ABSTRACT.
Plasmodium falciparum and Plasmodium vivax are co-endemic in Ethiopia. This study investigated whether mixed infections were missed by microscopy from a 2017 therapeutic efficacy study at two health facilities in Ethiopia. All patients (N = 304) were initially classified as having single-species P. falciparum (n = 148 samples) or P. vivax infections (n = 156). Dried blood spots were tested for Plasmodium antigens by bead-based multiplex assay for pan-Plasmodium aldolase, pan-Plasmodium lactate dehydrogenase, P. vivax lactate dehydrogenase, and histidine-rich protein 2. Of 304 blood samples, 13 (4.3%) contained both P. falciparum and P. vivax antigens and were analyzed by polymerase chain reaction for species-specific DNA. Of these 13 samples, five were confirmed by polymerase chain reaction for P. falciparum/P. vivax co-infection. One sample, initially classified as P. vivax by microscopy, was found to only have Plasmodium ovale DNA. Plasmodium falciparum/P. vivax mixed infections can be missed by microscopy even in the context of a therapeutic efficacy study with multiple trained readers.
Ethiopia is one of the few African countries where both Plasmodium falciparum and Plasmodium vivax species are co-endemic at substantial proportions, and are accounted for in malaria diagnostic and treatment guidelines.1 Plasmodium falciparum accounts for ≈60% of all cases; P. vivax accounts for most of the remaining cases in Ethiopia.2 To prevent morbidity and mortality resulting from malaria, the Ethiopian Ministry of Health aims to achieve malaria elimination by 2030.3 To facilitate malaria elimination and prevent severe disease, accurate diagnosis and effective treatment of all malaria cases are essential. The Ministry of Health requires malaria diagnosis by either microscopy or rapid diagnostic test (RDT).3 Microscopy allows for laboratory technicians to distinguish between different malaria species; however, the accuracy is limited by the sample’s parasite density and the technician’s expertise.4 The detection limit of microscopy and RDTs is generally considered to be ≈100 parasites/µL.5
Persons may become infected with multiple malaria parasites simultaneously.6 However, mixed infections are likely underreported because they are often difficult to detect by microscopy and unable to be detected by many RDTs.4,7 Ayalew et al.4 reported that only about 45% of microscopists from Ethiopian hospitals and health centers accurately identified a P. falciparum/P. vivax mixed infection. RDTs detecting histidine-rich protein 2 (HRP2) and/or pan-Plasmodium lactate dehydrogenase antigens are unable to distinguish between a P. falciparum and P. falciparum/P. vivax mixed infection.7 Plasmodium falciparum/P. vivax combination-test RDTs are available that can detect HRP2 and P. vivax-specific LDH (PvLDH) and therefore can identify P. falciparum/P. vivax mixed infections; however, they cannot detect P. malariae or P. ovale. Accurate diagnosis of multiple malaria species is important for effective treatment and public health surveillance in a region.
Tests more sensitive than microscopy and RDTs are available for detecting Plasmodium infection, including polymerase chain reaction (PCR) and bead-based multiplex antigen detection assays, but these methods are limited to laboratory settings because they require specialized equipment and skilled technicians.8 The bead-based antigen assay simultaneously detects multiple Plasmodium antigens from a blood sample and provides sensitivity comparable to PCR.9 To identify malaria mixed infections, samples from a 2017 therapeutic efficacy study (TES) in Ethiopia were analyzed using a bead-based multiplex antigen detection assay and PCR to assess samples initially classified as single-species P. falciparum or P. vivax infections by microscopy. Persons presenting with symptoms of malaria and diagnosed via microscopy with a parasite density of 500 to 100,000 asexual parasites/μL of blood for P. falciparum or more than 250 asexual parasites/μL of blood for P. vivax were eligible for the study. Only single-species P. falciparum or P. vivax infections were eligible for inclusion. RDTs (P. falciparum/pan-Plasmodium) were also conducted at the time of enrollment, but the results did not affect enrollment procedures. Treatment of malaria was provided according to the study protocol with artemether–lumefantrine or dihydroartemisinin–piperaquine for P. falciparum infection and chloroquine or dihydroartemisinin–piperaquine for P. vivax infection. Individuals presenting with P. vivax and testing normal for glucose-6-phosphate dehydrogenase were offered primaquine radical cure at the end of the 42-day follow-up period. The TES protocol was approved by the Ethiopian Public Health Institute, the National Ethical Committee (3.10/171/2016) and the Food, Medicine and Health Care Administration and Control Authority in Ethiopia (02/6/9-1/81). In addition, the study was reviewed by Columbia University (AAAQ9414) and the CDC Human Subjects office (no. 6892.0) and was conducted consistent with applicable U.S. federal law and CDC policy.
