNP8EF and SNP8ER or primers RNP8EF and RNP8ER (each 3′ 15 bases are complementary to each other), respectively, were annealed and the 5′ overhang was blunted by Taq DNA polymerase. Then, their BamHI-EcoRI fragments were cloned into pGEX2T. For the longer peptide of the Sudan subtype, the fragment generated with SNP8EF and SNP8ER was gradually elongated by successive PCRs with primers SN8EF, SN8EF2+, SN8ER2+, SN8ER3+, SN8ER4+, SN8ER5+, and SN8ER6+. To obtain a partial cDNA encoding the 109 aa of the Reston subtype, randomly primed cDNA (Ready-To-Go RT-PCR beads; Amersham Pharmacia) from the clinical specimens was amplified by PCR with primers RES-N8F and RES-N8R. The nucleotide sequences were confirmed by using an automated sequencer (Applied Biosystems, Foster City, Calif.). The partial NP polypeptides were purified with glutathione Sepharose 4B, as recommended by the supplier (Amersham Pharmacia).
TABLE 1.
PCR primers used in the present study
| Primer | Sequence (5′ to 3′) | NP region amplified (subtype) |
|---|---|---|
| EBO(Z)NP/F | CAAGGATCCGAGTATGGATTCTCG | From aa 1 (Zaire) |
| EBO(Z)NP/R | ATGGATCCATGCTCATTCACTGAT | To aa 739 (Zaire) |
| EBO NP8F | CCGGGATCCACTCAAGAGGCCA | From aa 631 (Zaire) |
| EBO NP8R | TGCGAATTCACTGATGATGTTGC | To aa 739 (Zaire) |
| EN8/2-1R | AAAAGAATTCACTCATCCTTCATCA | To aa 674 (Zaire) |
| EN8/1.5R | CTAGAATTCAATCCTTCATCATATGATAAT | To aa 673 (Zaire) |
| EN8/2-2R | CAGGAATTCACTTCATCATATGATAATAC | To aa 672 (Zaire) |
| EN8/2.5R | CTGAATTCACATCATATGATAATACAAAAC | To aa 671 (Zaire) |
| EN8/2-14F | GGATCCATGTATCGCCACATTC | From aa 651 (Zaire) |
| EN8/2-15F | GGATCCGAGATGTATCGCCAC | From aa 650 (Zaire) |
| EN8/2-16F | GGATCCGAGGAGATGTATCGC | From aa 649 (Zaire) |
| EN8/2-17F | GGATCCTTTGAGGAGATG | From aa 648 (Zaire) |
| SNP8EF | CGCGGATCCCTAGAAGAAACATATTATCATCTCCTAAAAACACAGGGTCCATTTG | From aa 648 (Sudan) |
| SNP8ER | GCTGAATTCAATCACTCATTAGGTGATAATAATTGATTGCCTCAAATGGACCCTGTG | To aa 673 (Sudan) |
| RNP8EF | CGCGGATCCTTAGGAGAGACATATCACCATCTGCTGAGAACTCAAGGTCCATTTG | From aa 648 (Reston) |
| RNP8ER | GCTGAATTCAATCCTTCATCATGTGATAATAATTGATAGCTTCAAATGGACCTTGAG | To aa 673 (Reston) |
| SN8EF+ | GGATCCCTCCCAATCAACTCTAAAAAGAGTTCCGCACTAGAAGAAACATATTATCATC | From aa 638 (Sudan) |
| SN8ER+ | GAATTCAGCCACTTTCAGTGCTAAAAGCAATGGGTTCATCACTCATTAGGTGATAATAATTG | To aa 683 (Sudan) |
| SN8EF2+ | GGATCCTCTGAAAGTGAAGCTCTCCCAATCAACTCTA | From aa 633 (Sudan) |
| SN8ER2+ | GAATTCTCAAAGATATATTCCTTGCCACTTTCAGTGCTAA | To aa 688 (Sudan) |
| SN8ER3+ | GAATTCATTCAAGGGAGTCTGGAAAGATATATTCCTTGCC | To aa 693 (Sudan) |
| SN8ER4+ | GAATTCACTTCTCTAAGGCCTCCTTCTCACTCAACCACGGCGGGTAGGCTTCTTCAAGGGAGTCTGGAAAGAT | To aa 708 (Sudan) |
| SN8ER5+ | GAATTCATACCGGCCAGAGGAATTGCTGGCCATCAATGACCAGATAACGATTTTCCTTCTCTAAGGCCTCCTT | To aa 724 (Sudan) |
| SN8ER6+ | GAATTCAGTCATGTTGAAGAACGGCAAGGAACTTGTCCCGTAGGCTCATTACCGGCCAGAGGAATTG | To aa 738 (Sudan) |
| RES-N8F | GCTGGATCCTCACAATTGAATGAAGACC | From aa 631 (Reston) |
| RES-N8R | GTGGAATTCTTACTGATGGTGCTGCAA | To aa 739 (Reston) |
ELISA.
