Significance
Making immunotherapy more widely effective requires improved understanding of the pathways that regulate the interactions between tumor and immune cells. This work identified a previously unappreciated difference in these pathways between heterogenous epigenetic states that coexist in neuroblastoma. The generally rarer mesenchymal subpopulation has more active inflammatory sensing and signaling, revealing a potential immunologic vulnerability of these chemotherapy-resistant cells.
Keywords: neuroblastoma, immunotherapy, epigenetic states, Toll-like receptors, MYCN
Abstract
Immunotherapy has revolutionized cancer treatment, but many cancers are not impacted by currently available immunotherapeutic strategies. Here, we investigated inflammatory signaling pathways in neuroblastoma, a classically “cold” pediatric cancer. By testing the functional response of a panel of 20 diverse neuroblastoma cell lines to three different inflammatory stimuli, we found that all cell lines have intact interferon signaling, and all but one lack functional cytosolic DNA sensing via cGAS–STING. However, double-stranded RNA (dsRNA) sensing via Toll-like receptor 3 (TLR3) was heterogeneous, as was signaling through other dsRNA sensors and TLRs more broadly. Seven cell lines showed robust response to dsRNA, six of which are in the mesenchymal epigenetic state, while all unresponsive cell lines are in the adrenergic state. Genetically switching adrenergic cell lines toward the mesenchymal state fully restored responsiveness. In responsive cells, dsRNA sensing results in the secretion of proinflammatory cytokines, enrichment of inflammatory transcriptomic signatures, and increased tumor killing by T cells in vitro. Using single-cell RNA sequencing data, we show that human neuroblastoma cells with stronger mesenchymal signatures have a higher basal inflammatory state, demonstrating intratumoral heterogeneity in inflammatory signaling that has significant implications for immunotherapeutic strategies in this aggressive childhood cancer.
In order to be effective, immunotherapies require cytotoxic immune cells to traffic to and enter the tumor microenvironment, recognize tumor cells, and kill them. These requirements provide a number of potential mechanisms for intrinsic or acquired immune evasion. For instance, with immune checkpoint blockade (ICB), resistant tumors tend to have lower preexisting infiltration with cytotoxic T cells (1) (trafficking and entry) and lower mutation rates (2–4) (tumor recognition). The level of tumor cell inflammatory signaling has the potential to affect trafficking and entry of immune cells through the secretion of cytokines and tumor recognition through antigen presentation. Indeed, lower baseline inflammatory signaling has also been associated with ICB resistance (4, 5), and response can be restored in some tumor models by activating tumor cell inflammatory signaling (6, 7). Increasing inflammatory signaling has the potential to enhance other immunotherapies as well, including adoptive cellular immunotherapies and antibody-based therapeutics that require antibody-dependent cellular cytotoxicity for function.
Understanding which inflammatory sensing pathways are intact in a given tumor is an important initial step in determining how to activate cellular inflammatory signaling. These sensing pathways include the recognition of interferon, which can be secreted from both immune and nonimmune cells, as well as the detection of molecular patterns typically found in microbial infection. Such patterns are recognized by a large group of receptors, including Toll-like receptors (TLRs), often collectively referred to as pattern recognition receptors. These include proteins that detect double-stranded RNA (dsRNA; TLR3, MDA5, and RIG-I), single-stranded RNA breakdown products (TLR7 and TLR8), CpG-rich (TLR9) or cytosolic DNA (cGAS), and lipopolysaccharides (LPS; TLR4), among others (8). In tumors, products capable of stimulating such sensors can be released by dead and dying cells (9), produced by inhibition of splicing (10) or by activation of endogenous retroviral elements in response to therapy (11), exposed by breakdown of the nuclear envelope (12), or transferred through exosomes from tumor or stromal cells (13–15). Some of these receptors have been shown to be critical to an immunologic response to anticancer therapy, including TLR3 with chemotherapy (16) and cGAS with radiation therapy (12, 17). Exogenous agonists of these pathways are being incorporated into clinical trials, particularly in combination with ICB. Thus, knowledge about the responsiveness of these pathways in cancer may contribute to our understanding of the basal inflammatory state of the tumor, suggest potential avenues to alter that state, and identify biomarkers for which an agonist might be more effective in a given tumor.
Neuroblastoma accounts for ∼15% of childhood cancer mortality in addition to significant morbidity among survivors (18, 19). Improved therapies are needed, particularly for high-risk disease, including the subset with amplification of MYCN (v-myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog) (19, 20). Neuroblastoma is a classically “cold” tumor with a lack of T cell infiltration, very low major histocompatibility complex class I (MHC-I) expression, and a low tumor mutation burden (21–24), all of which are even more pronounced in MYCN-amplified disease (25–28). Published studies to date have shown little to no efficacy with ICB (29, 30). Chimeric antigen receptor (CAR) T cell therapies have demonstrated limited efficacy, thought to be in part due to poor penetration or function in the tumor microenvironment (31). However, an antibody against the disialoganglioside GD2 is an immunotherapy that is Food and Drug Administration approved for use in neuroblastoma despite significant off-tumor toxicity due to GD2 expression on pain nociceptors. This therapy improves survival when used in combination with cytokines in patients in first remission (32) and has a ∼40% response rate when used in combination with cytokines and chemotherapy in relapsed disease (33, 34). This demonstrates that the immune system can play a role in treating neuroblastoma, but expanding the benefit of immunotherapy to a greater percentage of neuroblastoma patients remains an important challenge.
One recent advance in the understanding of neuroblastoma biology is the discovery that neuroblastoma cells can exist in two different epigenetic states defined by super enhancer elements enriched in one state or the other (35–37). There is a less-differentiated state described as either mesenchymal or neural crest cell–like and a more differentiated state described as adrenergic or sympathetic noradrenergic (herein referred to as MES and ADRN, respectively). These states are thought to coexist within human tumors and can spontaneously interconvert, although cell lines tend to stably adopt one state or the other, with some exceptions. In human primary neuroblastomas, the ADRN state comprises the majority of the tumor at the time of diagnosis (35, 36). Evidence from human tumors (35, 38) and mouse models (39) indicates that the MES state may be enriched in relapse and in metastatic disease (40), and MES cell lines are more chemoresistant in vitro. Hence, the ability to specifically target the MES subpopulation may prove important in both treating and preventing relapsed disease. To our knowledge, connections have not been made between ADRN and MES states and inflammatory signaling or response to immunotherapy.
To identify ways to increase inflammatory signaling in neuroblastoma, here we define the responsiveness of a large panel of neuroblastoma cell lines to different inflammatory stimuli, test how those differences impact tumor–immune interactions, and identify determinants of response, revealing immune vulnerabilities of neuroblastoma.
Results
Neuroblastoma Lacks Basal Inflammatory Signaling.
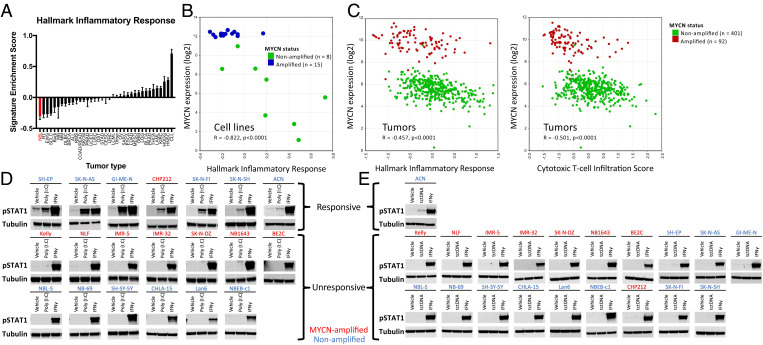
We first asked if the immunologically “cold” (21–23) nature of neuroblastoma correlated with a lack of tumor cell inflammatory signaling. We compared expression of the hallmark inflammatory response signature (41) across a large number of tumor cell lines using RNA-sequencing (RNA-seq) data from the Cancer Cell Line Encyclopedia (CCLE) project (42). Analysis of 991 cell lines across 37 tumor types showed that neuroblastoma has the lowest expression of this signature as well as several other inflammatory signaling signatures (Fig. 1A and SI Appendix, Fig. S1), demonstrating that neuroblastoma cells have strongly suppressed inflammatory signaling. We then asked how MYCN amplification status impacts this signature. Similar to published reports (25), we found that inflammatory signaling is further decreased in the MYCN-amplified subset of both cell lines and tumors compared to those without MYCN amplification, which corresponds to less infiltration with cytotoxic T cells in tumors (Fig. 1 B and C).
Fig. 1.
Neuroblastoma cells lack basal inflammatory signaling and respond heterogeneously to inflammatory stimuli. (A) Relative enrichment score of the hallmark inflammatory response signature across cell lines in the CCLE, with neuroblastoma shown in red. Abbreviations are provided in SI Appendix, Fig. S1. Error bars represent SEM between cell lines of the indicated tumor type. (B) Comparison of MYCN expression and relative enrichment score of the hallmark inflammatory response signature in 23 neuroblastoma cell lines. Data are from GSE28019, obtained from and analyzed in R2 (https://hgserver1.amc.nl:443/). (C) Comparison of MYCN expression and relative enrichment score of either the hallmark inflammatory response signature (Left) or a cytotoxic T cell infiltration score (Right, from ref. 82) in 498 neuroblastoma tumors. Data are from ref. 83, obtained from and analyzed in R2 (https://hgserver1.amc.nl:443/). (D) Western blot showing change in pSTAT1 levels when 20 neuroblastoma cell lines were treated with either 30 μg/mL poly (I:C) or 20 ng/mL IFNγ for 24 h. (E) Western blot showing change in pSTAT1 levels when 20 neuroblastoma cell lines were treated with either tctDNA (1 μg/mL), IFNγ, or transfection reagent control (vehicle) for 6 h. Western blots are representative of results from at least three separate experiments.
