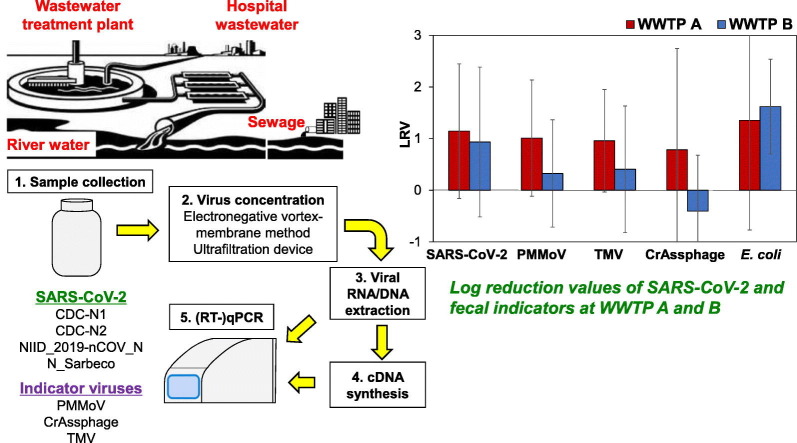
Abstract
The applicability of wastewater-based epidemiology (WBE) has been extensively studied throughout the world with remarkable findings. This study reports the presence and reduction of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at two wastewater treatment plants (WWTPs) of Nepal, along with river water, hospital wastewater (HWW), and wastewater from sewer lines collected between July 2020 and February 2021. SARS-CoV-2 RNA was detected in 50%, 54%, 100%, and 100% of water samples from WWTPs, river hospitals, and sewer lines, respectively, by at least one of four quantitative PCR assays tested (CDC-N1, CDC-N2, NIID_2019-nCOV_N, and N_Sarbeco). The CDC-N2 assay detected SARS-CoV-2 RNA in the highest number of raw influent samples of both WWTPs. The highest concentration was observed for an influent sample of WWTP A (5.5 ± 1.0 log10 genome copies/L) by the N_Sarbeco assay. SARS-CoV-2 was detected in 47% (16/34) of the total treated effluents of WWTPs, indicating that biological treatments installed at the tested WWTPs are not enough to eliminate SARS-CoV-2 RNA. One influent sample was positive for N501Y mutation using the mutation-specific qPCR, highlighting a need for further typing of water samples to detect Variants of Concern. Furthermore, crAssphage-normalized SARS-CoV-2 RNA concentrations in raw wastewater did not show any significant association with the number of new coronavirus disease 2019 (COVID-19) cases in the whole district where the WWTPs were located, suggesting a need for further studies focusing on suitability of viral as well as biochemical markers as a population normalizing factor. Detection of SARS-CoV-2 RNA before, after, and during the peaking in number of COVID-19 cases suggests that WBE is a useful tool for COVID-19 case estimation in developing countries.
Keywords: CrAssphage, Nepal, SARS-CoV-2, Wastewater-based epidemiology, Wastewater treatment plant
Graphical abstract
1. Introduction
Wastewater-based epidemiology (WBE) has been one of the most promising surveillance tools for monitoring the status, trend of infection, possible future outbreaks of coronavirus disease-2019 (COVID-19), to gather the information on disease surveillance, epidemiological models, genetic diversity, and geographic distribution. Previous studies documented that WBE is a non-invasive early warning surveillance tool (Daughton, 2020; Mallapaty, 2020; Naddeo and Liu, 2020), while other studies reported that application of WBE can detect the presence of both asymptomatic (Kitajima et al., 2020; Bivins et al., 2020) and pre-symptomatic cases in the community (Bivins et al., 2020). Considerable number of studies have been conducted throughout the world and these studies successfully quantified the RNA of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from untreated (Ahmed et al., 2020a; Hata et al., 2021; La Rosa et al., 2020b; Medema et al., 2020; Nemudryi et al., 2020; Schmitz et al., 2021; Sherchan et al., 2020, Sherchan et al., 2021; Wu et al., 2020a) and treated wastewater (Haramoto et al., 2020; Randazzo et al., 2020, Randazzo et al., 2020; Westhaus et al., 2020), and river water (Guerrero-Latorre et al., 2020), including a few studies from developing countries, such as India (Kumar et al., 2020) and Bangladesh (Ahmed et al., 2021). SARS-CoV-2 is transmitted through respiratory droplets, while clinical manifestation of SARS-CoV-2 infection in enterocytes of small intestine suggests that it can be shed through feces (Wölfel et al., 2020; Zhang et al., 2020). Evidence of the presence of SARS-CoV-2 RNA in stool of infected patients (Tang et al., 2020; Wang et al., 2020; Wölfel et al., 2020; Wu et al., 2020; Xiao et al., 2020; Zhang et al., 2020) and from other body fluids (Peng et al., 2020) has been well documented. According to Wu et al. (2020) and Zheng et al. (2020), SARS-CoV-2 can be detected from stool even after recovery from respiratory symptoms. Importantly, there is no sufficient information on viability of SARS-CoV-2 in wastewater. Because of the severe dilution of viruses in wastewater samples, several concentration methods, such as ultrafiltration (Ahmed et al., 2020d), polyethylene glycol precipitation (Kumar et al., 2020; Torii et al., 2022), ultracentrifugation (Sherchan et al., 2020), and electronegative membrane filtration (Haramoto et al., 2020), have been applied for quantification of SARS-CoV-2 RNA. Moreover, studies are underway to develop optimized protocols to detect SARS-CoV-2 RNA in wastewater (Ahmed et al., 2020d; Kitajima et al., 2020).
In Nepal, the first imported case of COVID-19 was recorded on January 13, 2020. After an occurrence of the first outbreak, the number of reported cases is increasing until today with the total number cases 819,019 as of October 21, 2021 (MoHP Nepal, 2021). Direct discharge of wastewater produced by hospitals, industries, household, and improperly treated effluents to nearby river is common in developing countries such as Nepal. The Kathmandu Valley has five centralized wastewater treatment plants (WWTPs), of which, only two WWTPs are currently working [i.e., WWTP A in Kathmandu (activated sludge process) and WWTP B in Lalitpur (non-aerated lagoons)] (Green et al., 2003). The sewerage system covers only 15% of the houses (National Planning Commission, 2005), whereas already existing WWTPs are very simple with no mechanized parts and under poor management, resulting in major problems of managing wastewater produced by the entire city (Leclerc et al., 2002).