Each well of microwell immunoplates (Falcon, Franklin Lakes, N.J.) were coated with 100 ng of purified rNP or partial NP in PBS at 4°C overnight, followed by blocking with 5% nonfat milk. Sample (100 μl) was added to each well, and bound antibody was detected with horseradish peroxidase (HRPO)-labeled anti-mouse immunoglobulin G (IgG; Zymed, San Francisco, Calif.) and 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) substrate solution (Boehringer, Mannheim, Germany).
Establishment of MAbs.
BALB/c mice were immunized three or four times with the purified rNP. The spleen cells were obtained 3 days after the last immunization and were fused with P3/Ag568 cells by using polyethylene glycol (Gibco BRL). The culture supernatants of hybridoma cells were screened by ELISA with purified rNP as the antigen. MAbs were purified from the culture supernatants with an MAbTrap GII antibody purification kit, as instructed by the manufacturer (Amersham Pharmacia). The isotypes of the MAbs were determined with a Mouse Monoclonal Antibody Isotyping kit (Gibco BRL).
Polyclonal antibody.
Rabbit polyclonal antibody was prepared by subcutaneous injection of purified rNP emulsified in aluminum adjuvant (Pierce, Rockford, Ill.). The titer of this antiserum was >1:10,000 in an immunofluorescence assay in which rNP-expressing cells were used as antigen (16).
Antigen-capture ELISA.
Purified MAb was coated on microwell immunoplates (Falcon) at a concentration of 100 ng/well in 100 μl of PBS for 2 h at room temperature (RT), followed by blocking with 5% nonfat milk in PBS for 1 h at RT. After the plates were washed with PBS containing 0.05% Tween 20 (PBST), 100-μl samples containing rNP or clinical specimens were added and the plates were incubated for 1 h at RT. The plates were then washed with PBST, and 100 μl of rabbit polyclonal antibody diluted 1:1,000 in 0.5% nonfat milk was added to each well. After 1 h of incubation at RT, the plates were again washed with PBST and HRPO-labeled anti-rabbit IgG (Zymed) was added. The plates were incubated for 1 h at RT. After another extensive wash with PBST, 100 μl of ABTS substrate solution (Boehringer) was added and the optical density (OD) was measured at 405 nm (OD405) after 30 min of incubation at RT.
Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses were performed with a 10% acrylamide gel and an Immobilon nylon membrane (Millipore, Bedford, Mass.), respectively. Ascitic fluid containing MAb was used at a dilution of 1:1,000. The bound antibody was detected with HRPO-labeled anti-mouse IgG (Zymed) and peroxydase substrate (Wako Pure Chemical, Tokyo, Japan). A polyclonal antibody to GST was purchased from Amersham Pharmacia.
RESULTS
Development of MAbs.