Neuroblastoma Cell Lines Respond Heterogeneously to Inflammatory Stimuli.
With the rationale that increasing cellular inflammatory signaling would increase human leukocyte antigen I (HLA-I) expression and lead to cytokine secretion that could drive T cell recruitment, we next analyzed the response of a panel of 20 diverse neuroblastoma cell lines to different inflammatory stimuli (Fig. 1 D and E), including exogenous interferon gamma (IFNγ), polyinosinic:polycytidylic acid (poly [I:C]; a dsRNA mimetic), and transfected calf thymus DNA (tctDNA; cytosolic DNA to activate cGAS). All of the cell lines tested generated a robust pSTAT1 response to exogenous IFNγ, consistent with previous reports of the effect of IFNγ on neuroblastoma cells (24, 43, 44). Only one cell line responded strongly to cytosolic DNA, consistent with essentially undetectable expression of cGAS across neuroblastoma cell lines in the CCLE, in a different neuroblastoma RNA-seq data set (45), and in RNA-seq data from neuroblastoma xenografts (46) (SI Appendix, Fig. S2 A–C). Similarly, Western blotting for cGAS in our cell line panel showed that only the single responsive cell line had detectable expression (SI Appendix, Fig. S2D). Response to the dsRNA mimetic poly (I:C), however, was heterogenous, with the majority lacking response, while a subset showed robust responses. Intrigued by this heterogeneity, we decided to pursue the response to poly (I:C) further.
dsRNA-Responsive Cell Lines Demonstrate Widespread Inflammatory Changes and Increase Susceptibility to T Cell Killing.
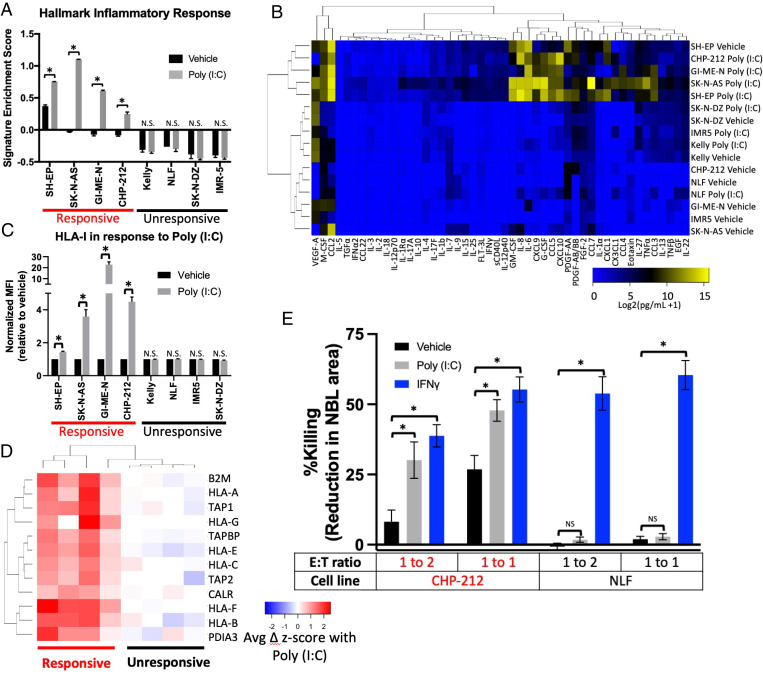
We next demonstrated using a subset of cell lines that the cells that responded to poly (I:C) with phosphorylation of STAT1 also responded by increasing transcription of IFN-response genes by qPCR, increasing nuclear localization of nuclear factor κ-light chain enhancer of activated B cells (NF-κB), increasing phosphorylation of IFN regulatory factor 3 (IRF3), and increasing activation of an NF-κB reporter construct (SI Appendix, Fig. S3 A–C). Unresponsive cell lines showed none of these changes. We then performed transcriptomic analysis using 3′ RNA sequencing (QuantSeq) on duplicate samples of four dsRNA-responsive and four unresponsive cell lines treated with poly (I:C) or a vehicle control. Responsive cell lines showed a marked up-regulation of inflammatory signaling pathways, while unresponsive cell lines remained unchanged (Fig. 2A and SI Appendix, Fig. S4).
Fig. 2.
dsRNA-responsive cell lines respond by multiple metrics. (A) Relative enrichment of the hallmark inflammatory response signature in the indicated cell lines treated with vehicle or 30 μg/mL poly (I:C) for 24 h as measured by QuantSeq. (B) Heat map depicting the level of the indicated cytokines in the supernatant of the indicated neuroblastoma cell lines after treatment with vehicle or poly (I:C) for 24 h. (C) Change in surface expression of HLA-I, measured by flow cytometry, in the indicated neuroblastoma cell lines after treatment with vehicle or poly (I:C) for 24 h. MFI, mean fluorescence intensity. (D) Heat map depicting the change in transcript expression of the indicated antigen processing and presentation genes after treatment with poly (I:C) for 24 h as measured by QuantSeq. (E) Change in killing of neuroblastoma cells exogenously expressing MART-1 after culture with MART-1 transgenic TCR-transfected T cells. Neuroblastoma cells were treated with the indicated agonists for 24 h then washed and cultured with the T cells. E:T ratio is the effector (T cell)-to-target (neuroblastoma) ratio. Killing was calculated by microscopy-based detection of change in cell area. A two-tailed paired t test between biological replicates (A and C) or an ANOVA (E) were run; *P < 0.05. Error bars represent SEM between biological replicates. N.S., not significant.
We then asked how poly (I:C) changed cytokine secretion, myeloid cell migration, antigen presentation, and immune killing in neuroblastoma cell lines. We analyzed the concentration of 38 cytokines and chemokines in the supernatant of four responsive and four unresponsive cell lines cultured with and without poly (I:C). The profiles of the four responsive cell lines were similar once treated, while the unresponsive treated cell lines had profiles similar to the untreated lines (Fig. 2B). A group of inflammatory cytokines were particularly increased in responsive cell lines when treated, including interleukin-6 (IL-6), IL-8, C-C motif chemokine ligand 5 (CCL5), C-X-C motif chemokine ligand 9 (CXCL9), CXCL10, granulocyte colony-stimulating factor (G-CSF), and granulocyte–macrophage colony-stimulating factor (GM-CSF) (SI Appendix, Fig. S5). Consistent with this finding, conditioned medium from only responsive cell lines treated with poly (I:C) increased migration of the THP-1 monocytic cells (SI Appendix, Fig. S3D). Responsive cell lines also increased HLA-I surface expression, as measured by flow cytometry, while unresponsive cell lines showed no change (Fig. 2C), although all the cell lines responded to IFNγ (SI Appendix, Fig. S3E). Responsive lines markedly increased expression at the RNA level of the entire antigen processing and presentation pathway (Fig. 2D). To study well-controlled T cell–mediated cytotoxicity independent of endogenous tumor cell antigen expression, we expressed melanoma-associated antigen recognized by T cells 1 (MART-1) in one responsive (CHP-212) and one unresponsive (NLF) HLA-A2 neuroblastoma cell line (SI Appendix, Fig. S3F) and cocultured them with T cells transfected with a T cell receptor (TCR) targeting MART-1 (47). We found that the responsive cells were killed more easily by T cells even prior to stimulation, and killing was enhanced by treatment with either poly (I:C) or IFNγ. In contrast, the unresponsive cells were resistant at baseline, and killing was enhanced only by IFNγ, not by poly (I:C) (Fig. 2E and SI Appendix, Fig. S3G). We also tested the ability of primary natural killer (NK) cells to kill these two cell lines, with or without pretreatment of the cell lines with poly (I:C) (SI Appendix, Fig. S3H). We found less dramatic differences in pretreatment susceptibility than with T cells. Similarly, poly (I:C) only modestly increased killing of the responsive cell line (significant at one NK cell dose but not the other) and did not increase killing of the unresponsive line. Hence, dsRNA sensing is only active in a subset of neuroblastoma cells lines; in those lines, it drives an inflammatory transcriptome, increases immune cell recruitment, augments antigen processing and presentation, and increases killing of neuroblastoma cells by T cells and, to a less extent, NK cells.
Poly (I:C)–Responsive Cell Lines Have Multiple Active dsRNA Sensors and Respond to Additional TLR Agonists.
