Significance
In persons living with HIV-1 who start antiretroviral therapy, virus in the blood decreases rapidly to below the detection limit. The decrease occurs in two phases: a rapid initial decrease in the first weeks, followed by a second, slower phase occurring over the next few months. These decay processes are important because infected cells that remain may become part of the stable latent reservoir that prevents cure. The decay in virus levels in blood presumably reflects the loss of infected cells, but the relationship between the decay of free virus and of infected cells has been unclear. Here, we have analyzed this question using an assay that distinguishes between cells with intact and defective forms of the viral genome.
Keywords: HIV, viral dynamics, latent reservoir, IPDA, 2LTR circles
Abstract
In persons living with HIV-1 (PLWH) who start antiretroviral therapy (ART), plasma virus decays in a biphasic fashion to below the detection limit. The first phase reflects the short half-life (<1 d) of cells that produce most of the plasma virus. The second phase represents the slower turnover (t1/2 = 14 d) of another infected cell population, whose identity is unclear. Using the intact proviral DNA assay (IPDA) to distinguish intact and defective proviruses, we analyzed viral decay in 17 PLWH initiating ART. Circulating CD4+ T cells with intact proviruses include few of the rapidly decaying first-phase cells. Instead, this population initially decays more slowly (t1/2 = 12.9 d) in a process that largely represents death or exit from the circulation rather than transition to latency. This more protracted decay potentially allows for immune selection. After ∼3 mo, the decay slope changes, and CD4+ T cells with intact proviruses decay with a half-life of 19 mo, which is still shorter than that of the latently infected cells that persist on long-term ART. Two-long-terminal repeat (2LTR) circles decay with fast and slow phases paralleling intact proviruses, a finding that precludes their use as a simple marker of ongoing viral replication. Proviruses with defects at the 5′ or 3′ end of the genome show equivalent monophasic decay at rates that vary among individuals. Understanding these complex early decay processes is important for correct use of reservoir assays and may provide insights into properties of surviving cells that can constitute the stable latent reservoir.
For persons living with HIV-1 (PLWH), lifelong adherence to antiretroviral therapy (ART) is critical for maintaining suppression of viral replication and forestalling the development of fatal immunodeficiency. Following initiation of ART, plasma virus levels decay rapidly to below the limit of detection of clinical assays (1–6). Because antiretroviral drugs block new infection of susceptible cells, but not virus production by cells that have an integrated viral genome, this decay must reflect the loss of productively infected cells, cells that were infected prior to the initiation of ART. Productively infected cells could die from viral cytopathic effects, cytolytic host effector mechanisms, or virus-independent T cell turnover. In principle, the decay of plasma virus could also be explained by transition to a nonproductive or latent state of infection. Importantly, the decay is biphasic, indicating the presence of two populations of productively infected cells with different half-lives. Most of the plasma virus is produced by cells that decay very rapidly, with a half-life of less than 1 d. Perelson et al. (4) showed that after most of these cells have decayed, the slope changes, reflecting the slower elimination of a second population of productively infected cells. This population decays with a variable half-life (mean ∼ 2 wk). Although this biphasic decay is a consistent feature of the response to ART, there is still uncertainty about the nature, anatomic location, and fate of the cells responsible for virus production during the first and second phases of decay (referred to here as first- and second-phase cells, respectively). The differences between these two populations have never been elucidated.
The first and second phases of decay bring viremia down to below the limit of detection of clinical assays (typically 20 to 50 copies of HIV-1 RNA per mL of plasma) within months of ART initiation, initially raising hope for eradication. However, a latent form of the virus persists in resting memory CD4+ T cells (7–14). Initial studies used a quantitative viral outgrowth assay (QVOA) to demonstrate that latently infected resting CD4+ T cells purified from PLWH on long-term suppressive ART could be induced to produce replication-competent virus by global T cell activation (8, 9). Longitudinal studies using the QVOA demonstrated that the half-life of the latent reservoir in resting CD4+ T cells is 44 mo in PLWH who are adherent to ART. This half-life is long enough to guarantee lifetime persistence of HIV-1 despite ART (12–14). Strategies targeting the latent reservoir in resting CD4+ T cells are a major focus of HIV cure research (15–17). In addition to resting CD4+ T cells, other cell types may contribute to HIV-1 persistence (18–20).