Wastewater can contain various types of pathogens, depending on the incidence of disease in the community and animal population (Leclerc et al., 2002). Resource-poor countries such as Nepal cannot afford clinical testing materials to fulfill the demand of individual testing, thus WBE can generate essential data required to track the trend of disease.
The timely emergence of new variants of SARS-CoV-2 is a major public health concern throughout the world. Among the various variants, those classified as Variants of Concern (VOC) by the World Health Organization (WHO) pose a serious threat as they are associated with increase in transmissibility or increase in virulence or decrease in effectiveness of public health measures, including diagnostics, therapeutics, and vaccines (WHO, 2020). During the study period, only three variants, namely, α, β, and γ were classified as VOC. Since all three VOC display N501Y spike mutation (ECDC, 2021), it was selected for mutation-specific quantitative polymerase chain reaction (qPCR) run. Although the WHO recommends strengthening sequencing capacities for surveillance of SARS-CoV-2 variants, sequencing approach requires extensively trained manpower, and is time consuming compared to qPCR. Similarly, digital PCR has also been used to monitor variants of SARS-CoV-2 (Heijnen et al., 2021). All the mentioned approaches are not feasible in developing countries such as Nepal. Therefore, the applicability of environmental surveillance to track VOC using a mutation-specific qPCR assay was also analyzed. Only two functional WWTPs exist in the Kathmandu Valley, and both WWTPs were enrolled for the detection of SARS-CoV-2. Since two WWTPs are not sufficient to represent the presence of SARS-CoV-2 in the Kathmandu Valley, river water, wastewater from sewer lines, and a hospital wastewater were collected as alternatives of raw wastewater in regions where coverage of WWTP was low. The primary objective of this study was to determine the presence of SARS-CoV-2 RNA along with N501Y mutation in the collected water samples. Furthermore, analysis for Escherichia coli was performed using the most probable number (MPN) method utilizing Colilert reagent (IDEXX Laboratories, Westbrook, CA, USA). In addition, pepper mild mottle virus (PMMoV), tobacco mosaic viruses (TMV), and crAssphage, all considered as human fecal markers due to their presence in human feces, were also quantified using their respective qPCR assays (García-Aljaro et al., 2017; Hamza et al., 2019; Kitajima et al., 2018; Tandukar et al., 2020) and evaluated as a factor to normalize SARS-CoV-2 RNA concentration in untreated wastewater.
2. Materials and methods
2.1. Collection of water samples and analysis of E. coli
A total of 84 grab water samples were collected from two WWTPs, a hospital, sewer lines, and rivers in the Kathmandu Valley between July 2020 and February 2021. Oxidation ditch system is installed in WWTP A to treat 32.4 million liters per day (MLD), whereas WWTP B has a non-aerated lagoon facility with a design capacity of 1.1 MLD. Forty (20 each of influent and effluent) and 28 samples (14 each of influent and effluent) were collected from WWTP A and B, respectively. The variation in sample numbers collected from two WWTPs was due to the inability to collect wastewater samples from WWTP B during repair and maintenance. Decrease in sampling frequency after September was due to lockdown imposed by the government.
Thirteen river water samples were collected from the Bagmati River and its tributaries from densely populated midstream and downstream regions of the valley.
Two wastewater samples from sewer lines were also collected from a single site (latitude 27° 41′ 44″ and longitude 85° 18′ 22″) in October 2020 and January 2021. In addition, one wastewater sample was collected from sewer line of a tertiary care hospital of the Kathmandu Valley in September 2020. In the hospital an on-site treatment plant was lacking, and generated wastewater was directly discharged into the municipal sewerage system.
All the water samples were collected in autoclaved 100-mL plastic bottles taking precautionary measures and transported to the laboratory on icepacks within 2 h. Soon after the arrival at the laboratory, samples were processed for E. coli by MPN method using the Colilert reagent (IDEXX Laboratories) according to the manufacturers' instructions.
2.2. Virus concentration
In this study, viruses present in water samples were concentrated using the electronegative membrane-vortex (EMV) method (Haramoto et al., 2020). As described previously (Haramoto et al., 2020; Tandukar et al., 2020), 1 mL of 2.5 mol/L MgCl2 was added to 100 mL of a water sample, which was then filtered through a mixed cellulose-ester membrane (pore size, 0.8 μm; diameter, 47 mm; Merck Millipore, Billerica, MA, USA). The membrane was placed into a 50-mL plastic tube containing 10 mL of elution buffer [0.2 g/L Na4P2O7 10H2O, 0.3 g/L C10H13N2O8Na3 3H2O, and 0.1 mL/L Tween 80 (polyoxyethylene (20) sorbitan monooleate)] and a football-shaped stirring bar, and vortexed vigorously. The eluate was recovered in a new 50-mL plastic tube. This step was repeated using the 5-mL elution buffer to obtain a final volume of approx. 15 mL. The eluate was centrifuged at 2000 ×g for 10 min at 4 °C, and the recovered supernatant was further filtered using a disposable membrane filter unit (pore size, 0.45 μm; diameter, 25 mm; Advantec, Tokyo, Japan). The filtrate was subsequently concentrated using a Centriprep YM-50 ultrafiltration device (Merck Millipore) to obtain a viral concentrate.
Due to the unavailability of the Centriprep YM-50, Amicon Ultra-15 ultrafiltration device (Merck Millipore) was used for samples collected between November 2020 to February 2021 (n = 16). In this case, elution step was conducted using a total of 12 mL of the elution buffer (8 mL and 4 mL for the first and second elution steps, respectively). After that, the whole suspension was centrifuged at 2000 ×g for 10 min at 4 °C. The supernatant was then concentrated using Amicon Ultra-15 ultrafiltration device, which was centrifuged at 5000 ×g for 15 min at 25 °C, to obtain viral concentrate. Prior to the use of Amicon Ultra-15, the recovery efficiency by Amicon Ultra-15 was compared to that by Centriprep YM-50 for the recovery of Pseudomonas bacteriophage Φ6, a surrogate for enveloped viruses, from raw wastewater. Comparison testes between the two ultrafiltration devices revealed comparable recovery of Φ6 (data not shown).