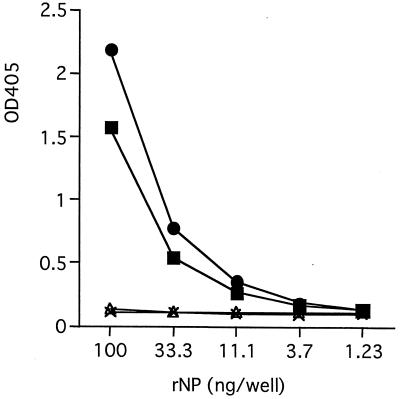
Twelve hybridoma clones secreting rNP-reactive IgG antibodies were established. These antibodies were examined in the antigen-capture ELISA after purification. As shown in Fig. 1, two MAbs, MAbs 3-3D and 2-11G, were reactive in this format. None of the rest of the MAbs showed any specific reaction even at higher concentrations of rNP (up to 2,700 ng/well), as represented by MAbs 2-1G and 3-7D (Fig. 1 and data not shown). The isotypes of MAbs 3-3D and 2-11G were identified as IgG1.
FIG. 1.
Reactivity of each MAb in the antigen-capture ELISA format. Purified MAbs (●, MAb 3-3D; ▪, MAb 2-11G; ▵, MAb 2-1G; ×, MAb 3-7D) were coated onto the microplates at a concentration of 100 ng/well, and their ability to capture rNP was examined with various amounts of rNP in the antigen-capture ELISA format.
Antigen-capture ELISA.
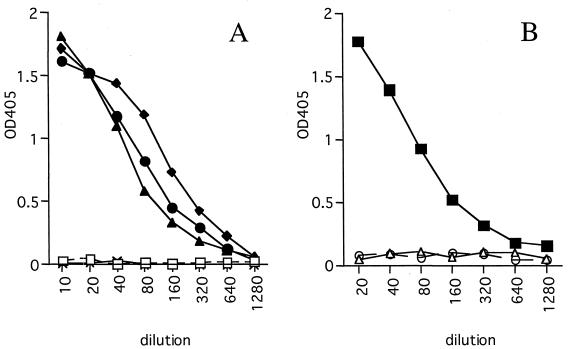
The sensitivity of the antigen-capture ELISA with MAbs 3-3D and 2-11G was examined. The minimal amount of rNP detected in this ELISA with either MAb was approximately 33 ng/well (Fig. 1). To confirm the reactivities of these MAbs with authentic NP, clinical specimens from monkeys infected with Ebola virus of the Reston subtype were used. Although the rNP used to develop the MAbs was derived from subtype Zaire, only subtype Reston-infected clinical specimens were available to us. As shown in Fig. 2, antigen-capture ELISA with MAb 3-3D detected Ebola virus NP in Reston subtype-positive tissue (Fig. 2A) and serum (Fig. 2B) samples. Two liver samples from two animals showed a positive reaction (OD405, >0.3) up to dilutions of 1:160 or 1:320. A spleen sample showed a positive result up to a dilution of 1:160, and a serum sample was positive up to a dilution of 1:320. All the similarly processed samples from uninfected monkeys showed negative results (OD405, <0.1), even at the lowest dilution examined (1:10 for tissue samples and 1:20 for serum samples). Positive controls consisting of rNP run concomitantly in this assay showed positive results (OD405, >0.3) at concentrations of 125 ng/well or higher. The other MAb, MAb 2-11G, failed to react with Reston subtype-infected samples (data not shown). These results indicate that the antigen-capture ELISA system with MAb 3-3D is highly sensitive and can be applicable to clinical specimens derived from Ebola virus subtype Reston-infected monkeys.
FIG. 2.
Reactivities of Reston subtype-infected clinical specimens in the antigen-capture ELISA. (A) Two livers (⧫ and ●) and a spleen (▴) from two infected monkeys were examined in the antigen-capture ELISA. A liver (×) and a spleen (□) from an uninfected monkey were used to show the specificity of the reaction. (B) Sera from an infected (▪) and two uninfected (○ and ▵) monkeys were examined by the antigen-capture ELISA.
Epitope mapping of capture MAb 3-3D.