Two study sites were included in the TES: Pawe (Benishangul Gumuz region) and Arba Minch (Southern Nations, Nationalities, and Peoples’ region). For samples with dried blood spots (DBS) available for laboratory evaluation (N = 304), microscopy diagnosis identified 148 with single-species P. falciparum (136 from Pawe and 12 from Arba Minch) and 156 samples with single-species P. vivax infections (60 from Pawe and 96 from Arba Minch). Slides were read by a study site microscopist and then re-examined blinded by a WHO-certified level-1 and level-2 microscopist.
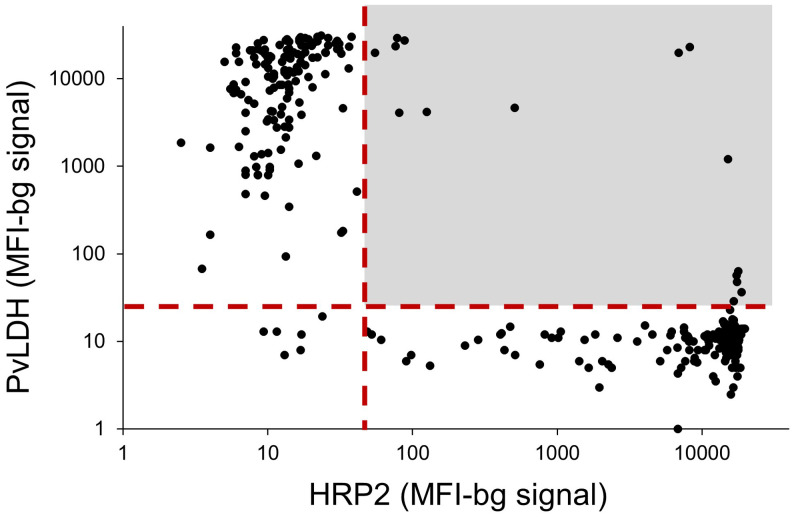
All 304 DBS samples were analyzed by bead-based multiplex antigen assay for pan-Plasmodium aldolase, pan-Plasmodium lactate dehydrogenase, PvLDH, and HRP2. The PvLDH and HRP2 antigens are species-specific markers for P. vivax and P. falciparum, respectively. From the multiplex antigen screening of all samples, the majority showed species-specific antigen concordance with the participants’ microscopy results: 145/148 P. falciparum infections (98.0%) and 153/156 P. vivax infections (98.1%). However, 15 of the 304 samples (4.9%) showed the presence of both species-specific antigens, nine of which were identified previously as P. falciparum and six as P. vivax by microscopy. A scatterplot for these two species-specific antigen levels and designation of the double positives is shown in Figure 1.
Figure 1.
Scatterplot of Plasmodium falciparum histidine-rich protein 2 (HRP2) assay signal compared with Plasmodium vivax lactate dehydrogenase (PvLDH) assay signal for 304 dried blood spot samples. The vertical dashed red line designates the threshold for the antigen positivity signal for the HRP2 antigen; the horizontal dashed red line designates the threshold for the antigen positivity signal for PvLDH. The lightly shaded region denotes values where dried blood spot samples were positive for both species-specific antigens. MFI-bg = median fluorescence intensity minus background fluorescence. This figure appears in color at www.ajtmh.org.