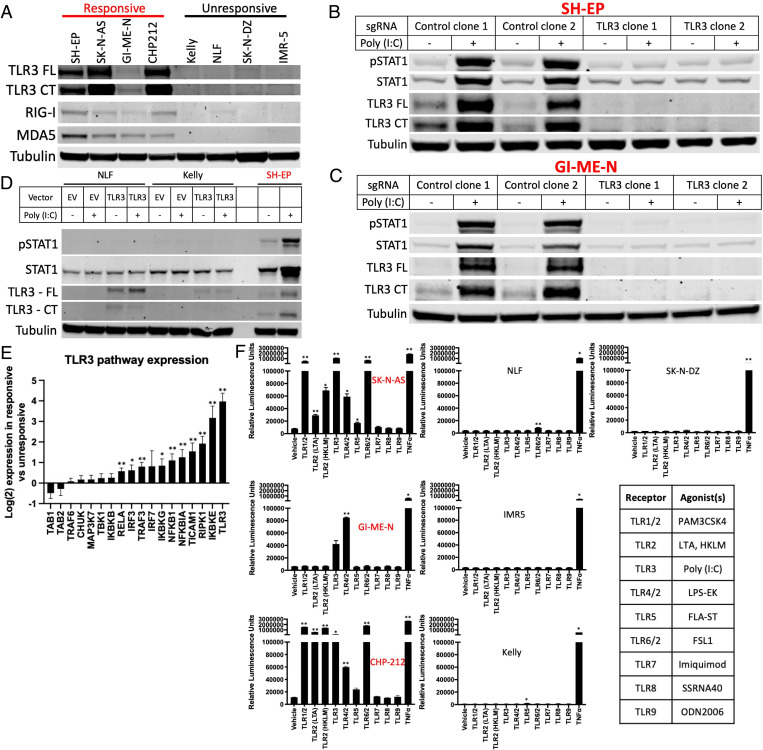
We next asked whether the defect in dsRNA sensing was limited to the expression of a single dsRNA sensor in unresponsive cell lines or whether this was indicative of a broader defect in inflammatory sensing of dsRNA and other stimuli, resulting in a widely insensitive state. We found that the dsRNA sensors TLR3, RIG-I, and MDA5 were all more highly expressed in responsive cell lines at the RNA (SI Appendix, Fig. S6 A–C) and protein (Fig. 3A) levels, although the expression of TLR3 was uniquely undetectable at the RNA level in unresponsive cell lines. We therefore used CRISPR-Cas9 to knockout TLR3 in two responsive cell lines and found that this fully abrogated response to poly (I:C) (Fig. 3 B and C), demonstrating that sensing of exogenous dsRNA requires TLR3 in this context. This is consistent with TLR3 localization in endosomes and a role in sensing extracellular dsRNA, while RIG-I and MDA-5 detect cytosolic dsRNA (8). However, we found that poly (I:C)–unresponsive cell lines were also unresponsive to transfection of poly (I:C) (SI Appendix, Fig. S6D), demonstrating defective sensing by RIG-I and MDA-5. In contrast, responsive cell lines with or without TLR3 knocked out were responsive to transfected poly (I:C) (SI Appendix, Fig. S6E), demonstrating that intracellular dsRNA sensing is also active only in poly (I:C)–responsive lines. We then exogenously expressed TLR3 in two unresponsive cell lines and found that this did not result in a response to poly (I:C), despite restoring TLR3 protein to a level similar to that found in a responsive cell line (Fig. 3D), demonstrating that TLR3 expression is necessary but not sufficient for response to dsRNA. Consistent with a broader suppression of signaling not limited to a single node, analysis of the expression of other members of the Kyoto Encyclopedia of Genes and Genomes (KEGG) TLR3 signaling pathway showed that transcripts for 10 of the 18 genes were significantly higher in the responsive lines (Fig. 3E). We then analyzed the chromatin accessibility of these 10 genes using assay for transposase-accessible chromatin with sequencing (ATAC-seq). We found that three genes (TLR3, IKBKE, and IKBKG) were significantly more accessible in the responsive cell lines (SI Appendix, Fig. S6F) than in unresponsive cells, supporting differences in the underlying epigenetic state between these cells influencing their responsiveness.
Fig. 3.
dsRNA-responsive cell lines require TLR3 and respond to additional inflammatory stimuli. (A) Western blot depicting the expression of TLR3, RIG-I, and MDA5 in four poly (I:C)–responsive and four unresponsive cell lines. Both the full length (FL) and an active C-terminal fragment (CT) of TLR3 are shown. (B and C) Western blots showing the change in pSTAT1, total STAT1, and TLR3 after treatment with 30 μg/mL of poly (I:C) for 24 h with or without CRISPR-Cas9–mediated knockout of TLR3 in the SH-EP (B) and GI-ME-N (C) cell lines. (D) Western blot showing change in pSTAT1, total STAT1, and TLR3 after treatment with poly (I:C) for 24 h with or without expression of exogenous TLR3. (E) Difference in transcript expression of genes in the TLR3 signaling pathway between responsive and unresponsive cell lines as measured by QuantSeq. (F) Relative luminescence in the indicated cell lines transfected with an NF-κB reporter after treatment with a vehicle control or the indicated TLR agonists for 6 h. Table showing the agonists used in the adjacent experiments. Western blots are representative of results from at least three separate experiments. Either a two-tailed t test (E) or an ANOVA (F) was performed: *P < 0.05 and **P < 0.01. Error bars represent SEM between biological replicates.
Changes in multiple components of the TLR3-signaling pathway, many of which are shared with other TLR-signaling pathways, suggested that additional TLR-signaling pathways may also be differentially active. To test this further, we used an NF-κB reporter assay to measure the response to nine additional TLR agonists in three responsive cell lines and four unresponsive cell lines. While the responsive cell lines were also able to respond strongly to some additional stimuli (although heterogenous), the unresponsive lines did not show any strong responses (Fig. 3F). Together, these data pointed toward a transcriptomic and epigenetic state broadly unresponsive to inflammatory stimuli, including intra- and extracellular dsRNA and additional TLR agonists.
MYCN Plays a Minor Role in dsRNA Response.
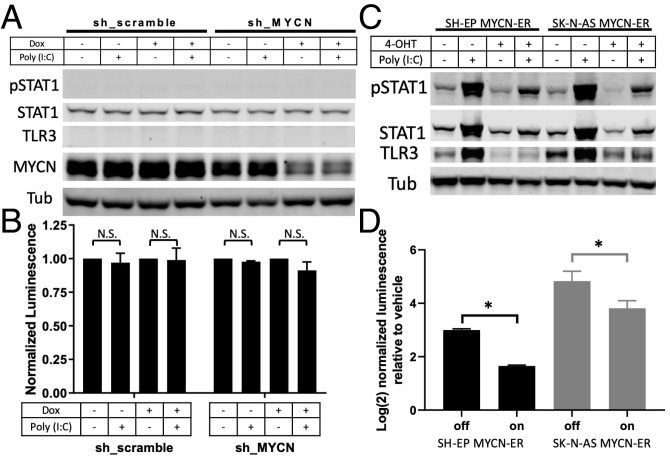
Our first hypothesis for the driver of this insensitive state was MYCN amplification status. Amplified tumors have distinct transcriptomes (48) and, as discussed, have lower markers of immune infiltration and inflammatory signaling (25–28). While six nonamplified lines responded to poly (I:C) and six others did not, only one of eight amplified lines (CHP-212) showed any response. However, as shown in SI Appendix, Fig. S7A, this responsive amplified line does have the high level of MYCN transcript and the robust MYCN signature (49) that is typical of MYCN-amplified cell lines. We tested whether knocking down MYCN in unresponsive amplified lines could restore response to poly (I:C). We used either an inducible short hairpin RNA (shRNA) in BE2(c) cells (50) or short interfering RNAs (siRNAs) in Kelly and NLF cells and found no change in response (Fig. 4 A and B and SI Appendix, Fig. S7B). While we achieved >80% knockdown in all three settings, given the supraphysiologic levels of MYCN signaling in such amplified lines, it is difficult to ascertain whether this knockdown resulted in a sufficiently “low” MYCN state. Therefore, to further test this hypothesis, we used two responsive nonamplified lines stably expressing a MYCN-estrogen receptor (ER) construct (51, 52), which allowed acute activation of MYCN signaling upon application of an ER agonist. As shown in Fig. 4 C and D, acute MYCN activation only modestly decreased responsiveness to poly (I:C) and did not result in an insensitive state. Hence, we surmise that MYCN expression alone is not sufficient to explain the difference between responsive and unresponsive cell lines.
Fig. 4.
MYCN is insufficient to fully change dsRNA-sensing state. (A) Western blot showing changes in pSTAT1, STAT1, TLR3, and MYCN when BE2(c) cells expressing an inducible shRNA, either a scramble control or targeting MYCN, were treated with either vehicle or doxycycline (dox) for 72 h, at which point they were then treated with either vehicle or 30 μg/mL poly (I:C) for 24 h (with continued dox or vehicle). (B) Change in luminescence detected after the cells used in A were transfected with an NF-κB reporter and then treated as described in A. N.S., not significant. (C) Western blot showing changes in pSTAT1, STAT1, and TLR3 when SH-EP or SK-N-AS cells expressing a MYCN-ER construct were treated with vehicle or 4-hydroxytamoxifen (4-OHT) for 24 h, at which point they were treated with either vehicle or poly (I:C) for 24 h (with continued 4-OHT or vehicle). (D) Change in luminescence detected after the cells used in C were transfected with an NF-κB reporter and then treated as described in C. Each bar shows the increase comparing vehicle to poly (I:C) treatment in the specified cell line and MYCN-ER on/off condition. Western blots are representative of results from at least three separate experiments. A two-tailed paired t test between biological replicates was performed; *P < 0.001. Error bars represent SEM between biological replicates.
MES State Has Active dsRNA Response.