Prior to and immediately following initiation of ART, the frequency of latently infected cells detected by QVOA is substantially higher than frequencies observed in PLWH on long-term ART (21). In principle, several different types of decay processes occurring over the first 6 to 12 mo of treatment could reduce the frequency of latently infected cells to the more stable frequencies observed in PLWH on long-term ART. Early studies by Jerome Zack and Mario Stevenson demonstrated that infected resting CD4+ T cells could harbor linear, unintegrated HIV-1 DNA in a state of preintegration latency (22, 23). Following cellular activation, linear unintegrated HIV-1 DNA can be integrated and transcribed, allowing production of virus (22, 23). The half-life of linear, unintegrated forms of the viral genome is not clear, with some studies suggesting that these forms are labile (22, 24–26). Some reverse-transcribed viral genomes can undergo homology-dependent or end-to-end ligation, generating one-long-terminal repeat or two-long-terminal repeat (2LTR) circles, respectively (reviewed in ref. 27). The stability of these forms is also controversial, but they are clearly replication-defective (27–31). Following integration of linear viral cDNA, decay dynamics depend on dynamics of the infected host cells, which can be eliminated by viral cytopathic effects, immune cytolytic effector mechanisms, and normal contraction-phase death of previously activated CD4+ T cells (32, 33).
While the QVOA provides a definitive minimal estimate of the frequency of latently infected cells, it underestimates reservoir size because not all proviruses in resting CD4+ T cells are induced upon one round of maximum T cell activation (34–36). Many replication-competent proviruses require multiple rounds of stimulation for induction. As an alternative to the QVOA, many studies use PCR-based assays to measure proviral DNA. However, the vast majority of HIV-1 proviruses are defective due to apolipoprotein B messenger RNA editing enzyme, catalytic polypeptide-like (APOBEC)-mediated hypermutation or large internal deletions (34, 37–39). PCR-based assays do not distinguish between defective and intact proviruses (40, 41). Although infected cell dynamics have been explored using PCR-based assays (42), the results likely reflect the dynamics of defective proviruses (41). The recently developed intact proviral DNA assay (IPDA) uses two carefully chosen amplicons to probe informative regions of individual proviruses to provide better discrimination between intact and defective proviruses (41, 43). This assay has proven useful in evaluating the long-term dynamics of cells with intact and defective proviruses, demonstrating differences in decay rates that may reflect some vulnerability of cells with intact proviruses to immune effector mechanisms (41, 44, 45).
In this study, we use the IPDA to explore the decay of intact and defective proviruses at early time points following initiation of ART. We identify decay processes occurring over intermediate time scales, but with pronounced differences between intact and defective proviruses. Of particular importance is the second-phase decay because infected cells that survive second-phase decay may down-regulate HIV-1 gene expression and enter the stable latent reservoir. Our findings also provide insight into mechanisms for the elimination of the cells with intact viral genomes and into the proper use of assays for the latent reservoir.
Results
Characteristics of Study Participants.
We studied viral decay processes in 17 PLWH prior to and at very early times following initiation of ART (SI Appendix, Table S1). All participants were male with a median age of 31.5 y. The median nadir CD4+ T cell count was 331 cells per μL. Twelve individuals were ART-naïve. The remaining 5 had interrupted ART for >6 mo and were reinitiating ART. Plasma HIV-1 RNA, IPDA, and two-long-terminal repeat (2LTR) circle measurements were taken at the time of ART initiation (time 0) and then every 2 wk for the next 3 mo. Subsequently, samples were obtained once a month for a median total follow-up time of 12.4 mo. A total of 231 IPDA measurements across the cohort were performed. On average, 13.6 samples per participant were studied. All participants were on regimens including two nucleoside/nucleotide reverse-transcriptase inhibitors and an integrase strand transfer inhibitor (InSTI); one participant’s regimen also included a protease inhibitor (SI Appendix, Table S1).
Biphasic Decay of Plasma HIV-1 RNA.
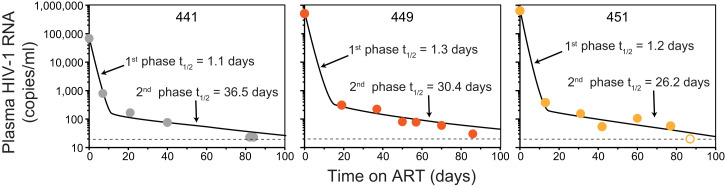
In each participant, initiation of ART resulted in similarly rapid decline in plasma HIV-1 RNA levels (Fig. 1 and SI Appendix, Tables S2 and S3). We used a mixed-effect modeling approach to fit either a single-phase or two-phase decay model to plasma HIV-1 RNA levels. The decay of plasma virus was best fit by a two-phase decay model. This biphasic decay was most easily visualized in participants who had high baseline levels of viremia, such that two phases of decay could be discerned before plasma HIV-1 RNA levels fell below the limit of detection (Fig. 1), but similar decay was observed in all 17 participants (SI Appendix, Tables S2 and S3). An initial first-phase decay of two to three logs was followed by a slower second phase, consistent with previous observations (1–4). Except for one participant, who had persistent low-level viremia (no. 458), plasma HIV-1 RNA levels dropped to below the limit of detection in a median time of 1.8 mo (range 0.43 to 6.44 mo). The half-life of the first phase of plasma virus decay was 1.28 d (95% CI: 1.04 to 1.57 d). Because plasma virus levels decay so rapidly after initiation of ART, frequent early sampling is required to measure the first-phase half-life precisely. Therefore, it is likely that these half-life values are maximal estimates. With more frequent early sampling, half-lives of less than 1 d would likely have been obtained, consistent with previous reports (1–4). The median fraction of plasma HIV-1 RNA that decayed in the first phase was 99.92%. The first-phase half-life was independent of the starting level of plasma virus (R2 = 0.012).