2.3. RNA/DNA extraction and reverse transcription (RT)
In this study, murine norovirus (MNV) was used as a molecular process control to estimate the efficiency of RNA extraction and RT-qPCR (Haramoto et al., 2018). One microliter of MNV S7-PP3 strain, kindly provided by Dr. Yukinobu Tohya (Nihon University, Fujisawa, Japan), was added to 140 μL of the viral concentrate. The mixture was processed for viral RNA extraction with QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) in QIAcube automated platform (QIAGEN) to obtain a 60-μL viral RNA extract. A High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to obtain a 60-μL cDNA from 30 μL of viral RNA, following the manufacturer's protocol. One-hundred forty microliters of PCR-grade water was also inoculated with 1 μL of MNV and subjected to RNA extraction and RT-qPCR for MNV as an inhibitory control (IC) to calculate the efficiency.
Viral DNA (200 μL) was also extracted from 200 μL of the viral concentrate using QIAamp DNA mini kit (QIAGEN).
2.4. qPCR
N_Sarbeco, NIID_2019-nCOV_N, CDC-N1, and CDC-N2 assays (Centers for Disease Control and Prevention, 2020; Corman et al., 2020; Shirato et al., 2020) were used for the detection of SARS-CoV-2 RNA. A N501Y mutation-specific qPCR assay that targets nucleotide at spike receptor binding domain (adenine replaced by uracil at position 23,063 with reference to NC_045512.2) was also tested to detect N501Y mutation (Bedotto et al., 2021). In addition, MNV was tested as a molecular process control (Kitajima et al., 2010), while crAssphage (Stachler et al., 2017), PMMoV (Haramoto et al., 2013; Zhang et al., 2006), and TMV (Balique et al., 2013) were tested as indicator viruses to compare their reductions with SARS-CoV-2. RT-qPCR assays for SARS-CoV-2 were performed in a 25-μL qPCR mixture containing 12.5 μL of Probe qPCR Mix with UNG (Takara Bio, Kusatsu, Japan), 0.1 μL each of 100-μM forward and reverse primers, 0.05 μL of 100-μM TaqMan probe, and 2.5 μL of template cDNA. Sequences of primers and probes used in this study are summarized in Table 1 . The thermal conditions were as follows: 25 °C for 10 min and 95 °C for 30 s, followed by 45 cycles of 95 °C for 5 s and 60 °C for 60 s (for N_Sarbeco, NIID_2019-nCOV_N, PMMoV, TMV, and MNV), or 60 °C for 30 s (for CDC-N1 and CDC-N2, N501Y, and crAssphage).
Table 1.
Primers and probes of qPCR assays used in this study.
| Assay | Function | Name | Sequence (5′–3′)a | Product length (bp) | Reference |
|---|---|---|---|---|---|
| CDC-N1 | Forward Primer | 2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | 72 | CDC, 2020 |
| Reverse Primer | 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | |||
| Probe | 2019-nCoV_N1-P | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | |||
| CDC-N2 | Forward Primer | 2019-nCoV_N1-F | TTACAAACATTGGCCGCAAA | 67 | CDC, 2020 |
| Reverse Primer | 2019-nCoV_N1-R | GCGCGACATTCCGAAGAA | |||
| Probe | 2019-nCoV_N1-P | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | |||
| N_Sarbeco | Forward Primer | N_Sarbeco_F1 | CACATTGGCACCCGCAATC | 128 | Corman et al., 2020 |
| Reverse Primer | N_Sarbeco_R1 | GAGGAACGAGAAGAGGCTTG | |||
| Probe | N_Sarbeco_P1 | FAM-ACTTCCTCAAGGAACAACATTGCCA-BHQ1 | |||
| NIID_2019-nCOV_N | Forward Primer | NIID_2019-nCOV_N_F2 | AAATTTTGGGGACCAGGAAC | 158 | Shirato et al., 2020 |
| Reverse Primer | NIID_2019-nCOV_N_R2ver3 | GCACCTGTGTAGGTCAAC | |||
| Probe | NIID_2019-nCOV_N_P2 | FAM-ATGTCGCGCATTGGCATGGA-BHQ1 | |||
| N501Y | Forward Primer | Pri_IHU_N501Y_F1 | ATCAGGCCGGTAGCACAC | 156 | Bedotto et al., 2021 |
| Reverse Primer | Pri_IHU_N501Y_R1 | AAACAGTTGCTGGTGCATGT | |||
| Probe | Pro_IHU_C_GB_1_MBP | FAM-CCACTTATGGTGTTGGTTACCAA-ZEN/IBFQ | |||
| PMMoVb | Forward Primer | PMMV-FP1 | GAGTGGTTTGACCTTAACGTTTGA | 68 | Zhang et al., 2006 |
| Reverse Primer | PMMV-FP1-rev | TTGTCGGTTGCAATGCAAGT | Haramoto et al., 2013 | ||
| Probe | PMMV-Probe1 | FAM-CCTACCGAAGCAAATG-NFQ-MGB | Zhang et al., 2006 | ||
| TMVc | Forward Primer | TMV_Mars_CPFwd1 | CAAGCTGGAACTGTCGTTCA | 125 | Balique et al., 2013 |
| Reverse Primer | TMV_Mars_CPRev1 | CGGGTCTAAYACCGCATTGT | |||
| Probe | TMV_Mars_CP1 | FAM-CAGTGAGGTGTGGAAACCTTCACCACA-TAMRA | |||
| CrAssphage | Forward Primer | CPQ_056_F1 | CAGAAGTACAAACTCCTAAAAAACGTAGAG | 125 | Stachler et al., 2017 |
| Reverse Primer | CPQ_056_R1 | GATGACCAATAAACAAGCCATTAGC | |||
| Probe | CPQ_056_P1 | FAM-AATAACGATTTACGTGATGTAAC-NFQ-MGB | |||
| MNVd | Forward Primer | MNV-S | CCGCAGGAACGCTCAGCAG | 128 | Kitajima et al., 2010 |
| Reverse Primer | MNV-AS | GGYTGAATGGGGACGGCCTG | |||
| Probe | MNV-TP | FAM-ATGAGTGATGGCGCA-MGB-NFQ |
FAM, 6-carboxyfluorescein; MGB, minor groove binder; NFQ, nonfluorescent quencher; BHQ1, black hole quencher 1; IBFQ, Iowa black fluorescent quencher.
PMMoV, pepper mild mottle virus.
TMV, tobacco mosaic virus.
MNV, murine norovirus.