Of the 15 blood samples containing both species-specific antigens,13 had enough blood remaining for DNA extraction for PCR assays to investigate potential mixed-species infections (Table 1). One additional sample identified as containing P. vivax by microscopy but PvLDH negative and HRP2 positive was also subjected to PCR testing. For these 14 samples, DNA was extracted from a single 6-mm DBS punch using the Qiagen DNA Mini Kit (QIAGEN, Germantown, MD), and photo-induced electron transfer–PCR with species-specific primers was performed as described previously.10 Of the seven microscopy-positive P. falciparum samples with DBS positive for PvLDH (and HRP2), all (100%) were also found to have P. vivax DNA. Of the six microscopy-positive P. vivax samples with DBS positive for HRP2 (and PvLDH), one (16.7%) was found to contain P. falciparum and P. vivax DNA. The sample initially identified with P. vivax infection by microscopy but PvLDH negative was found to contain P. ovale DNA, but neither P. falciparum nor P. vivax DNA. Plasmodium ovale is likely underdiagnosed in Ethiopia because the estimated seroprevalence for P. ovale is 3.1%.11 From the combination of antigen and DNA data, of the 304 clinical samples assessed, five (1.6%) were found to be P. falciparum/P. vivax mixed infections and one (0.3%) was a P. ovale infection. Most of the mixed infections (80%) were identified as P. falciparum by microscopy. Three samples identified as P. falciparum by microscopy and RDT, but noted to contain P. vivax DNA only, were likely P. vivax mono-infections in the setting of a recently cleared P. falciparum infection. Plasmodium falciparum diagnosis by microscopy was less accurate compared with P. vivax diagnosis as three of seven P. falciparum diagnoses were missing P. falciparum DNA compared with one of seven P. vivax diagnoses missing P. vivax DNA. Table 1 shows the assay results, study sites, demographics, and clinical information for these 14 samples.
Table 1.
Results from microscopy, antigen detection, and polymerase chain reaction assays for samples selected with suspicion of Plasmodium falciparum/Plasmodium vivax mixed infections
| Sample no. | Microscopy diagnosis | Study site | Treatment provided | Parasite density by microscopy (parasites/μL blood) | RDT | Gender | Age (y) | Hgb (g/dL) | HRP2 (pg/mL) | PvLDH (pg/mL) | PET-PCR Pf Ct* | PET-PCR Pv Ct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Pf | Pawe | AL | 26,820 | Pf | M | 11 | 10.8 | 1,881,181 | 871 | 26.9 | 36.6 |
| 2 | Pf | Pawe | AL | 17,480 | Pf | M | 17 | 10.1 | 1,118,668 | 655 | 28.2 | 39.9 |
| 3 | Pf | Pawe | AL | 6,834 | Pf | F | 16 | 15.8 | 1,162 | 54,941 | – | 29.4 |
| 4 | Pf | Pawe | AL | 18,246 | Pf | M | 17 | 14.0 | 106,889 | 14,333 | 24.9 | 35.8 |
| 5 | Pf | Pawe | AL | 78,238 | Pf | F | 11 | 12.4 | 913,114 | 778 | 23.3 | 39.3 |
| 6 | Pf | Pawe | AL | 8,142 | Pf | M | 20 | 12.9 | 166 | 47,572 | – | 30.0 |
| 7 | Pf | Arba Minch | AL | 2,480 | Pf | F | 19 | 13.7 | 28,354 | 25,353 | 44.4 | 27.7 |
| 8 | Pv | Pawe | CQ | 22,246 | Pv | F | 5 | 11.4 | 113 | 824,001 | 39.0 | 28.3 |
| 9 | Pv | Pawe | CQ | 16,038 | Pv | M | 15 | 14.8 | 160 | 27,616 | 41.4 | 27.4 |
| 10† | Pv | Pawe | CQ | 5,200 | Neg† | M | 15 | 14.1 | 100 | 0.0† | – | – |
| 11 | Pv | Pawe | CQ | 9,595 | Pv | F | 9 | 12.4 | 260 | 48,688 | – | 28.4 |
| 12 | Pv | Arba Minch | CQ | 48,320 | Pv | M | 20 | 16.1 | 156 | 24,225 | – | 27.7 |
| 13 | Pv | Arba Minch | DP | 27,200 | Pv | M | 23 | 12.8 | 22,272 | 830,205 | – | 27.5 |
| 4 | Pv | Arba Minch | CQ | 11,720 | Pv | M | 3 | 12.6 | 181 | 26,483 | 44.2 | 26.7 |
AL = artemether lumefantrine; CQ = chloroquine; Ct = cycle threshold value; DP = dihydroartemisinin–piperaquine; F = female; Hgb = hemoglobin; HRP2 = histidine-rich protein 2; M = male; PET-PCR = photo-induced electron transfer polymerase chain reaction; Pf = Plasmodium falciparum; Pv = Plasmodium vivax; PvLDH = P. vivax lactate dehydrogenase; RDT = rapid diagnostic test.