Fig. 1.
Decay of plasma HIV-1 RNA levels following initiation of ART in three representative study participants with high baseline plasma HIV-RNA measurements (68500, 265092, and 643000 copies per mL for 441, 449, and 451, respectively). Decay parameters for all study participants are given in SI Appendix, Tables S2 and S3. Data were fitted to a biexponential decay model (black lines), as described in Methods. Open symbol, plasma HIV-1 RNA value below the limit of detection.
After the first 2 wk of ART, the slope of the decay curve for plasma virus changes dramatically (Fig. 1), consistent with a much slower second phase, as described (4). The model-derived population estimate of the half-life of the second phase of plasma virus decay was 30.6 d (95% CI: 20.6 to 45.4 d; SI Appendix, Table S2). The rate of decay for the second phase (λ2 = 0.0227 d−1) was much slower (∼24-fold) than the mean rate of decay for the first phase (λ1 = 0.542 d−1). The second phase of decay reflects the turnover of a second population of productively infected cells that produce <1% of plasma virus. Although these cells decay more slowly than the cells that produce most of the plasma virus, their decay rate is still much faster than the decay rate of latently infected cells in PLWH on long-term ART (t1/2 = 3.7 y) (12–14). The first- and second-phase half-lives were not significantly different for donors starting their first ART regimen compared to those reinitiating ART after a treatment interruption (P = 0.75 and 0.34 for the first and second phases, respectively). These results confirm in PLWH taking modern ART regimens including InSTIs that plasma virus levels initially decay in a striking biexponential fashion following the initiation of treatment. Studies with more frequent sampling and ultrasensitive assays have identified additional complexities in the decay dynamics related to where the InSTIs act in the virus life cycle (26, 46–48). However, the overall pattern of biphasic decay (Fig. 1) is similar to that originally described by Perelson et al. (4).
Decay of Infected CD4+ T Cells following Initiation of ART.
The rapid first-phase decay of plasma virus should reflect the correspondingly rapid elimination of the productively infected cells that produce most of the plasma virus. However, it has been difficult to validate this prediction using standard PCR assays for proviral DNA because these assays mainly detect defective proviruses (34, 40). Therefore, we used the IPDA to measure the decay rate of cells carrying intact proviruses in the blood of PLWH who were initiating ART (Fig. 2A). The IPDA is a digital droplet PCR assay that distinguishes potentially intact proviruses from those carrying common fatal defects, including large deletions and APOBEC3-mediated hypermutation (41). It also provides direct quantitation of most defective proviruses. It has been successfully used to demonstrate differences in the long-term decay rates of cells with intact and defective proviruses (41, 44, 45). Here, we used the IPDA to examine infected cell decay immediately after initiation of ART.
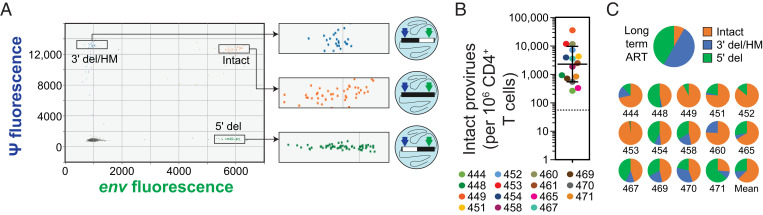
Fig. 2.