To draw a standard curve, six 10-fold serial dilutions of gBlocks gene fragments (Integrated DNA Technologies, Coralville, IA, USA) (for NIID_2019-nCOV_N, N_Sarbeco, and N501Y), 2019-nCoV_N Positive Control (Integrated DNA Technologies, cat. no. 10006625) (for CDC-N1 and CDC-N2), or artificially synthesized plasmid DNA (for MNV, PMMoV, TMV, and crAssphage) containing the amplification region sequences of the qPCR assays were used. These standards were considered positive controls, while molecular-grade water was used as a negative control. All samples, along with positive and negative controls, were tested in duplicate. Samples which showed cycle threshold values greater than 40 were considered as negative.
2.5. Reduction efficiency of viruses and WWTPs
Log10 reduction values (LRVs) of indicator viruses at the two WWTPs were calculated by subtracting the virus concentration in influent with that in effluent (log10 genome copies (GC)/L). Sample sets where viruses were detected in influent only were used for the calculation, and one tenth limit-of-detection (LOD) values were given to non-detect effluent samples: 2.4–3.3, 2.2–2.4, 2.5–3.5, and 2.9–3.1 log10 GC/L for CDC-N1, CDC-N2, NIID_2019-nCOV_N, and N_Sarbeco qPCR assays, respectively. Similar to indicator viruses, LRV of SARS-CoV-2 was calculated as the difference in concentration between influent and effluent wastewater samples. In those sample sets where SARS-CoV-2 RNA reduction was observed using more than one SARS-CoV-2 assay, an average of those values was used as LRV of SARS-CoV-2.
2.6. Statistical analysis
Pearson's correlation coefficient test was used to correlate the concentrations of both normalized and non-normalized-SARS-CoV-2 RNA concentrations in influent samples with the number of weekly new cases in the district where WWTP was located. Comparison of positive ratio of SARS-CoV-2 RNA in wastewater samples before and after September 2020 was conducted using Chi-square test, whereas LRVs in WWTP A were compared to those of WWTP B using analysis of variance (ANOVA). All the statistical analyses were performed using Microsoft Excel 2016 (Microsoft, Redmond, WA, USA) and a p-value of less than 0.05 was considered significant.
3. Results
3.1. RNA-extraction-RT-qPCR efficiency
MNV was used as a molecular process control to calculate the efficiency of RNA extraction-RT-qPCR. Extraction-RT-qPCR efficiency was calculated to be 53.3 ± 70.7% (n = 84), indicating good extraction efficiency (>10%) and the absence of serious inhibitions during RT-qPCR (Haramoto et al., 2018).
3.2. Detection of SARS-CoV-2 RNA in influents and effluents of WWTPs
As summarized in Table 2 , among the 68 water samples collected from two WWTPs, 38 (56%) samples were positive for SARS-CoV-2 RNA by at least one of the four qPCR assays tested: positive ratios were 53% (21/40) and 61% (17/28) for WWTP A and B, respectively.
Table 2.
Detection of SARS-CoV-2 RNA in water samples.
| Sample type (no. of tested samples) | SARS-CoV-2 |
N501Y variant |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDC-N1 |
CDC-N2 |
NIID_2019-nCOV_N |
N_Sarbeco |
Total |
||||||||
| No. of positives (%) | Conc. (log10 GC/L)a | No. of positives (%) | Conc. (log10 GC/L)a | No. of positives (%) | Conc. (log10 GC/L)a | No. of positives (%) | Conc. (log10 GC/L)a | No. of positives (%) | No. of positives (%) | Conc. (log10 GC/L)a | ||
| WWTP A | Influent (20) | 7 (35) | 5.0 ± 0.2 | 11 (55) | 5.1 ± 0.4 | 3 (15) | 5.1 ± 0.2 | 8 (40) | 4.8 ± 0.7 | 12 (60) | 1 (5) | 2.3 |
| Effluent (20) | 5 (25) | 5.2 ± 0.7 | 7 (35) | 5.3 ± 0.5 | 4 (20) | 5.3 ± 0.6 | 4 (20) | 5.5 ± 1.0 | 8 (40) | 0 (0) | ND | |
| WWTP B | Influent (14) | 4 (29) | 5.4 ± 1.1 | 8 (57) | 5.1 ± 0.9 | 5 (36) | 5.3 ± 1.0 | 5 (36) | 5.3 ± 1.0 | 10 (71) | 0 (0) | ND |
| Effluent (14) | 6 (43) | 5.3 ± 0.8 | 8 (57) | 5.0 ± 0.9 | 6 (43) | 5.3 ± 0.8 | 6 (43) | 5.4 ± 0.8 | 8 (57) | 0 (0) | ND | |
| River water (13) | 5 (38) | 5.0 ± 0.6 | 7 (47) | 4.0 ± 1.2 | 4 (31) | 5.1 ± 0.6 | 6 (46) | 5.0 ± 0.4 | 9 (69) | 0 (0) | ND | |
| Hospital wastewater (1) | 0 (0) | NDb | 1 (100) | 2.6 | 0 (0) | NDb | 0 (0) | ND | 1 (100) | 0 (0) | ND | |
| Sewer lines (2) | 2 (100) | 4.9 ± 0.1 | 2 (100) | 3.9 ± 0.3 | 1 (50) | 4.8 | 1 (50) | 4.9 | 2 (100) | 0 (0) | ND | |
Mean ± standard deviation.
ND, not detected.
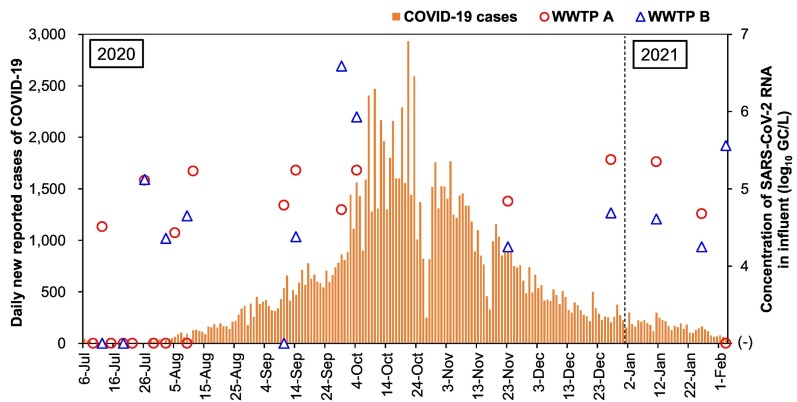
In July 2020, 30% (3/10) of the influent samples were found positive for SARS-CoV-2 RNA by at least one qPCR assay. When a sample was found positive for SARS-CoV-2 RNA by more than one qPCR assay, a mean value of SARS-CoV-2 RNA concentrations by these assays was calculated, as shown in Fig. 1 . Between July and August 2020, 44% (7/16) of the influent samples were tested positive at least by one assay with concentrations ranging from 4.4–5.2 log10 GC/L. On the other hand, SARS-CoV-2 RNA was consistently detected after September 2020 in majority (17/18) of the samples by at least one qPCR assay. During the study period, the CDC-N2 assay detected SARS-CoV-2 RNA in the highest number of influent samples of both WWTP A and B [55% (11/20) and 57% (8/14)] with concentrations of 5.0 ± 0.2 and 5.4 ± 1.0 log10 GC/L, respectively.