Since only authentic Reston subtype NP and rNP of subtype Zaire were available to us, it was unclear if this antigen-capture system works with NPs of other subtypes of Ebola virus. To address this question, we attempted to map the epitope recognized by MAb 3-3D. Eight overlapping peptides, each of which consisted of approximately 100 aa, were prepared to cover entire the amino acid sequence of the Zaire subtype NP. MAb 3-3D was reactive with the peptide corresponding to aa 631 to 739 by both Western blotting and ELISA. Pepscan analysis using overlapping 10-aa peptides (10 aa with 9 aa overlaps) derived from this 109-aa region was performed but failed to show a specific reaction (data not shown), suggesting that the epitope is not linear. To define the minimum epitope, we first determined the amino acid residue at the C terminus required for recognition. As shown in Fig. 3A, aa 673 was required to show full reactivity by Western blotting. With aa 673 at the C terminus, the upstream 26 aa was required to show full reactivity, while weaker reactivity was observed with a peptide as short as 23 aa (Fig. 3B). These peptides, however, did not react with MAb 3-3D in the ELISA, with or without the GST tag (data not shown). These results suggest that MAb 3-3D recognizes a conformational epitope formed by a short stretch of 26 aa between aa 648 and 673 on NP.
FIG. 3.
Epitope mapping of MAb 3-3D by Western blotting. (A) Bacterial lysates containing GST-fusion peptides corresponding to aa positions 631 to 671, 672, 673, or 674 of NP (subtype Zaire) were examined for their reactivities to MAb 3-3D. (B) Bacterial lysates containing GST-fusion peptides corresponding to aa positions 648, 649, 650, or 651 to 673 of NP (subtype Zaire) were examined for their reactivities to MAb 3-3D. To show the similar levels of expression of each fusion peptide, a duplicate membrane was stained with anti-GST antibody.
Cross-reactivity of MAb 3-3D with other subtypes of Ebola virus.
We synthesized oligonucleotides and expressed GST-fusion peptides corresponding to the 26-aa epitope from subtypes Sudan and Reston. The cross-reactivity of MAb 3-3D to these two peptides was examined by Western blotting. Unexpectedly, MAb 3-3D did not react with either peptide (data not shown), despite the cross-reactivity to the Reston subtype in clinical specimens that was found. When longer peptides consisting of 109 aa (subtypes Zaire and Reston) or 106 aa (subtype Sudan) residues at the C termini were examined, MAb 3-3D reacted to these three peptides by both Western blotting (data not shown) and ELISA (Fig. 4). These results suggest that the ELISA with MAb 3-3D detects NP of subtype Sudan as well as those of subtypes Zaire and Reston.
FIG. 4.
Reactivity of MAb 3-3D to the epitope region on NPs from three subtypes of Ebola virus. GST-fusion peptides (100 ng/well) corresponding to aa positions 631 to 739 of the Zaire (▪) and Reston (●) subtypes or aa 633 to 738 of the Sudan (▴) subtype were coated onto microplates and examined for their reactivities with MAb 3-3D by ELISA. An irrelevant GST-fusion peptide (◊) was used as a negative control.
DISCUSSION
We have developed an antigen-detection ELISA for Ebola virus using a novel MAb to NP. Although there was a reported antigen-detection ELISA system for Ebola virus, the information on that system is limited (6). Our new system detected Ebola virus subtype Reston antigen in liver, spleen, and serum specimens from naturally infected monkeys. The background levels in samples from uninfected monkeys were remarkably low. The sensitivity of this system was determined to be as low as 30 ng/100 μl when purified rNP was used. The results indicate that this system is useful in practice, at least for the detection of Reston subtype infection in monkeys. Furthermore, it is highly probable that this system detects Ebola virus subtype Zaire infection as well, since the rNP from subtype Zaire was used to prepare antibodies. We also showed that capture MAb 3-3D was cross-reactive with Sudan subtype-derived peptides in the ELISA, suggesting cross-reaction of this system with the Sudan subtype. We could not examine whether the ELISA detects the forth subtype, subtype Côte d'Ivoire (8), since neither clinical materials nor NP sequence data for this subtype were available to us. From a comparison of the glycoprotein genes of the four subtypes of Ebola virus, subtype Côte d'Ivoire is genetically more closely related to subtype Zaire than to the other two subtypes (3, 5). Combined with the results showing cross-reaction of MAb 3-3D with the other two subtypes, this ELISA system may work with subtype Côte d'Ivoire as well, although further studies with authentic NP including that of subtype Côte d'Ivoire are needed to determine the cross-reactivity.