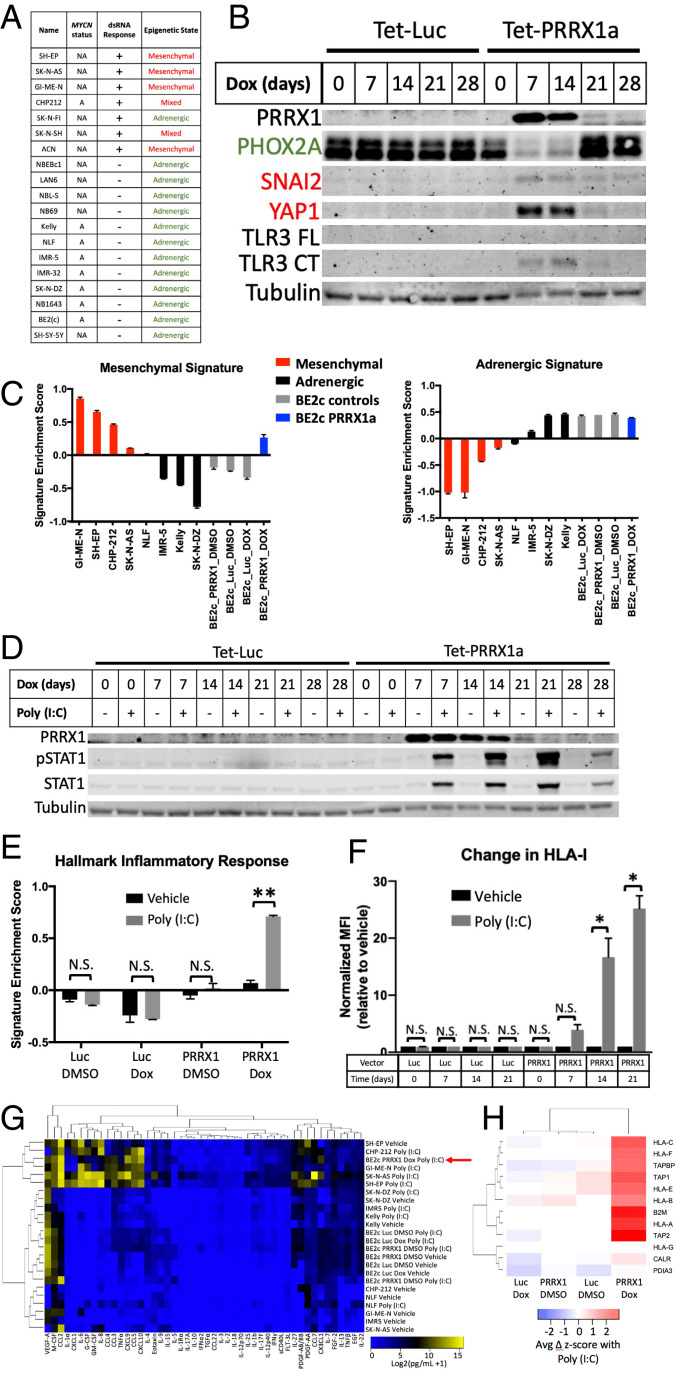
Given the recently described MES and ADRN states of neuroblastoma cells (35–37), we next hypothesized that these epigenetic states were associated with dsRNA and other TLR responses. Based on published categorizations of our cell lines (35, 36) supplemented with additional RNA-seq data (45) for unclassified lines (SI Appendix, Fig. S8A), we found that these states were highly predictive of poly (I:C) responsiveness (Fig. 5A), strongly correlated with expression of TLR3 itself in cell lines and tumors (SI Appendix, Fig. S8B), and predictive of histone 3 lysine 27 (H3K27) acetylation near the TLR3 promoter (SI Appendix, Fig. S8C). All of the unresponsive cell lines were ADRN, while all but one responsive cell lines were MES. Interestingly, SH-EP and SH-SY5Y cell lines are subclones from the same parent line (53) and have been classified as MES and ADRN, respectively (35). Concordantly, SH-EP is responsive to poly (I:C), while SH-SY5Y is unresponsive. Lastly, the only MYCN-amplified line that responds to poly (I:C) is the only MYCN-amplified line in the MES state.
Fig. 5.
Epigenetic state determines dsRNA responsiveness. (A) Table showing the cell lines used, their MYCN amplification status (A, amplified; NA, nonamplified), their dsRNA response, and their epigenetic state. Epigenetic state was determined based on prior reports (SH-EP, SK-N-AS, GI-ME-N, ACN, SH-SY-5Y, IMR32, BE2c, and SK-N-FI from ref. 35; CHP-212, SK-N-SH, NB-69, NBEBc1, and SK-N-DZ from ref. 36) or from an expression signature-based comparison with characterized lines as shown in SI Appendix, Fig. S8A. (B) Western blot showing changes in the adrenergic marker PHOX2A, the mesenchymal markers SNAI2 and YAP1, and TLR3 after inducible expression of luciferase (Luc) or PRRX1 for the indicated number of days. Both the FL and an active CT of TLR3 are shown. (C) Relative enrichment of the mesenchymal (Left) and adrenergic (Right) expression signatures in the indicated cell lines as measured by QuantSeq. Inducible BE2(c) cells expressing either Luc or PRRX1 were treated with vehicle or dox for 14 d prior to analysis. (D) Western blot showing changes in pSTAT1 and total STAT1 when BE2(c) cells inducibly expressing either Luc or PRRX1 were treated with vehicle or dox for the indicated number of days and treated with vehicle or 30 μg/mL poly (I:C) for 24 h. (E) Relative enrichment of the hallmark inflammatory response signature in BE2(c) cells expressing inducible Luc or PRRX1 treated with vehicle or dox for 14 d and treated with vehicle or poly (I:C) for 24 h, as measured by QuantSeq. (F) Change in surface expression of HLA-I, measured by flow cytometry, in BE2(c) cells expressing inducible Luc or PRRX1 treated with dox for the indicated number of days and treated with vehicle or poly (I:C) for 24 h. (G) Heat map depicting the level of the indicated cytokines in the supernatant of the indicated neuroblastoma cell lines after treatment with vehicle or poly (I:C) for 24 h. Inducible BE2(c) cells were treated with either vehicle (DMSO) or dox for 14 d prior to poly (I:C) treatment. The red arrow highlights the BE2(c) cells with PRRX1 turned on and treated with poly (I:C). (H) Heat map depicting the change in transcript expression of the indicated antigen processing and presentation genes after treatment with poly (I:C) for 24 h as measured by QuantSeq. Inducible BE2(c) cells were treated with either vehicle (DMSO) or dox for 14 d prior to poly (I:C) treatment. Western blots are representative of results from at least three separate experiments. A two-tailed paired t test between biological replicates was performed; *P < 0.05 and **P < 0.02. Error bars represent SEM between biological replicates. N.S., not significant.
To determine whether the MES epigenetic state was simply correlated with dsRNA responsiveness or whether it was causally related, we sought to determine if altering the epigenetic state would also result in a change in dsRNA responsiveness. As has been described (35), we inducibly expressed the mesenchymal transcription factor PRRX1 (or luciferase as a control) in the ADRN BE2(c) cell line. As shown in Fig. 5B, this resulted in a loss of the adrenergic marker PHOX2A and new expression of the mesenchymal markers SNAI2 and YAP1. QuantSeq data after a 14-d PRRX1 induction showed a strong increase in the MES signature score (35) to a level comparable to native MES lines (Fig. 5C), although only a modest decrease in the ADRN score was evident. This switch also resulted in TLR3 protein expression (Fig. 5B), an increase in transcript levels of TLR3 and multiple other genes in the TLR3 pathway (SI Appendix, Fig. S9A), and a stark conversion from unresponsive to dsRNA to fully responsive (Fig. 5D). Similar to our findings with Kelly and NLF cells, expression of TLR3 alone did not restore response to poly (I:C) in BE2(c) cells (SI Appendix, Fig. S9B). After PRRX1 expression, responsiveness measured by STAT1 phosphorylation was accompanied by up-regulation of inflammatory expression signatures (Fig. 5E and SI Appendix, Fig. S10), secretion of inflammatory cytokines (Fig. 5G and SI Appendix, Fig. S11), increased HLA-I surface expression (Fig. 5F), and increased expression of antigen processing and presentation genes (Fig. 5H), all of which were absent in the cell line without induction of transgene expression or expressing a luciferase control. To confirm that the connection between cell state and dsRNA response is not limited to this single cell line, we expressed PRRX1 in the ADRN SH-SY5Y cell line with a similar result (SI Appendix, Fig. S9 C and D).
MES State Activates Additional Sensing Pathways and Has an Elevated Basal Inflammatory State.
While these findings indicate that cells in the MES state are responsive to stimulation of TLR3, we next asked whether the MES state is also associated with the broader active inflammatory sensing we identified in poly (I:C)–responsive cell lines. The first evidence to suggest that this is the case is that PRRX1 expression led to full restoration of the pSTAT1 response to poly (I:C), unlike exogenous TLR3 expression in insensitive lines, which did not lead to a response (Figs. 3D and 5D, and SI Appendix, Fig. S9 B and D). We next looked at the correlation between the MES gene signature (35) and the transcripts of receptors, adaptors/signaling proteins, and effector transcription factors involved in TLR and other pattern recognition receptor signaling in addition to how these transcripts changed with PRRX1 expression. In bulk tumors, nearly all of the examined transcripts were highly correlated with the MES signature, although this could be confounded by expression in stromal and immune cells. However, in addition to TLR3, several other transcripts involved in pattern recognition signaling were also highly correlated in two different cell line transcriptomic datasets (SI Appendix, Table S1 and Fig. S12A) and also increased with PRRX1 expression in our cells (SI Appendix, Fig. S12B). This included STING, which is activated by cGAS signaling, and the NF-κB subunit RELB as well as TLR4 and TLR6. TLR4 is notably the receptor for the ligand LPS that was active in all three mesenchymal cell lines that were tested for response (Fig. 3F). Concordantly, PRRX1 expression in BE2(c) cells resulted in a new sensitivity to LPS (Fig. 6A).
Fig. 6.