Fig. 1.
Number of daily new reported cases of COVID-19 and the concentrations of SARS-CoV-2 RNA in influent samples of WWTP A and B. The new cases data were extracted from the Nepal government COVID-19 dashboard (https://covid19.mohp.gov.np/).
For the effluent samples, 53% (18/34) were positive by at least one qPCR assay, with concentrations ranging from 4.4 to 6.5 log10 GC/L (Table 2). Positive ratios of SARS-CoV-2 RNA were comparable between before and after the wastewater treatment. Before September 2020, two (18%) effluent samples of WWTP A were found positive for SARS-CoV-2 RNA, while all the five effluent samples of WWTP B were negative. A significant increase in the positive ratio of SARS-CoV-2 RNA (89%, 16/18) was observed after September (Chi-square test, p < 0.05).
Fig. 1 shows the number of daily new reported cases of COVID-19 and the concentrations of SARS-CoV-2 RNA in the influent samples at WWTP A and B. At WWTP A, the highest concentration of SARS-CoV-2 RNA (6.5 log10 GC/L) was observed on November 1, followed by September 14 (6.2 log10 GC/L), when the number of daily new reported cases of COVID-19 was rapidly increasing. SARS-CoV-2 RNA was always detected after the peak observed in October, except for one sample collected at WWTP A in February 2021.
3.3. Detection of SARS-CoV-2 RNA in river water, HWW, and wastewater collected from sewer lines
Based on sample type, 69% (9/13), 100% (1/1), and 100% (2/2) samples were positive for SARS-CoV-2 RNA by at least one qPCR assay for river water, HWW, and wastewater collected from sewer lines, respectively. As shown in Table 2, the concentration of SARS-CoV-2 RNA in river water and wastewater collected from sewer lines were [(4.0 ± 1.2 to 5.0 ± 0.6) log10 GC/L and (3.9 ± 0.3 to 4.9 ± 0.1) log10 GC/L] by at least one assay, respectively. For the HWW sample, SARS-CoV-2 RNA was detected by CDC-N2 assay only with concentration of 2.6 log10 GC/L.
3.4. Detection of indicators in water samples
As summarized in Table 3 , the positive ratios of the indicator viruses tested were 83% (70/84) each for PMMoV and TMV, and 89% (75/84) for crAssphage. In the WWTP A, the highest concentration in the influents and effluents was observed for TMV (8.8 ± 0.5 and 7.9 ± 1.2 log10 GC/L, respectively), while in WWTP B, it was obtained for TMV in the influents (8.5 ± 0.8 log10 GC/L) and for crAssphage in the effluents (8.3 ± 1.1 log10 GC/L). For the river water samples, the highest concentration was observed for crAssphage (8.5 ± 0.5 log10 GC/L), while in the sewage samples it was observed for TMV (9.1 log10 GC/L). Both the positive ratios and concentrations of these indicator viruses were greater than those of SARS-CoV-2 RNA. E. coli was also analyzed in these samples, where it was detected in 100% (84/84) of the samples, with concentrations of 5.3–11.0 log10 MPN/100 mL.
Table 3.
Detection of indicator viruses and E. coli in water samples.
| Sample type (no. of tested samples) | PMMoV |
TMV |
CrAssphage |
E. coli |
|||||
|---|---|---|---|---|---|---|---|---|---|
| No. of positives (%) | Conc. (log10 GC/L)a | No. of positives (%) | Conc. (log10 GC/L) | No. of positives (%) | Conc. (log10 GC/L) | No. of positives (%) | Conc. (log10 MPN/100 mL) | ||
| WWTP A | Influent (20) | 19 (95) | 7.9 ± 0.3 | 19 (95) | 8.8 ± 0.5 | 20 (100) | 8.3 ± 1.4 | 19 (95) | 8.1 ± 0.7 |
| Effluent (20) | 19 (95) | 7.0 ± 0.9 | 20 (100) | 7.9 ± 1.2 | 20 (100) | 7.5 ± 1.3 | 20 (100) | 6.8 ± 2.3 | |
| WWTP B | Influent (14) | 14 (100) | 7.6 ± 0.7 | 13 (93) | 8.5 ± 0.8 | 13 (93) | 7.9 ± 1.2 | 14 (100) | 7.8 ± 0.7 |
| Effluent (14) | 14 (100) | 7.3 ± 0.7 | 14 (100) | 8.1 ± 0.9 | 14 (100) | 8.3 ± 1.1 | 14 (100) | 6.2 ± 0.9 | |
| River water (13) | 7 (54) | 6.0 ± 1.0 | 9 (69) | 7.1 ± 1.3 | 5 (38) | 8.4 ± 1.1 | 13 (100) | 6.4 ± 0.6 | |
| Hospital wastewater (1) | 1 (100) | 6.3 | 1 (100) | 8.7 | 1 (100) | 6.3 | 1 (100) | 7.2 | |
| Sewage (2) | 1 (50) | 8.5 | 1 (50) | 9.1 | 1 (50) | 8.6 | 2 (100) | 7.4 ± 0.1 | |
Mean ± standard deviation.