IPDA analysis of CD4+ T cells from PLWH prior to ART initiation. (A) Representative baseline (pre-ART) IPDA dot plot showing fluorescent signal from the packaging signal (ψ) amplicon at the 5′ end of the genome (y axis) and the env amplicon at the 3′ end of the genome (x axis) for individual proviruses suspended in nanoliter-sized droplets (illustrated in diagrams to the Right of each expanded box). Proviruses with 5′ deletions (5′ del) appear in the lower right quadrant. Proviruses with 3′ deletions and/or APOBEC3-mediated hypermutation (3′ del/HM) appear in the upper left quadrant. Intact proviruses are directly counted as double-positive droplets in the upper right quadrant. Raw droplet counts are corrected for shearing between the amplicons. For assay details, see ref. 41. (B) Baseline frequency of intact proviruses in circulating CD4+ T cells prior to initiation of ART. Solid lines, geometric mean ± SD; dotted line, median frequency of intact proviruses among 400 PLWH on long-term ART for comparison, based on ref. 43. Baseline samples were not available for participants 441 and 447. (C) Mean fraction of intact, 5′ del, and 3′ del/HM proviruses in CD4+ T cells from PLWH. (C, Upper) Mean fractions from 400 PLWH on long-term ART (for comparison, based on ref. 43). (C, Lower) Fractions for individual participants in this study prior to ART. Baseline samples were not available for participants 441 and 447. For participant 461, defective proviruses could not be detected above the background level of single positive droplets generated by DNA shearing.
Prior to ART, Circulating CD4+ T Cells with Intact Proviruses Are Present at High Frequency and Outnumber Cells with Defective Proviruses.
At the time of ART initiation, the geometric mean frequency of cells with intact proviruses was 2,255 copies per 106 CD4+ T cells (Fig. 2B). This value is 42-fold higher than the median frequency observed in large-scale IPDA analysis of PLWH on long-term ART [54 copies per 106 CD4+ T cells, n = 400 (43)]. These results suggest that substantial decay in the number of cells with intact proviruses must occur during the initial months of ART, and the studies described below define the decay rates. In IPDA analysis of CD4+ T cells from PLWH on long-term ART, intact proviruses represented only a small fraction of all proviruses detected (8%) and were greatly outnumbered by defective proviruses (by an average of 12.5-fold) (41, 43). However, we found that at the time of ART initiation, intact proviruses represented the majority of proviruses detected (mean 63%; Fig. 2C), although there was substantial variation among participants. The high frequency of intact proviruses and the preponderance of intact over defective proviruses are both consistent with active viral replication occurring in the absence of ART.
Circulating CD4+ T Cells with Intact Proviruses Do Not Show First-Phase Decay Kinetics.
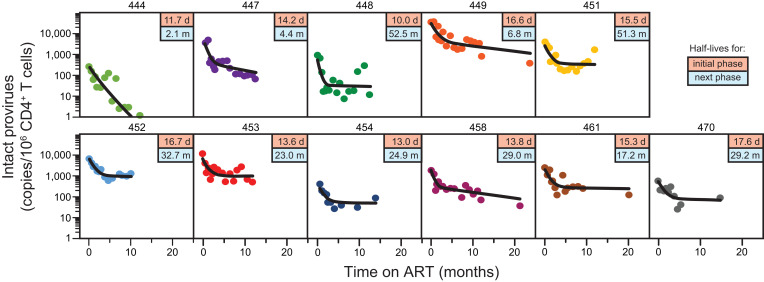
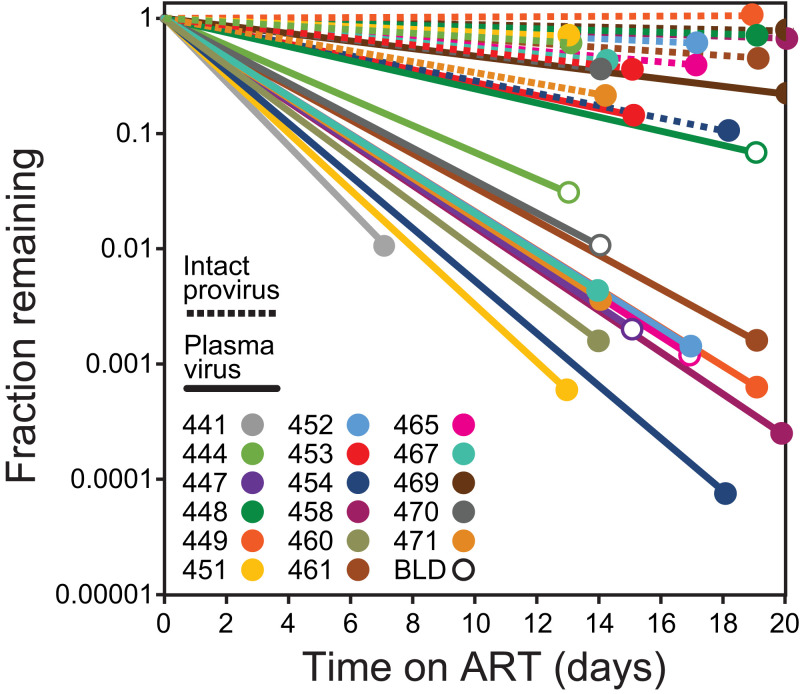
To assess the decay of CD4+ T cells with intact proviruses following the initiation of ART, we carried out IPDA measurements every 2 to 4 wk during the first year of treatment in all study participants. We found a striking biphasic pattern of decay of intact proviruses during the first year of ART (Fig. 3 and SI Appendix, Table S4 and Fig. S1). Using mixed-effect modeling, we determined the half-lives for each phase (SI Appendix, Tables S4 and S5). The population-level estimate for the half-life for the initial decay of cells with intact proviruses was 0.425 mo (12.9 d, 95% CI: 10.3 to 21.5 d; SI Appendix, Table S4). This is at least 10 times slower than the first-phase decay rate of plasma virus in the same participants (t1/2 < 1.28 d). This difference is also evident in the degree of decrease observed between ART initiation and the first on-ART time point 2 to 3 wk later (Fig. 4). During the first 2 to 3 wk of ART, plasma virus levels fell by an average of over 3 logs, while intact proviruses declined by less than 0.3 logs. These results demonstrate that the vast majority of circulating CD4+ T cells carrying intact proviruses are distinct from the rapidly decaying first-phase cells that produce most of the plasma virus. Our data strongly suggest that the first-phase cells are located in the tissues and not in the circulation.