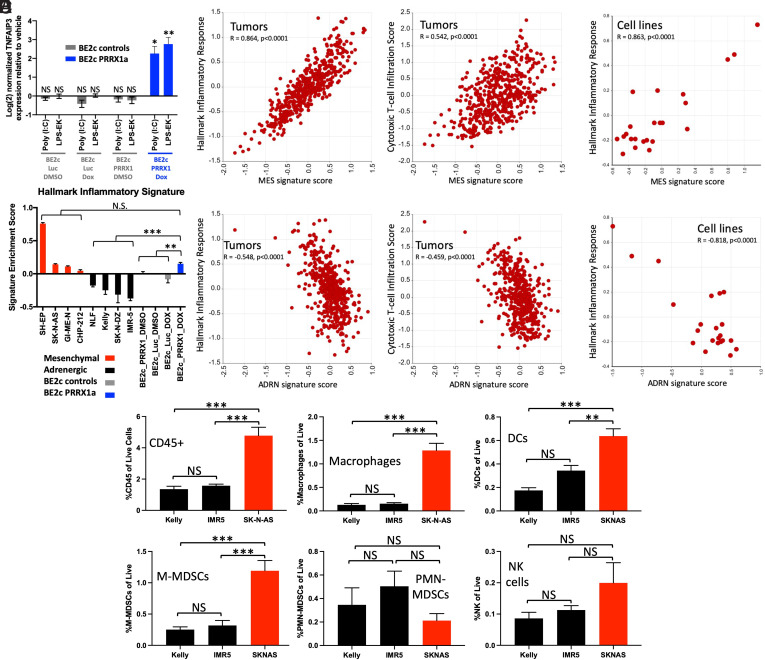
Epigenetic state controls additional TLRs and correlates with basal inflammatory state and basal immune infiltration. (A) Change in expression of TNFAIP3 as measured by qPCR after treatment with the indicated agonists for 6 h in BE2(c) cells after treatment with vehicle or dox for 14 d. (B) Comparison of the MES or ADRN signature scores (35) to the enrichment of the hallmark inflammatory response signature and to a cytotoxic T cell infiltration signature (82) in 498 neuroblastoma tumors (83). Data were obtained from and analyzed in R2 (https://hgserver1.amc.nl:443/). (C) Comparison of the MES or ADRN signature scores to the enrichment of the hallmark inflammatory response signature in 23 neuroblastoma cell lines (data from GSE28019). Data were obtained from and analyzed in R2 (https://hgserver1.amc.nl:443/). (D) Relative enrichment of the hallmark inflammatory response signature in the indicated neuroblastoma cell lines as measured by QuantSeq. BE2(c) cells expressing inducible Luc control or PRRX1 were treated with vehicle or dox for 14 d. (E) Relative infiltration of xenograft tumors from the indicated cell lines with the indicated immune cell types, as measured by flow cytometry and presented as percent of live cells. DC, dendritic cell. An ANOVA comparing biological replicates was performed; *P < 0.05, **P < 0.02, and ***P < 0.001. Error bars represent SEM between biological replicates. NS, not significant.
We next examined the relationship between MES and ADRN signatures and the basal inflammatory state in neuroblastoma by looking at correlations with inflammatory signaling gene sets. In tumors, which are thought to be a heterogenous mix of the two states, we found that the MES signature score was highly correlated with both an inflammatory signaling gene set and with a transcriptional signature of cytotoxic T cell infiltration (Fig. 6B). This was also true for inflammatory signaling in cell lines (Fig. 6C), and the reverse was true for the ADRN signature. PRRX1 expression in BE2(c) cells resulted in an increase in the inflammatory gene set to a level indistinguishable from the MES lines (Fig. 6D). These strong correlations were not driven by overlapping genes in the gene sets; for example, of the 200 total genes in the hallmark inflammatory signaling gene set, 5 are in the 485-gene MES gene signature and 3 are in the 369-gene ADRN gene signature. There is no overlap between the genes in the MES and ADRN signature and the cytotoxic T cell infiltration signature. Furthermore, even without treatment with poly (I:C), responsive MES neuroblastoma cell lines have higher surface HLA-I expression (SI Appendix, Fig. S12C), and PRRX1 expression increases it in BE2(c) cells (SI Appendix, Fig. S12D). Similarly, the transcripts of HLA-I and multiple other genes involved in antigen presentation and processing are both higher in the poly (I:C)–responsive MES lines and increased by PRRX1 expression (SI Appendix, Fig. S12 E and F). Finally, using ATAC-seq, we found that that three of the ten genes involved in antigen processing and presentation that increased at the transcript level were significantly more accessible in responsive lines than in unresponsive cell lines (SI Appendix, Fig. S12G). Hence, the MES epigenetic state not only has activated TLR3 signaling but also shows activation of additional immune sensing pathways and has an elevated basal inflammatory state.
To address the question of whether tonic stimulation of TLRs might drive an enhanced state of inflammatory signaling and sensing, we used CRISPR-Cas9 to knockout TLR3, MYD88, or both in an MES cell line background (SH-EP MYCN-ER). MYD88 is a coreceptor involved in signaling through all TLRs other than TLR3 (54). MYD88-knockout cells, but not TLR3-knockout or double-knockout cells, continued to respond to poly (I:C) (SI Appendix, Fig. S13A), demonstrating that tonic stimulation of a TLR other than TLR3 is not required for active sensing of dsRNA. To test whether tonic signaling through TLRs might drive the differences in basal inflammatory transcriptional signatures between ADRN and MES cells, we identified the 50 genes that were most significantly altered between ADRN and MES cell lines from the 200-gene hallmark inflammatory response gene set. Using QuantSeq, we found that these genes are largely unaltered by knocking out TLR3 or both TLR3 and MYD88 (SI Appendix, Fig. S13B). These observations indicate that increased inflammatory sensing and higher transcriptional signatures of inflammation in MES cell lines are not a result of tonic stimulation of TLRs.
Immune Infiltration of Xenograft Tumors from MES and ADRN Cell Lines.
Given that MES cell lines have elevated expression of inflammatory sensors and a higher basal inflammatory state, we asked if these features would persist in vivo and trigger immune infiltration compared to ADRN cell lines. We implanted two MES cell lines (SK-N-AS and GI-ME-N) and two ADRN lines (Kelly and IMR-5) into CB17 severe combined immunodeficiency (SCID) mice, which lack T and B cells but retain NK and myeloid cells. The MES SH-EP cell line is not tumorigenic (55, 56) and was not tested, and the GI-ME-N cells also did not form tumors in SCID mice. Tumors formed from the MES SK-N-AS cells and the two ADRN cell lines. Using a human-specific TLR3 antibody, we found that only SK-N-AS tumors expressed TLR3 in vivo (SI Appendix, Fig. S14A), consistent with our cell culture findings. Notably, there were stark differences between the basal immune infiltration of tumors from MES and ADRN cell lines. The SK-N-AS tumors had more than three times as many CD45 cells, including more dendritic cells, macrophages, and monocytic myeloid-derived suppressor cells (M-MDSCs), compared to the ADRN line–derived tumors (Fig. 6E). We injected the tumors with poly (I:C) to test if there were differences in tumor-intrinsic responses. We found that treatment increased human TLR3 only in the MES SK-N-AS–derived tumors (SI Appendix, Fig. S14A), consistent with the feed-forward signaling we see in cell lines (Figs. 3 B–D and 4C). Similarly, immunohistochemical staining for pSTAT1 was only detected after poly (I:C) treatment in the MES SK-N-AS–derived tumors (SI Appendix, Fig. S14B). Nontumor cells in the immune microenvironment have active dsRNA sensing and are predicted to respond to poly (I:C). Consistently, poly (I:C) increased infiltration of immune cells in tumors from both MES and ADRN cell lines (SI Appendix, Fig. S14C). These findings demonstrate that the increased basal inflammatory signaling in the MES SK-N-AS cell line observed in vitro corresponds to increased immune cell infiltration in vivo compared to ADRN cell lines. Local injection of poly (I:C) increased TLR3 and pSTAT1 only in tumors from the MES SK-N-AS line, but immune cell infiltration increased in tumors from both MES and ADRN cell lines.
Single-Cell RNA Sequencing Shows that MES Signature in Human Tumor Cells Correlates with Inflammatory Signature In Vivo.
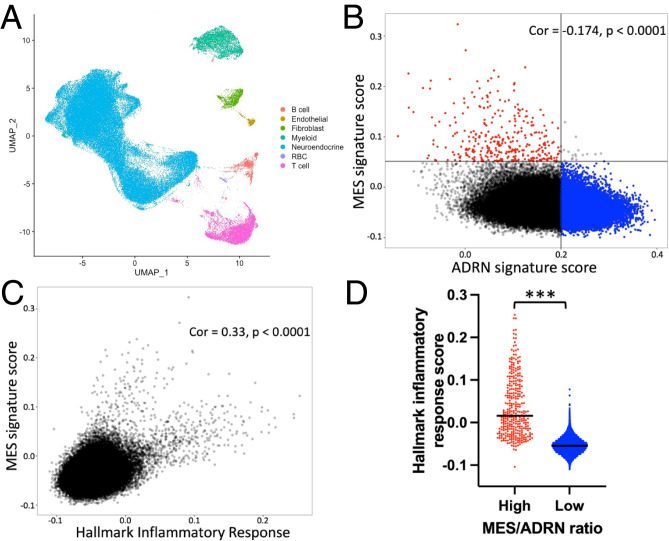
Lastly, we asked whether MES/ADRN states are associated with inflammatory signaling in human neuroblastoma cells in vivo. To address this, we analyzed single-cell RNA sequencing (scRNAseq) data from 10 untreated adrenal high-risk neuroblastomas that were recently published (57). Classification by cell identity marker genes demonstrated that the majority of cells showed a neuroendocrine phenotype with smaller numbers of immune cells and stromal cells (Fig. 7A). We then used the same approach taken by the authors of this study to infer chromosomal copy number variations (CNVs) from the transcriptomic data individually for each sample, which allowed for separation of presumed tumor and nontumor cells (SI Appendix, Fig. S15A). Similar to the original study, we did not identify a transcriptionally distinct population of pure tumor cells with mesenchymal markers. However, there is substantial transcriptional variability within the tumor compartment, including within individual tumors (SI Appendix, Fig. S15A). We therefore asked whether cells with stronger ADRN or MES signatures could be identified. Indeed, the two signatures were negatively correlated overall and in every tumor (Fig. 7B and SI Appendix, Fig. S15B). We then compared the MES signature to the hallmark inflammatory signature and found that these signatures were strongly positively correlated overall and in every individual tumor (Fig. 7C and SI Appendix, Fig. S15C). If the cells with the highest MES score and lowest ADRN score are compared to the cells with the lowest MES and highest ADRN score, the difference in the inflammatory signature is even more striking (Fig. 7D). This demonstrates that in human neuroblastoma tumors, cells with stronger MES signatures have higher levels of basal inflammatory transcripts compared to those with stronger ADRN signatures. Similar to cell lines with higher MES character, we postulate that these cells are thus likely to be uniquely vulnerable to inflammatory stimuli and therefore may be more susceptible to immune-based therapies. Given the reports of chemotherapy resistance in this subset of cells, immune targeting of these cells has the potential to improve outcomes and decrease relapse in neuroblastoma.