3.5. LRVs of SARS-CoV-2 and indicators during wastewater treatment process
Fig. 2 shows the LRVs of SARS-CoV-2 and indicators (three viruses and E. coli) at the tested WWTPs. Low LRVs of E. coli (WWTP A: 1.35 ± 2.12; WWTP B: 1.62 ± 0.92) were observed. LRVs of SARS-CoV-2 were 1.14 ± 1.30 (n = 12) and 0.93 ± 1.45 (n = 11) at WWTP A and B, respectively. There was no significant difference in the LRVs of SARS-CoV-2 RNA between WWTP A and B (ANOVA, p > 0.05). Among the four indicators, the LRVs of crAssphage [0.78 ± 1.97 at WWTP A (n = 20) and −0.40 ± 1.10 at WWTP B (n = 13)] were significantly lower than those of SARS-CoV-2 (ANOVA, p < 0.05).
Fig. 2.
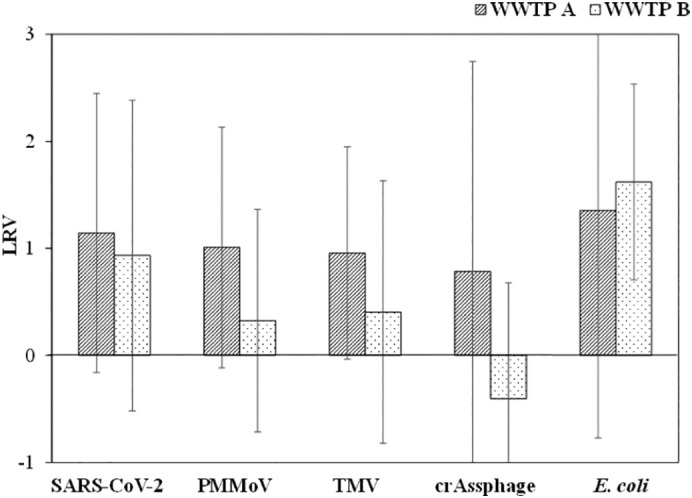
LRVs of SARS-CoV-2 and indicators at WWTP A and B.
3.6. Association between new COVID-19 cases and SARS-CoV-2 RNA concentration in wastewater
Moderate correlation was obtained between the number of weekly new cases of COVID-19 in the district and SARS-CoV-2 RNA concentrations (0.47–0.50; p < 0.05) based on CDC-N1 and CDC-N2 assays, whereas no significant correlations (p > 0.05) were observed using N_Sarbeco, and NIID_2019-nCOV_N assays. For all four assays, SARS-CoV-2 RNA concentrations in influent samples normalized by those of crAssphage, PMMoV, and TMV did not correlate with the weekly number of new cases in the whole district (p > 0.05).
4. Discussion
In this study, four qPCR assays (CDC-N1, CDC-N2, N_Sarbeco, and NIID_2019-nCOV_N) targeting the nucleoprotein (N) gene were tested for the detection of SARS-CoV-2 RNA from the water samples. Out of 84 samples tested, 39 (46%) water samples were found positive for SARS-CoV-2 RNA by at least one of the four qPCR assays. The high positive ratio observed in this study could be attributed to the peaking of COVID-19 transmission between September and October 2020, the method used to concentrate viruses prior to RNA extraction, and/or the use of four qPCR assays. Electronegative membrane filtration with addition of magnesium chloride and polyethylene glycol-based separation method has been recommended for detection of SARS-CoV-2 RNA in wastewater (Lu et al., 2020). The same method was also applied for environmental surveillance of SARS-CoV-2 elsewhere in Japan (Haramoto et al., 2020) and Australia (Ahmed et al., 2020a). Similarly, the use of multiple qPCR assays targeting different signature genes was also recommended to improve the detection of SARS-CoV-2 RNA in wastewater. Almost all of these previous studies were carried out in developed countries with good water, sanitation, and hygiene (WASH) practices. In contrast to the study conducted by Haramoto et al. (2020), in this study, 69% (9/13) of river water samples tested were positive for SARS-CoV-2 RNA by at least one qPCR assay. The urban rivers in the Kathmandu Valley receive untreated sewage, and these rivers have poor microbiological quality (Tandukar et al., 2018; Thakali et al., 2020; Sthapit et al., 2020). Similar to the study conducted in Ecuador (Guerrero-Latorre et al., 2020), high detection frequency of SARS-CoV-2 RNA in river water suggests that sampling of river water can be carried out in polluted urban rivers to implement WBE in developing countries where insufficient treatment of wastewater is carried out before discharge into rivers.
All hospital and sewage samples tested were also positive for SARS-CoV-2 RNA by one or more qPCR assays. However, it should be noted that the presence of SARS-CoV-2 RNA does not guarantee the presence of infectious SARS-CoV-2 particles. To date, no studies have reported the presence of infectious SARS-CoV-2 particles, and more studies that clearly define SARS-CoV-2 survival time in wastewater or water systems to estimate risk and leave no possibility for fecal oral transmission of SARS-CoV-2 are required (Tiwari et al., 2021). SARS-CoV-2 RNA was detected in HWW by CDC-N2 assay with concentration of 2.6 log10 GC/L. HWW can contain a mixture of chemicals such as pharmaceuticals and disinfectants in greater quantities compared to municipal wastewater that can lead to nucleic acid fragmentation, resulting in detection by a single assay only. In Nepal, direct discharge of HWW to nearby sewage pipeline without prior treatment is practiced, and the use of an on-site treatment plant for HWW management has been recommended (Thakali et al., 2021).
Twenty wastewater samples were collected between July 2020 and February 2021. In the wastewater samples, for all four assays, the highest concentration (6.6 log10 GC/L) was obtained in September. SARS-CoV-2 RNA was consistently detected in wastewater between September 2020 and February 2021. The highest concentration (6.1 log10 GC/L) of SARS-CoV-2 RNA in influent of WWTP A was observed on October 4, 2020 using the CDC-N2 assay. On the other hand, the highest concentration in effluent (6.5 log10 GC/L) was observed using the N_Sarbeco assay on January 11, 2021 when the number of daily new cases in the Kathmandu Valley had decreased. This result highlights the discrepancies between clinical and wastewater surveillance. Surveillance via clinical testing can be affected by the number of symptomatic cases, availability of testing facilities, and the affordability of the PCR tests. In resource limited settings of Nepal, PCR test is expensive and therefore, prone to under reporting of the actual number of cases. One of the limitations of our study is that only grab sampling was performed as per our convenience since lockdowns were imposed in the Kathmandu Valley. The inability to collect wastewater samples periodically such as weekly sampling hinders the opportunity to assess whether WBE can accurately predict the future surge in COVID-19 cases (Ahmed et al., 2020c). Furthermore, grab samples may be less representative of a community compared to 24-h composite samples, and intra-day variability in SARS-CoV-2 RNA concentration in influent samples has been observed (Bivins et al., 2021).