Of particular interest, MAb 3-3D required longer peptides for recognition of heterologous subtypes. Even with the homologous Zaire subtype, the peptide required for reactivity with MAb 3-3D was shorter in Western blotting than in ELISA. The reason for this apparent discrepancy is not clear. Steric interference by fused GST was not involved, since the cleavage of GST did not affect the results. A possible explanation may be that the 26-aa peptide from subtype Zaire formed an adequate epitope structure due to denaturation and renaturation during SDS-PAGE and Western blotting, while the peptides from subtypes Reston and Sudan did not.
One of the advantages of our new ELISA system is that the highest level of security containment is not required, since no live virus except that in the clinical specimens is involved. This means that an ordinarily equipped laboratory can reproduce the system as long as the secondary polyclonal antibody, which can also be prepared by use of noninfectious recombinant protein, is available. The disadvantage is that it uses only one MAb that recognizes at least three of four subtypes of Ebola virus. Although Ebola virus is genetically stable (15, 19) and so far only four subtypes have been reported, there is no guarantee that this MAb cross-reacts with all the minor variants that might appear in the future. To minimize such a risk, it may be necessary to use a mixture of MAbs with different specificities.
ACKNOWLEDGMENTS
We thank C. J. Peters for providing NP cDNA from a clone of Ebola virus subtype Zaire. We also thank staff members of RITM of the Philippines for assistance.
This work was supported by grants from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Baize S, Leroy E M, Georges-Courbot M C, Capron M, Lansoud-Soukate J, Debre P, Fisher-Hoch S P, McCormick J B, Georges A J. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 2.Dowell S F, Mukunu R, Ksiazek T G, Khan A S, Rollin P E, Peters C J. Transmission of Ebola hemorrhagic fever: a study of risk factors in family members, Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179(Suppl. 1):S87–S91. doi: 10.1086/514284. [DOI] [PubMed] [Google Scholar]
- 3.Feldmann H, Kiley M P. Classification, structure, and replication of filoviruses. Curr Top Microbiol Immunol. 1999;235:1–5. doi: 10.1007/978-3-642-59949-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Fisher-Hoch S P, Brammer T L, Trappier S G, Hutwagner L C, Farrar B B, Ruo S L, Brown B G, Hermann L M, Perez-Oronoz G I, Goldsmith C S, Hanes M A, McCormic J B. Pathogenic potential of filoviruses: role of geographic origin of primate host and virus strain. J Infect Dis. 1992;166:753–763. doi: 10.1093/infdis/166.4.753. [DOI] [PubMed] [Google Scholar]
- 5.Georges-Courbot M C, Sanchez A, Lu C Y, Baize S, Leroy E, Lansout-Soukate J, Tevi-Benissan C, Georges A J, Trappier S G, Zaki S R, Swanepoel R, Leman P A, Rollin P E, Peters C J, Nichol S T, Ksiazek T G. Isolation and phylogenetic characterization of Ebola viruses causing different outbreaks in Gabon. Emerg Infect Dis. 1997;3:59–62. doi: 10.3201/eid0301.970107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek T G, Rollin P E, Jahrling P B, Johnson E, Dalgard D W, Peters C J. Enzyme immunosorbent assay for Ebola virus antigens in tissues of infected primates. J Clin Microbiol. 1992;30:947–950. doi: 10.1128/jcm.30.4.947-950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ksiazek T G, Rollin P E, Williams A J, Bressler D S, Martin M L, Swanepoel R, Burt F J, Leman P A, Khan A S, Rowe A K, Mukunu R, Sanchez A, Peters C J. Clinical virology of Ebola hemorrhagic fever (EHF): virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179(Suppl. 1):S177–S187. doi: 10.1086/514321. [DOI] [PubMed] [Google Scholar]