A Ct value less than 40.0 is considered DNA positive; a negative sign indicates no amplification
Sample had a negative assay signal for PvLDH; found to contain Plasmodium ovale DNA.
Seven samples identified initially as P. falciparum infections by microscopy were later found to contain PvLDH antigens and P. vivax DNA. Because LDH clears from circulation quickly after parasite clearance,12 the concomitant PvLDH antigenemia and P. vivax DNA are not surprising. However, of the six samples with potential P. falciparum/P. vivax mixed infection based on a P. vivax microscopy result and the presence of HRP2 antigens, only one contained P. falciparum DNA. This could represent a newly acquired P. falciparum infection without a high-enough parasitemia to be identified easily by microscopy. In addition, the one sample with P. ovale DNA also contained HRP2 antigens. Unlike DNA, HRP2 can persist in the circulation for months after a P. falciparum infection and may be detected coincidentally during an ongoing or recent P. vivax (or P. ovale) infection.13,14 In some cases, there may be sufficient antigen levels in a DBS sample to be detected, but not enough DNA to be detected by PCR.15 The bead-based antigen assay provides a more efficient method to screen samples for potential mixed infection compared with testing all samples using PCR, which is more expensive and time-consuming.
In different areas of Ethiopia, the proportion of P. falciparum/P. vivax mixed infections is estimated to be between 0.5% to 5.0% of malaria cases.16,17 The value reported here (1.6%) is likely an underestimate because mixed infections were excluded from the TES. Missing mixed infections by microscopy is consistent with findings in other settings.18,19 Mixed infections may be missed by microscopic examination of blood films with low parasite density of one species or difficulty differentiating the lower density species from the dominant species.20 In either case, refresher training with a focus on identifying P. falciparum/P. vivax mixed infections would be beneficial. However, often, a highly sensitive method, such as PCR, is needed to uncover mixed infections.19
In areas where P. falciparum and P. vivax are endemic, uncovering P. falciparum/P. vivax mixed infections is important for the success of malaria control programs, elimination efforts, and effective clinical treatment. The recommended treatment of P. falciparum/P. vivax mixed infections in Ethiopia is artemether–lumefantrine plus radical cure with primaquine (0.25 mg/kg for 14 days).3 For P. falciparum, the recommended treatment is artemether–lumefantrine and single-dose primaquine (0.25 mg/kg); whereas for P. vivax, chloroquine and radical cure with primaquine (0.25 mg/kg for 14 days) is recommended.1 If a mixed infection is misdiagnosed as a P. falciparum or P. vivax mono-infection, then the given treatment would be inadequate. Although microscopy is the gold standard, missing mixed infections may lead to treatments that would fail to clear the presenting blood-stage infection or would miss clearing hypnozoites, either of which could impede malaria elimination efforts.
ACKNOWLEDGMENTS
We sincerely thank the study participants, data collectors, and laboratory technicians who assisted in the therapeutic efficacy study.
REFERENCES
- 1. Ethiopia Ministry of Health , 2017. National Malaria Guidelines, 4th edition. Addis Ababa, Ethiopia: Ethiopian Federal Ministry of Health.