On January 18, 2021, the first case of α variant was reported in Nepal (The Himalayan Times Online, 2021; Poudel, 2021). The N501Y mutant was detected in a wastewater sample collected from WWTP A on February 3, 2021. Despite the delay in reporting of the circulation of variant of SARS-CoV-2 by 2 weeks, it displays the applicability of WBE to monitor variants of SARS-CoV-2 in developing countries without having to depend upon other countries or international organizations. Therefore, future studies should focus on developing WBE to detect variants of SARS-CoV-2 spread in a community.
The LRVs of SARS-CoV-2 at the WWTP B were significantly higher than those of crAssphage (ANOVA; p < 0.05), while those of PMMoV and TMV did not differ significantly from SARS-CoV-2 (ANOVA; p > 0.05). Enveloped viruses have a higher affinity to solid particles (Ye et al., 2016), resulting in the possibility of maximum retention in sludge (Peccia et al., 2020; Balboa et al., 2021); therefore, the presence of SARS-CoV-2 in effluent is considered to be a rare occurrence. Nevertheless, similar to previous studies (Balboa et al., 2021; Haramoto et al., 2020; Randazzo et al., 2020, Randazzo et al., 2020), the results of our study demonstrate that inadequately treated wastewater can release SARS-CoV-2 RNA in the environment.
In addition to quality assurance measure, fecal markers are quantified to help account for fluctuation in fecal matter concentration in raw wastewater. Fecal marker-normalized SARS-CoV-2 RNA concentrations have been shown to associate with clinical case numbers, thereby increasing confidence in WBE for SARS-CoV-2 (Reynolds et al., 2022; Wilder et al., 2021). Such fecal markers suitable for use as a normalization factor should be resilient against degradation in sewer network. In this study, crAssphage DNA was adjudged to persist through wastewater treatment; however, neither crAssphage, TMV, or PMMoV-normalized SARS-CoV-2 RNA concentrations improved the association with new clinical cases in our study. Similar findings of weak association between new clinical cases and crAssphage and PMMoV-normalized SARS-CoV-2 RNA concentrations have been reported from Ireland and USA (Reynolds et al., 2022; Wilder et al., 2021). The lack of association in our study could be due to the possible difference in the number of new cases in the actual catchment area compared to the number of cases in the whole district. More studies focusing on the suitability of viral fecal markers as well as chemical markers as a normalizing factor are recommended to accurately predict near-term COVID-19 case levels as well as to enable comparisons between different cities.
5. Conclusions
This study successfully detected SARS-CoV-2 RNA in influent and effluent of WWTPs, river water, HWW, and wastewater from sewer lines in the Kathmandu Valley, suggesting that WBE can complement clinical testing for tracking the spread of COVID-19 in a community. High positive ratio of 94% (17/18) was obtained from the influent samples collected after September 2020. N501Y mutation was also detected in one of the influent samples, highlighting the applicability of WBE in detecting variants of SARS-CoV-2. Despite the persistence nature of crAssphage, crAssphage-normalized SARS-CoV-2 RNA concentrations in raw wastewater did not provide any significant association with the number of new COVID-19 cases in the whole district where the WWTPs were located, suggesting a need for further studies focusing on the suitability of viral as well as biochemical markers as a population normalizing factor.
CRediT authorship contribution statement
Sarmila Tandukar: Formal analysis, Investigation, Methodology, Visualization, Writing-original draft.
Niva Sthapit: Investigation.
Ocean Thakali: Investigation.
Bikash Malla: Investigation.
Samendra P. Sherchan: Supervision.
Bijay Man Shakya: Investigation.
Laxman P. Shrestha: Supervision.
Jeevan B. Sherchand: Supervision.
Dev Raj Joshi: Investigation.
Bhupendra Lama: Investigation.
Eiji Haramoto: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Visualization, Writing-reviewing and editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors acknowledge staff members at both WWTPs for their kind support in wastewater sampling and all the members of Public Health Research Laboratory, Institute of Medicine, and Tribhuvan University.
Funding sources
This study was supported by the Nepal Academy of Science and Technology (NAST) through COVID-19 Technology Development, the Japan Science and Technology Agency (JST) through J-RAPID program (grant number JPMJJR2001) and Accelerating Social Implementation for SDGs Achievement (aXis) (grant number JPMJAS2005), and the Japan Society for the Promotion of Science (JSPS) through Fund for the Promotion of Joint International Research (Fostering Joint International Research (B)) (grant number JP18KK0297) and Grant-in-Aid for Scientific Research (B) (grant number JP20H02284).