Fig. 7.
scRNAseq in human neuroblastoma shows that inflammatory signature correlates with MES signature. (A) UMAP plot showing cell type distributions in the 10 pooled samples. (B) Correlation (Cor) between the MES and ADRN gene signatures (35) in the cells determined to be tumor based on CNV. Cells are shown from all 10 samples. The quadrant in red is defined as high MES/ADRN ratio, and the quadrant in blue is defined as low MES/ADRN ratio. (C) Correlation between the hallmark inflammatory response signature (41) and the MES signature in the cells determined to be tumor based on CNV. Cells are shown from all 10 samples. (D) Graph depicting the hallmark inflammatory signature score for each cell with high MES/ADRN ratio (red) versus low MES/ADRN ratio (blue) as defined in B. The black bar indicates the mean. A two-tailed t test between indicated groups was performed; ***P < 0.001.
Discussion
Immunotherapy has had a transformative effect on cancer therapy. This includes neuroblastoma, as antibodies against GD2 improve survival when combined with immune-stimulating cytokines, although a cure remains elusive in large numbers of patients. Other immunotherapies, such as checkpoint inhibitors and CAR T cells, have had limited efficacy. Here, we show that neuroblastoma is a particularly difficult challenge, even among cold tumors. Using cell line transcriptomic data, we found that neuroblastoma cells are the most deficient in inflammatory signaling among the 37 cancer histologies examined. While such analysis will be more definitively performed once widespread scRNAseq data are available across tumor types, the current data provide further evidence that neuroblastoma cells lack substantial inflammatory signaling at baseline. However, there is reason for optimism that altering that cold state may allow improved entry, persistence, and activity of activated immune cells (6, 7). As a precursor to enhancing immunotherapy in neuroblastoma, we investigated the pathways that could be stimulated in order to activate inflammatory signaling in neuroblastoma cells.
Multiple prior studies have shown that neuroblastoma cell lines increase HLA-I upon stimulation with IFNγ (24, 43, 44). Consistent with this finding, we found that all cell lines generated an increase in pSTAT1 upon treatment with IFNγ. A prior clinical study treated neuroblastoma patients with IFNγ (58, 59), and a more recent analysis (44) showed that this increased T cell infiltration and HLA-I expression in human tumors and had similar effects in mouse models. This study was done prior to the development of checkpoint inhibitors, but such an approach could be revisited.
We found that only one of the 20 cell lines tested had detectable expression of cGAS or responded to transfected DNA. Sensing by cGAS has been implicated in driving inflammatory responses to chromothripsis (12), an important process in poor-prognosis neuroblastoma (60). Silencing of cGAS expression in neuroblastoma may result from selective pressure imposed by the immune-stimulating effects of chromothripsis or of extrachromosomal circular DNA, which is found widely in neuroblastoma, including as a frequent mechanism of MYCN amplification (61). Given the essential role of cGAS–STING signaling in mediating a productive interaction between radiation therapy and ICB (17), our findings suggest that radiation therapy could be optimized by strategies to activate cGAS–STING in neuroblastoma to improve the systemic immune response.
We focused on the heterogenous responses of neuroblastoma cells to the dsRNA mimetic poly (I:C). Response to poly (I:C) has previously been explored in neuroblastoma, with most studies focused on activation of cell death (62–65). Rather than induction of cell death, we concentrated on activation of inflammatory signaling, as this was our goal and since we saw little to no death after short treatments (<48 h). A prior study showed that in some neuroblastoma cell lines, poly (I:C) can increase PD-L1 and MHC-I surface expression, secretion of inflammatory cytokines, and activation of T cells (66). Consistent with this study, we found a subset of neuroblastoma cell lines that responded to treatment with poly (I:C), which resulted in multiple changes associated with an improved immune response. This included activation of an inflammatory transcriptome, secretion of immune-recruiting cytokines, activation of THP-1 cell migration by conditioned medium, and increased antigen presentation. Concordantly, poly (I:C) increased the in vitro killing of neuroblastoma cells by T cells, and to a lesser extent NK cells, but only in a responsive line.
Prior studies in neuroblastoma had noted some differences in TLR3 expression and responses to poly (I:C) (63–66), but none linked these differences to neuroblastoma epigenetic states. In retrospect, the responsive cell lines in these studies are from the MES subtype, consistent with our results. Our use of a large cell line panel allowed us to connect MES and ADRN states with dsRNA sensing. By showing that inducing a shift toward the MES state in an unresponsive ADRN cell line is sufficient to convert it to a poly (I:C)–responsive state, we show that epigenetic state is more relevant than MYCN amplification status and connect these epigenetic states to inflammatory sensing. This connection had not been previously made, to our knowledge. However, perhaps more importantly than differences in sensing themselves, this connection led us to the finding of a higher basal inflammatory state in MES cells. This state has increased expression of antigen processing and presentation genes, more surface expression of HLA-I, and higher levels of transcriptomic signatures that correlate with immunotherapy responses (4, 5).
Using a published scRNAseq dataset (57), we were able to show that within human tumors, neuroblastoma cells with stronger MES signatures have higher basal levels of inflammatory signaling. This observation suggests that MES cells are likely to be more responsive to inflammatory stimuli and more sensitive to immunotherapies. Similar to the findings of the group that originally published this dataset, we did not identify a distinct population of tumor cells with mesenchymal markers. However, the tumor cells have substantial transcriptional heterogeneity, including cells with higher MES and lower ADRN signatures. As this dataset is composed exclusively of untreated adrenal primary tumors, it may be that MES populations become more distinct after therapy or at relapse.
The optimal approach to translating knowledge about the status of neuroblastoma immune sensing pathways remains an open question. Future work will be required to understand the relative contributions of tumor cell-intrinsic sensing and of activation of sensors in nontumor cells. As mentioned, IFNγ has been used in neuroblastoma patients in the past, although such a treatment has potential systemic drawbacks. STING agonists have generated significant interest across oncology (67), including neuroblastoma (68). A more stable analog of poly (I:C) was used systemically in neuroblastoma patients (69), without significant efficacy and with marked systemic side effects, although a more recent study using a different dosing scheme showed little toxicity and some efficacy in children with brain tumors (70) and is currently being used widely in clinical trials. These studies did not account for the potential presence of cell states with differential therapeutic responses.
Our data suggest that an understanding of MES and ADRN states may contribute to the choice of the optimum immunostimulatory pathway to use in neuroblastoma and the most advantageous timing. For example, therapies that aim to target STING or TLRs may be more effective at later stages of therapy when responsive MES cells compose a higher percentage of remaining tumor cells. Indeed, the higher basal inflammatory state of residual MES cells may in part explain the effectiveness of anti-GD2 antibody immunotherapy when used in a minimal residual disease state (32). Comparatively, there is a relatively poor response when used alone in the setting of bulk tumor (71), which is likely to contain a significant portion of immunologically quiescent adrenergic cells. However, responses are much better when anti-GD2 therapy is combined with chemotherapy (33, 34), which may allow for targeting of both states simultaneously.
We found that tumor xenografts derived from an MES cell line had markedly increased basal immune infiltration in vivo compared to ADRN line–derived tumors, consistent with the elevated inflammatory state in MES cell lines. To extend our findings, immunocompetent animal models are needed. The lack of tractable immunocompetent models of MES and ADRN states is a limitation of the current study and in the field more broadly. The most widely used neuroblastoma genetically engineered mouse model is the tyrosine hydroxylase (TH)-MYCN model, with MYCN expressed from the TH promoter (72). There is some evidence that cells from these tumors can assume an MES-like state in the setting of treatment resistance (39). However, TH expression is much lower in the MES state (38), which in the TH-MYCN model may decrease expression of MYCN and diminish tumorigenesis. Further work is first needed to explore the MES and ADRN states in the TH-MYCN and/or other neuroblastoma models.
In conclusion, our studies uncover an unforeseen link between neuroblastoma epigenetic states and inflammatory signaling. Only the MES state has functional dsRNA sensing and active signaling through other TLRs. In responsive cells, dsRNA leads to increased inflammatory signaling, antigen presentation, and antitumor immune cytotoxicity. Given the distinct transcriptional and regulatory networks of MES and ADRN cells and the resistance of the MES state to cytotoxic chemotherapy, it is likely that curing neuroblastoma will require simultaneous targeting of both states with separate, but perhaps synergistic, therapies. Our data suggest that immune-directed therapies, particularly harnessing T cells, may be able to exploit the elevated basal inflammatory signaling and sensing of the MES state.
Materials and Methods
Additional information is provided in SI Appendix, Supplementary Materials and Methods and Tables S2 and S3.
Cell Culture.
Cell lines were generous gifts from Drs. Marie Arsenian-Henriksson, Michael Hogarty, John Maris, Michael Milone, Alessandro Gardini, and Linda Valentijn or purchased from either the German Collection of Microorganisms and Cell Cultures or the Interlab Cell Line Collection (specific details in SI Appendix, Table S2). Cells were maintained at 37 °C and 5% CO2 in a humidified incubator and cultured with medium components listed in SI Appendix, Table S2. Cell identities were verified by short tandem repeat profiling and routinely tested negative for mycoplasma contamination.
CRISPR-Cas9.
LentiV_Cas9_neo and the LRG2.1T vectors were gifts from Dr. Junwei Shi. The indicated cells were infected with lentivirus prepared from the LentiV_Cas9_Neo vector and then selected with G418. Guide RNAs were designed using https://www.benchling.com/, purchased from Integrated DNA Technologies, phosphorylated and annealed, and then digested and ligated into the LRG2.1T vector containing green fluorescent protein (GFP). Lentivirus was prepared, and stable Cas9-expressing cells were infected. Single GFP-positive cells were flow sorted into the wells of a 96-well plate in order to isolate single-cell clones. Sequences of single guide RNAs (sgRNAs) are listed in SI Appendix, Table S3.