- 2. Taffese HS Hemming-Schroeder E Koepfli C Tesfaye G Lee MC Kazura J Yan GY Zhou GF , 2018. Malaria epidemiology and interventions in Ethiopia from 2001 to 2016. Infect Dis Poverty 7: 1--9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ethiopian Ministry of Health , 2017. National Malaria Elimination Road Map. Addis Ababa, Ethiopia: Federal Ministry of Health. [Google Scholar]
- 4. Ayalew F Tilahun B Taye B , 2014. Performance evaluation of laboratory professionals on malaria microscopy in Hawassa Town, southern Ethiopia. BMC Res Notes 7: 1--8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization , 2013. WHO Evidence Review Group on Malaria Diagnosis in Low Transmission Settings. Available at: https://www.who.int/malaria/mpac/mpac_mar2014_diagnosis_low_transmission_settings_report.pdf. Accessed November 15, 2021.
- 6. Mayxay M Pukrittayakamee S Newton PN White NJ , 2004. Mixed-species malaria infections in humans. Trends Parasitol 20: 233–240. [DOI] [PubMed] [Google Scholar]
- 7. Moody A , 2002. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev 15: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. World Health Organization , 2017. A Framework for Malaria Elimination. Geneva, Switzerland: WHO. [Google Scholar]
- 9. Rogier E et al. 2017. Bead-based immunoassay allows sub-picogram detection of histidine-rich protein 2 from Plasmodium falciparum and estimates reliability of malaria rapid diagnostic tests. PLoS One 12: e0172139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Escobar DF Lucchi NW Abdallah R Valenzuela MT Udhayakumar V Jercic MI Chenet SM , 2020. Molecular and epidemiological characterization of imported malaria cases in Chile. Malar J 19: 1--9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Assefa A et al. 2019. Multiplex serology demonstrates cumulative prevalence and spatial distribution of malaria in Ethiopia. Malar J 18: 1--14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Plucinski MM McElroy PD Dimbu PR Fortes F Nace D Halsey ES Rogier E , 2019. Clearance dynamics of lactate dehydrogenase and aldolase following antimalarial treatment for Plasmodium falciparum infection. Parasit Vectors 12: 1--6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plucinski MM et al. 2018. Posttreatment HRP2 clearance in patients with uncomplicated Plasmodium falciparum malaria. J Infect Dis 217: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Commons RJ Simpson JA Thriemer K Hossain MS Douglas NM Humphreys GS Sibley CH Guerin PJ Price RN , 2019. Risk of Plasmodium vivax parasitaemia after Plasmodium falciparum infection: a systematic review and meta-analysis. Lancet Infect Dis 19: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Plucinski MM et al. 2019. Screening for Pfhrp2/3-deleted Plasmodium falciparum, non-falciparum, and low-density malaria infections by a multiplex antigen assay. J Infect Dis 219: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Argaw MD Woldegiorgis AG Abate DT Abebe ME , 2016. Improved malaria case management in formal private sector through public private partnership in Ethiopia: retrospective descriptive study. Malar J 15: 1--11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tadesse F Fogarty AW Deressa W , 2017. Prevalence and associated risk factors of malaria among adults in East Shewa Zone of Oromia Regional State, Ethiopia: a cross-sectional study. BMC Public Health 18: 1--8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fançony C Gamboa D Sebastião Y Hallett R Sutherland C Sousa-Figueiredo JC Nery SV , 2012. Various pfcrt and pfmdr1 genotypes of Plasmodium falciparum cocirculate with P. malariae, P. ovale spp., and P. vivax in northern Angola. Antimicrob Agents Chemother 56: 5271–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Snounou G Viriyakosol S Jarra W Thaithong S Brown KN , 1993. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol 58: 283–292. [DOI] [PubMed] [Google Scholar]
- 20. Richie TL , 1988. Interactions between malaria parasites infecting the same vertebrate host. Parasitology 96: 607–639. [DOI] [PubMed] [Google Scholar]