Editor: Damia Barcelo
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., Brien J.W.O., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., Hossen F., Hossain M.S., Islam M.S., Uddin M.M., Islam M.N., Bahadur N.M., Didar-Ul-Alam M., Reza H.M., Jakariya M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: variation along the sewer network. Sci. Total Environ. 2021;776 doi: 10.1016/j.scitotenv.2021.145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balique F., Colson P., Barry A.O., Nappez C., Ferretti A., Moussawi K.A., Ngounga T., Lepidi H., Ghigo E., Mege J.L., Lecoq H., Raoult D. Tobacco mosaic virus in the lungs of mice following intra-tracheal inoculation. PloS One. 2013;8(1) doi: 10.1371/journal.pone.0054993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedotto M., Fournier P., Houhamdi L., Colson P., Raoult D. Implementation of an in-house real-time reverse transcription-PCR assay to detect the emerging SARS-CoV-2 N501Y variants. J. Clin. Virol. 2021;140 doi: 10.1016/j.jcv.2021.104868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Cassi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bivins A., North D., Whu Z., Shaffer M., Ahmed W., Bibby K., et al. Within- and between-day variability of SARS-CoV-2 RNA in municipal wastewater during periods of varying COVID-19 prevalence and positivity. ACS ES&T Water. 2021;1(9):2097–2108. doi: 10.1021/acsestwater.1c00178. [DOI] [Google Scholar]
- Centers for Disease Control and Prevention 2019-novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes. 2020. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide COVID-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC SARS-CoV-2 variants of concern as of 30 April 2021. 2021. https://www.ecdc.europa.eu/en/covid-19/variants-concern
- García-Aljaro C., Ballesté E., Muniesa M., Jofre J. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 2017;10:1775–1780. doi: 10.1111/1751-7915.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H., Poh S.C., Richards A. Massachusetts Institute of Engineering; 2003. Wastewater Treatment in Kathmandu, Nepal. Master of Engineering in Civil and Environmental Engineering. [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza H., Rizk N.M., Gad M.A., Hamza I.A. Pepper mild mottle virus in wastewater in Egypt: a potential indicator of wastewater pollution and the efficiency of the treatment process. Arch. Virol. 2019;164:2707–2713. doi: 10.1007/s00705-019-04383-x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79(23):7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.-2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima K. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1101/2020.06.04.20122747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Elsinga G., de Graff M., Molenkamp R., Koopmans M.P.G., Medema G. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Oka T., Takagi H., Tohya Y., Katayama H., Takeda N., Katayama K. Development and application of a broadly reactive real-time reverse transcription-PCR assay for detection of murine noroviruses. J. Virol. Methods. 2010;169(2):269–273. doi: 10.1016/j.jviromet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Rachmadi A.T., Iker B.C., Haramoto E., Gerba C.P. Temporal variations in genotype distribution of human sapoviruses and Aichi virus 1 in wastewater in southern Arizona, United States. J. Appl. Microbiol. 2018;124:1324–1332. doi: 10.1111/jam.13712. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc H., Schwartzbrod L., Dei-Cas E. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 2002;28:371–409. doi: 10.1080/1040-840291046768. [DOI] [PubMed] [Google Scholar]
- Lu D., Huang Z., Luo J., Zhang X., Sha S. Primary concentration - the critical step in implementing the wastewater-based epidemiology for the COVID-19 pandemic: a mini-review. Sci. Total Environ. 2020;747 doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- MoHP Nepal 2021. https://covid19.mohp.gov.np/
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci.: Water Res. Technol. 2020;6(5):1213–1216. doi: 10.1039/D0EW90015J. [DOI] [Google Scholar]
- National Planning Commission Summary of Results on Poverty Analysis from Nepal Living Standards Survey (2003–04) National Planning Commission, Kathmandu. 2005 [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat.Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Liu J., Xu W., Luo Q., Chen D., Lei Z., Huang Z., Li X., Deng K., Lin B., Gao Z. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020;92(9):1676–1680. doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel A. UK variant responsible for surge in coronavirus cases in Nepal, Health Ministry says. The Kathmandu Post. 2021. https://kathmandupost.com/health/2021/04/08/uk-variant-responsible-for-surge-in-coronavirus-cases-in-nepal-health-ministry-says
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.2139/ssrn.3586696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds L.J., Sala-Comorera L., Khan M.F., Martin N.A., Whitty M., Stephens J.H., Nolan T.M., Joyce E., Fletcher N.F., Murphy C.D., Meijer W.G. Coprostanol as a population biomarker for SARS-CoV-2 wastewater surveillance studies. Water. 2022;14:225. doi: 10.3390/w14020225. [DOI] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Patel J., Ward L.M., Tandukar S., Uprety S., Schmitz B.W., Ahmed W., Simpson S., Gyawali P. Occurrence of SARS-CoV-2 RNA in six municipal wastewater treatment plants at the early stage of COVID-19 pandemic in the United States. Pathogens. 2021;10:798. doi: 10.3390/pathogens10070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73(4):304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative crAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51:9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sthapit N., Malla B., Ghaju Shrestha R., Tandukar S., Sherchand J.B., Haramoto E., Kazama F. Investigation of Shiga toxin-producing Escherichia coli in groundwater, river water, and fecal sources in the Kathmandu Valley, Nepal. Water Air Soil Pollut. 2020;231 doi: 10.1007/s11270-020-04920-4. [DOI] [Google Scholar]
- Tandukar S., Sherchan S.P., Haramoto E. Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Rep. 2020;10:3616. doi: 10.1038/s41598-020-60547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar S., Sherchand J.B., Bhandari D., Sherchan S.P., Malla B., Ghaju Shrestha R., Haramoto E. Presence of human enteric viruses, protozoa, and indicators of pathogens in the Bagmati River, Nepal. Pathogens. 2018;7(2) doi: 10.3390/pathogens7020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N., Wu W.J., Yuan C., Yu M.L., Li P., Yan J.B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child,China. Emerg. Infect. Dis. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakali O., Tandukar S., Brooks J.P., Sherchan S.P., Sherchand J.B., Haramoto E. The occurrence of antibiotic resistance genes in an urban river in Nepal. Water. 2020;12(2):450. doi: 10.3390/w12020450. [DOI] [Google Scholar]
- Thakali O., Malla B., Tandukar S., Sthapit N., Raya S., Furukawa T., Sei K., Sherchand J.B., Haramoto E. Release of antibiotic-resistance genes from hospitals and a wastewater treatment plant in the Kathmandu Valley,Nepal. Water. 2021;13:2733. doi: 10.3390/w13192733. [DOI] [Google Scholar]
- The Himalayan Times Nepal confirms the first three cases of new COVID variant. 2021. https://thehimalayantimes.com/nepal/nepal-confirms-the-first-three-cases-of-new-covid-variant
- Tiwari A., Phan N., Tandukar S., Ashoori R., Thakali O., Mousazadesh M., Hadi Dehghani M., Sherchan S.P. Persistence and occurrence of SARS-CoV-2 in water and wastewater environments: a review of the current literature. Environ. Sci. Pollut. Res. 2021:1–11. doi: 10.1007/s11356-021-16919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S., Oishi W., Zhu Y., Thakali O., Malla B., Yu Z., Zhao B., Arakawa C., Kitajima M., Hata A., Ihara M., Kyuwa S., Sano D., Haramoto E., Katayama H. Comparison of five polyethylene glycol precipitation procedures for the RT-qPCR based recovery of murine hepatitis virus, bacteriophage phi6, and pepper mild mottle virus as a surrogate for SARS-CoV-2 from wastewater. Sci. Total Environ. 2022;807(2) doi: 10.1016/j.scitotenv.2021.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the coronavirus disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linneman V., Brinkmanne M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2020;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Dud Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11:100100. doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V., Guggemos W., Seilmaier M., Zange S., Müller M., Niemeyer D., Kelly T., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized cases of coronavirus disease-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization Tracking SARS-CoV-2 variants. 2020. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4):e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4(1):0108–0118. doi: 10.1371/jour-nal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang S., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]