Cytokine Profiling.
Cytokine profiling was performed by the Human Immunology Core at the University of Pennsylvania using the Luminex FlexMap 3D system (EMD Millipore). A MILLIPLEX human cytokine 48-plex panel (Millipore Sigma HCYTA-60K-PX48) was run using supernatant from cells cultured in the presence or absence of 30 μg/mL poly (I:C) for 24 h. Samples were run in biological triplicate. Cytokine concentrations were obtained by comparison to a standard curve.
T Cell and NK Cell Killing Assays.
Primary human T cells were collected, transfected, and expanded as previously described (47) and stored as frozen aliquots on day 13 after bead activation. T cells were thawed, rested for 1 d, and activated with ImmunoCult Human CD3/CD28 T Cell Activator (Stem Cell Technologies) following the manufacturer’s protocol. T cell cultures were fed with fresh 10% Roswell Park Memorial Institute medium (RPMI) supplemented with human IL-2 (20 IU/mL; Stem Cell Technologies) every 2 to 3 d to maintain 1 × 106 cells/mL. T cells were rested for a minimum of 11 d before use in functional assays. Primary human NK cells were obtained from healthy donors in the Human Immunology Core at the University of Pennsylvania. Fresh NK cells were activated and expanded using NK MACS medium (Miltenyi Biotec, 130-114-429) supplemented with 5% AB human serum (Sigma Millipore, H4522) containing 500 IU/mL recombinant human IL-2 (Peprotech, 200-02) and 5 ng/mL recombinant human IL-15 (Peprotech, 200-15). At day 14 after expansion, expanded NK cells were cryopreserved in fetal bovine serum with 10% dimethyl sulfoxide (DMSO) and added directly to neuroblastoma cells after thawing and centrifugation to remove DMSO. Neuroblastoma cells were seeded in 96-well plates and then treated with vehicle, 30 μg/mL poly (I:C), or 20 ng/mL IFNγ for 24 h. Cells were then washed with medium, and T cells or NK cells were added at the indicated effector-to-target ratio. After 18 h, 12 nonoverlapping fields per well of a 96-well plate were captured with a 10× objective using a Nikon Ti-E Automated Inverted Microscope with NIS-Elements AR. The NIS Jobs template “Fixed Cells” was used to define image acquisition parameters. Tagged image format (TIF) images were processed and converted to simple segmentation images using the Ilastik 1.3.3 (interactive machine learning for [bio]image analysis) tool (73). The simple segmentation images were analyzed for GFP area in CellProfiler 4.07 (cell image analysis software) (74) and exported into Microsoft Excel.
Tumor Xenografts.
Cells were expanded in culture, dissociated, counted, and resuspended in 50% Matrigel in phosphate-buffered saline. They were then injected subcutaneously into the shaved flank of 6- to 8-wk-old male CB17 SCID mice (Taconic), with 4 × 106 cells in 100 μL total volume. Tumors were monitored with digital caliper measurements. Tumors that reached 500 mm3 were injected with either 50 μL of 1 mg/mL VacciGrade poly (I:C) or with saline control. Twenty-four hours after injection, mice were killed, and tumors were dissected. A portion was flash frozen and subsequently used for Western blots, a portion was fixed in formalin for subsequent use in immunohistochemistry (IHC), and a portion was immediately processed for flow cytometric analysis. All animal studies were approved by the Wistar Institute Animal Care and Use Committee (Institutional Animal Care and Use Committee protocol 201189).
QuantSeq.
RNA was extracted using the Qiagen RNeasy Plus mini kit (74134) after homogenization by centrifugation in QIAShredder (79656) microcentrifuge tubes. RNA quantity was determined using the Qubit 2.0 Fluorometer (Thermo Fisher Scientific), and the quality was validated using the TapeStation RNA ScreenTape (Agilent). Two hundred nanograms of DNase I–treated total RNA was used to prepare the library for Illumina sequencing using a QuantSeq 3′ mRNA-Seq Library Preparation kit (Lexogen). Library quantity was determined using qPCR (KAPA Biosystem). Overall library size was determined using the Agilent TapeStation and the DNA High Sensitivity D5000 ScreenTape (Agilent). Equimolar amounts of each sample library were pooled, denatured, and sequenced using high-output, single-read, 75-bp-cycle Illumina NextSeq 500 sequencing kits. Next-generation sequencing was done on a NextSeq 500 (Illumina).
Database and QuantSeq Analysis.
Data were downloaded from the Broad CCLE portal (https://sites.broadinstitute.org/ccle) , from GSE89413 (45), or from PedcBioPortal (https://pedcbioportal.kidsfirstdrc.org:443/saml/discovery?entityID=d3b-center.auth0.com&returnIDParam=idp) (46) as fragments per kilobase of exon per million mapped fragments (FPKM) values and converted to log2 (FPKM + 1). For QuantSeq, raw RNA-seq reads were trimmed using Trimmomatic v.0.39 (75), and data were aligned using the bowtie2 (76) algorithm against the hg38 human genome version. RSEM v1.2.12 software (77) was used to estimate read counts using gene information from Ensembl transcriptome version GRCh37.p13. Mesenchymal and adrenergic gene sets were taken from van Groningen and colleagues (35). Other gene sets were downloaded from the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/index.jsp). Each gene in each signature was z transformed across the samples included in the analysis, and the signature score was calculated as the average of the z-transformed gene scores. Other datasets were analyzed with tools built into the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl).
scRNAseq Analysis.
Count matrices for single-cell analysis were downloaded from GSE137804. Patients with tumors that were not classified as high-risk were excluded from the analysis. Of note, sample T71 was excluded as the clinical description provided does not fit standard Children’s Oncology Group–defined high-risk group status based on age, stage, and MYCN status. Gene expression matrices for each sample were analyzed using the Seurat package (78). DoubletFinder (79) was used to remove doublets. Cells with less than 500 features, 1,000 counts (unique molecular identifier [UMI]/cell), or greater than 10% mitochondrial counts were filtered out. The most variable genes were used to perform principal component analysis, and the top 20 principal components were included to perform Louvain clustering. Uniform manifold approximation and projection (UMAP) was generated using the Seurat package. Samples were integrated using Harmony (80). Cell types were annotated using the following gene markers: PHOX2B, DBH, TH, ISL1, GATA3, and HAND2 for neuroendocrine cells; HGBB and HGBD for red blood cells; CD2, CD3D, and CD247 for T cells; CD79A, PAX5, and BANK1 for B cells; CD14 and CD68 for myeloid cells; COL1A1, LAMA2, and COL3A1 for fibroblasts; and VWF, CALCRL, and FLT1 for endothelial cells. In order to identify malignant cells, inferCNV (81) was used. Specifically, inferCNV was applied for each sample using sympathoblasts, myeloid cells, and fibroblasts from fetal adrenal samples F6, F7, F106, and F107 as a reference. To determine the ADRN, MES, and inflammatory score of each cell, the AddModuleScore function in the Seurat R package was applied using previously published signatures (35, 41). Pearson correlations were calculated between adrenergic and mesenchymal score vectors and between inflammatory and MES score vectors.
Supplementary Material
Acknowledgments
We would like to thank Drs. Michael Milone, Linda Valentijn, and Alessandro Gardini for providing cell lines used in the study; Drs. Zachary Stine and Rebekah Brooks for careful reading of the manuscript and helpful discussions; Dr. Xue Zhang, Dr. Hamid Bassiri, and Chakkapong Burudpakdee for technical advice regarding tumor dissociation and immune profiling; and the Wistar Histotechnology Facility for IHC staining. This work was supported by Alex’s Lemonade Stand Young Investigator Awards (A.J.W. and G.P.-P.), a Hyundai Hope on Wheels Young Investigator Award (A.J.W.), NIH Grants K12 HD043245 and K12 CA076931 (A.J.W.), National Cancer Institute Grant K08 CA230223 (K.R.B.), Solving Kids’ Cancer (K.R.B.), Fishin’ For The Cure (K.R.B.), Pierce Phillips Charity (K.R.B.), Catherine Elizabeth Blair Memorial Foundation (K.R.B.), Ronan Thompson Foundation (K.R.B.), EVAN Foundation (K.R.B.), NIH Grant R50 CA211199 (A.K.), NIH Grants R35 CA220500, P01 CA217959, and U54 CA232568 (J.M.M.), the Giulio D’Angio Endowed Chair (J.M.M.), NIH Grants R01 CA057341 and R01 CA051497 (C.V.D.), and the Ludwig Institute for Cancer Research (C.V.D.).
Footnotes
Competing interest statement: C.V.D. is an advisor to the Barer Institute, Inc.
This article is a PNAS Direct Submission. W.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102358119/-/DCSupplemental.
Data Availability
The 3′ mRNA-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus under accession number GSE182052. Previously published data were used for this work (multiple sources of publicly available RNA-seq and chromatin immunoprecipitation-sequencing data were used and are cited in the text and/or figure legends).