- 8.Le Guenno B, Formentry P, Wyers M, Gounon P, Walker F, Boesch C. Isolation and partial characterisation of a new strain of Ebola virus. Lancet. 1995;345:1271–1274. doi: 10.1016/s0140-6736(95)90925-7. [DOI] [PubMed] [Google Scholar]
- 9.Matsuura Y, Possee R D, Overton H A, Bishop D H L. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J Gen Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 10.Miller R K, Baumgardner J Y, Armstrong C W, Jenkins S R, Woolard C D, Miller G B, Jr, Polk L D, Tavris D R, Hendricks K A, Taylor J P, Simpson D M, Schultz S, Sturman L, Debbie J G, Morse D L, Rollin P E, Jahrling P B, Ksiazek T G, Peters C J. Filovirus infections among persons with occupational exposure to nonhuman primates. Morb Mortal Wkly Rep. 1990;39:266–267. , 273. [PubMed] [Google Scholar]
- 11.Miller R K, Baumgardner J Y, Armstrong C W, Jenkins S R, Woolard C D, Miller G B, Jr, Rollin P E, Jahrling P B, Ksiazek T G, Peters C J. Filovirus infections in animal handlers. Morb Mortal Wkly Rep. 1990;39:221. [Google Scholar]
- 12.Miranda M E, Ksiazek T G, Retuya T J, Khan A S, Sanchez A, Fulhorst C F, Rollin P E, Calaor A B, Manalo D L, Roces M C, Dayrit M M, Peters C J. Epidemiology of Ebola (subtype Reston) virus in the Philippines, 1996. J Infect Dis. 1999;179(Suppl. 1):S115–S119. doi: 10.1086/514314. [DOI] [PubMed] [Google Scholar]
- 13.Peters C J, Sanchez A, Rollin P E, Ksiazek T G, Murphy F A. Filoviridae: Marburg and Ebola viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1161–1176. [Google Scholar]
- 14.Peters C J, LeDuc J W. An introduction to Ebola: the virus and the disease. J Infect Dis. 1999;179(Suppl. 1):iv–xvi. doi: 10.1086/514322. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez L L, De Roo A, Guimard Y, Trappier S G, Sanchez A, Bressler D, Williams A J, Rowe A K, Bertolli J, Khan A S, Ksiazek T G, Peters C J, Nichol S T. Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179(Suppl. 1):S170–S176. doi: 10.1086/514291. [DOI] [PubMed] [Google Scholar]
- 16.Saijo M, Niikura M, Morikawa S, Meyer R, Peters C J, Ksiazek T G, Kurane I. Enzyme-linked immunosorbent assays for detection of antibodies to Ebola and Marburg viruses using recombinant nucleoproteins. J Clin Microbiol. 2001;39:1–7. doi: 10.1128/JCM.39.1.1-7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saijo M, Niikura M, Morikawa S, Kurane I. Immunofluorescence method for detection of Ebola virus immunoglobulin G, using HeLa cells which express recombinant nucleoprotein. J Clin Microbiol. 2001;39:776–778. doi: 10.1128/JCM.39.2.776-778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez A, Kiley M P, Holloway B P, McCormick J B, Auperin D D. The nucleoprotein gene of Ebola virus: cloning, sequencing, and in vitro expression. Virology. 1989;170:81–91. doi: 10.1016/0042-6822(89)90354-1. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez A, Ksiazek T G, Rollin P E, Miranda M E, Trappier S G, Khan A S, Peters C J, Nichol S T. Detection and molecular characterization of Ebola viruses causing disease in human and nonhuman primates. J Infect Dis. 1999;179(Suppl. 1):S164–S169. doi: 10.1086/514282. [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization. World Health Organization recommended guidelines for epidemic preparedness and response: Ebola hemorrhagic fever (EHF). Geneva, Switzerland: World Health Organization; 1997. Part one: Background information on the organisms and the disease; pp. 1–3. [Google Scholar]