References
- 1.Tumeh P. C., et al. , PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi N. A., et al. , Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yarchoan M., Hopkins A., Jaffee E. M., Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristescu R., et al. , Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayers M., et al. , IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemek R. M., et al. , Sensitization to immune checkpoint blockade through activation of a STAT1/NK axis in the tumor microenvironment. Sci. Transl. Med. 11, eaav7816 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Ishizuka J. J., et al. , Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Junt T., Barchet W., Translating nucleic acid-sensing pathways into therapies. Nat. Rev. Immunol. 15, 529–544 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Apetoh L., et al. , Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 13, 1050–1059 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Bowling E. A., et al. , Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell 184, 384–403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiappinelli K. B., et al. , Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackenzie K. J., et al. , cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boelens M. C., et al. , Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 159, 499–513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., et al. , Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 30, 243–256 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Krenz B., et al. , MYC- and MIZ1-dependent vesicular transport of double-strand RNA controls immune evasion in pancreatic ductal adenocarcinoma. Cancer Res. 81, 4242–4256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sistigu A., et al. , Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 20, 1301–1309 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Harding S. M., et al. , Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Maris J. M., Recent advances in neuroblastoma. N. Engl. J. Med. 362, 2202–2211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodeur G. M., Seeger R. C., Schwab M., Varmus H. E., Bishop J. M., Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224, 1121–1124 (1984). [DOI] [PubMed] [Google Scholar]
- 21.Coughlin C. M., et al. , Immunosurveillance and survivin-specific T-cell immunity in children with high-risk neuroblastoma. J. Clin. Oncol. 24, 5725–5734 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Pugh T. J., et al. , The genetic landscape of high-risk neuroblastoma. Nat. Genet. 45, 279–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wölfl M., et al. , Expression of MHC class I, MHC class II, and cancer germline antigens in neuroblastoma. Cancer Immunol. Immunother. 54, 400–406 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raffaghello L., et al. , Multiple defects of the antigen-processing machinery components in human neuroblastoma: Immunotherapeutic implications. Oncogene 24, 4634–4644 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Layer J. P., et al. , Amplification of N-Myc is associated with a T-cell-poor microenvironment in metastatic neuroblastoma restraining interferon pathway activity and chemokine expression. OncoImmunology 6, e1320626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P., et al. , MYCN amplification is associated with repressed cellular immunity in neuroblastoma: An in silico immunological analysis of TARGET database. Front. Immunol. 8, 1473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei J. S., et al. , Clinically relevant cytotoxic immune cell signatures and clonal expansion of T-cell receptors in high-risk MYCN-not-amplified human neuroblastoma. Clin Cancer Res. 24, 5673–5684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernards R., Dessain S. K., Weinberg R. A., N-myc amplification causes down-modulation of MHC class I antigen expression in neuroblastoma. Cell 47, 667–674 (1986). [DOI] [PubMed] [Google Scholar]
- 29.Davis K. L., et al. , Nivolumab in children and young adults with relapsed or refractory solid tumours or lymphoma (ADVL1412): A multicentre, open-label, single-arm, phase 1-2 trial. Lancet Oncol. 21, 541–550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merchant M. S., et al. , Phase I clinical trial of ipilimumab in pediatric patients with advanced solid tumors. Clin. Cancer Res. 22, 1364–1370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards R. M., Sotillo E., Majzner R. G., CAR T cell therapy for neuroblastoma. Front. Immunol. 9, 2380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu A. L., et al. , Children’s Oncology Group, Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med. 363, 1324–1334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mody R., et al. , Irinotecan-temozolomide with temsirolimus or dinutuximab in children with refractory or relapsed neuroblastoma (COG ANBL1221): An open-label, randomised, phase 2 trial. Lancet Oncol. 18, 946–957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mody R., et al. , Irinotecan, temozolomide, and dinutuximab with GM-CSF in children with refractory or relapsed neuroblastoma: A report from the Children’s Oncology Group. J. Clin. Oncol. 38, 2160–2169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Groningen T., et al. , Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 49, 1261–1266 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Boeva V., et al. , Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 49, 1408–1413 (2017). [DOI] [PubMed] [Google Scholar]
- 37.van Groningen T., et al. , A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat. Commun. 10, 1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Wezel E. M., et al. , Mesenchymal neuroblastoma cells are undetected by current mRNA marker panels: The development of a specific neuroblastoma mesenchymal minimal residual disease panel. JCO Precis. Oncol. 3, PO.1800413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yogev O., et al. , In vivo modeling of chemoresistant neuroblastoma provides new insights into chemorefractory disease and metastasis. Cancer Res. 79, 5382–5393 (2019). [DOI] [PubMed] [Google Scholar]
- 40.van Nes J., et al. , A NOTCH3 transcriptional module induces cell motility in neuroblastoma. Clin. Cancer Res. 19, 3485–3494 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Liberzon A., et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghandi M., et al. , Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lampson L. A., George D. L., Interferon-mediated induction of class I MHC products in human neuronal cell lines: Analysis of HLA and beta 2-m RNA, and HLA-A and HLA-B proteins and polymorphic specificities. J. Interferon Res. 6, 257–265 (1986). [DOI] [PubMed] [Google Scholar]
- 44.Reid G. S., et al. , Interferon-gamma-dependent infiltration of human T cells into neuroblastoma tumors in vivo. Clin. Cancer Res. 15, 6602–6608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harenza J. L., et al. , Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci. Data 4, 170033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rokita J. L., et al. , Genomic profiling of childhood tumor patient-derived xenograft models to enable rational clinical trial design. Cell Rep. 29, 1675–1689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson L. A., et al. , Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J. Immunol. 177, 6548–6559 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajbhandari P., et al. , Cross-cohort analysis identifies a TEAD4-MYCN positive feedback loop as the core regulatory element of high-risk neuroblastoma. Cancer Discov. 8, 582–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valentijn L. J., et al. , Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc. Natl. Acad. Sci. U.S.A. 109, 19190–19195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henriksen J. R., et al. , Conditional expression of retrovirally delivered anti-MYCN shRNA as an in vitro model system to study neuronal differentiation in MYCN-amplified neuroblastoma. BMC Dev. Biol. 11, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valentijn L. J., et al. , Inhibition of a new differentiation pathway in neuroblastoma by copy number defects of N-myc, Cdc42, and nm23 genes. Cancer Res. 65, 3136–3145 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Ushmorov A., et al. , N-myc augments death and attenuates protective effects of Bcl-2 in trophically stressed neuroblastoma cells. Oncogene 27, 3424–3434 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Ross R. A., Spengler B. A., Biedler J. L., Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J. Natl. Cancer Inst. 71, 741–747 (1983). [PubMed] [Google Scholar]
- 54.O’Neill L. A., Bowie A. G., The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7, 353–364 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Schweigerer L., et al. , Augmented MYCN expression advances the malignant phenotype of human neuroblastoma cells: Evidence for induction of autocrine growth factor activity. Cancer Res. 50, 4411–4416 (1990). [PubMed] [Google Scholar]
- 56.Biedler J. L., Spengler B. A., Chang T. D., Ross R. A., Transdifferentiation of human neuroblastoma cells results in coordinate loss of neuronal and malignant properties. Prog. Clin. Biol. Res. 271, 265–276 (1988). [PubMed] [Google Scholar]
- 57.Dong R., et al. , Single-cell characterization of malignant phenotypes and developmental trajectories of adrenal neuroblastoma. Cancer Cell 38, 716–733 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Yang X., et al. , Induction of caspase 8 by interferon gamma renders some neuroblastoma (NB) cells sensitive to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) but reveals that a lack of membrane TR1/TR2 also contributes to TRAIL resistance in NB. Cancer Res. 63, 1122–1129 (2003). [PubMed] [Google Scholar]
- 59.Wexler L. H., et al. , “Adoptive immunotherapy of refractory neuroblastoma with tumor-infiltrating lymphocytes, interferon-gamma, and interleukin-2” in Proceedings of the American Society of Clinical Oncology Annual Meeting (1992), p. 368.
- 60.Molenaar J. J., et al. , Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Koche R. P., et al. , Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat. Genet. 52, 29–34 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chuang J. H., et al. , Preferential involvement of mitochondria in Toll-like receptor 3 agonist-induced neuroblastoma cell apoptosis, but not in inhibition of cell growth. Apoptosis 17, 335–348 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Chuang J. H., et al. , Differential toll-like receptor 3 (TLR3) expression and apoptotic response to TLR3 agonist in human neuroblastoma cells. J. Biomed. Sci. 18, 65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsu W. M., et al. , Toll-like receptor 3 expression inhibits cell invasion and migration and predicts a favorable prognosis in neuroblastoma. Cancer Lett. 336, 338–346 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Lin L. L., et al. , Downregulation of c-Myc is involved in TLR3-mediated tumor death of neuroblastoma xenografts. Lab. Invest. 96, 719–730 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Boes M., Meyer-Wentrup F., TLR3 triggering regulates PD-L1 (CD274) expression in human neuroblastoma cells. Cancer Lett. 361, 49–56 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Flood B. A., Higgs E. F., Li S., Luke J. J., Gajewski T. F., STING pathway agonism as a cancer therapeutic. Immunol. Rev. 290, 24–38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang-Bishop L., et al. , Potent STING activation stimulates immunogenic cell death to enhance antitumor immunity in neuroblastoma. J. Immunother. Cancer 8, e000282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lampkin B. C., Levine A. S., Levy H., Krivit W., Hammond D., Phase II trial of a complex polyriboinosinic-polyribocytidylic acid with poly-l-lysine and carboxymethyl cellulose in the treatment of children with acute leukemia and neuroblastoma: A report from the Children’s Cancer Study Group. Cancer Res. 45, 5904–5909 (1985). [PubMed] [Google Scholar]
- 70.Hartman L. L., et al. , Pediatric phase II trials of poly-ICLC in the management of newly diagnosed and recurrent brain tumors. J. Pediatr. Hematol. Oncol. 36, 451–457 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu A. L., et al. , Phase I trial of a human-mouse chimeric anti-disialoganglioside monoclonal antibody ch14.18 in patients with refractory neuroblastoma and osteosarcoma. J. Clin. Oncol. 16, 2169–2180 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Weiss W. A., Aldape K., Mohapatra G., Feuerstein B. G., Bishop J. M., Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 16, 2985–2995 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berg S., et al. , ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Kamentsky L., et al. , Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Korsunsky I., et al. , Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tirosh I., et al. , Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bindea G., et al. , Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Zhang W., et al. , Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 16, 133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3′ mRNA-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus under accession number GSE182052. Previously published data were used for this work (multiple sources of publicly available RNA-seq and chromatin immunoprecipitation-sequencing data were used and are cited in the text and/or figure legends).