Fig. 3.
Biphasic decay of circulating CD4+ T cells with intact proviruses. IPDA measurements (circles) and the best-fit biexponential decay model (black line) are shown for 11 participants for whom samples were obtained throughout the first year of ART. Boxed values in the upper right of each panel are the half-lives for the initial phase (top, in days) and the next phase (bottom, in months) based on the model. Similar results were obtained for an additional four participants sampled for shorter periods of time (SI Appendix, Fig. S1). Samples for early time points for participant 441 were not available. Participant 460 had an unusual IPDA dot plot pattern suggestive of multiple expanded clones (SI Appendix, Fig. S2) and was not included for that reason.
Fig. 4.
Comparison of initial decay rates for plasma virus and intact proviruses. Lines show fractional decrease in plasma virus (solid lines) and intact proviruses (dotted lines) between the baseline (pre-ART) timepoint and the first on-ART time point 2 to 3 wk later. For some participants, the first on-ART plasma virus measurement was below the limit of detection (BLD; open circles), and it is likely that the initial decay slope is even steeper than that shown. Samples were not available to measure early proviral decay in participants 441 and 447. Participant 460 had an unusual IPDA dot plot pattern suggestive of multiple expanded clones (SI Appendix, Fig. S2) and was not included in the analysis of proviral decay for that reason.
Three Distinct Phases of Decay during the First Year of ART.
Through the combined measurement of plasma virus and intact proviruses in circulating CD4+ T cells, we can define at least three distinct populations of infected cells with strikingly different decay rates during the first year of ART (Fig. 5). Over 99% of the plasma virus is produced by cells that turn over with a half-life measured in hours to days. As discussed above, these rapidly decaying cells are not evident among circulating CD4+ T cells carrying intact proviruses, which decay more slowly, with a half-life measured in weeks, similar to the decay rate of the cells responsible for the second-phase decay of viremia. The initial decay of circulating CD4+ T cells carrying intact proviruses (t1/2 = 12.9 d) is kinetically similar to the second-phase decay of plasma virus reported in the original viral dynamic studies of Perelson et al. (4) (t1/2 = 14.1 d). For clarity, we reserve the terms first and second phase for the decay of viremia, and we describe the biphasic decay of intact proviruses as having an “initial phase” and a subsequent “next phase.” The initial decay of circulating CD4+ T cells with intact provirus is actually slightly faster than the second-phase decay of plasma virus in the same study participants (t1/2 12.9 vs. 30.6 d; SI Appendix, Tables S2 and S4). Although this difference is significant (P < 0.0001), both decay processes are occurring on a time scale of weeks, potentially allowing immune selection to alter the composition of the surviving population. Importantly, our results are not confounded by defective proviruses, which are measured separately by the IPDA and which decay with different kinetics (see below). Interestingly, our results also suggest that the initial decay of circulating CD4+ T cells with intact proviruses is due, for the most part, to their elimination rather than transition to a nonproductive or latent state of infection since such a transition would not explain the observed decrease in the number of intact proviruses.
Fig. 5.
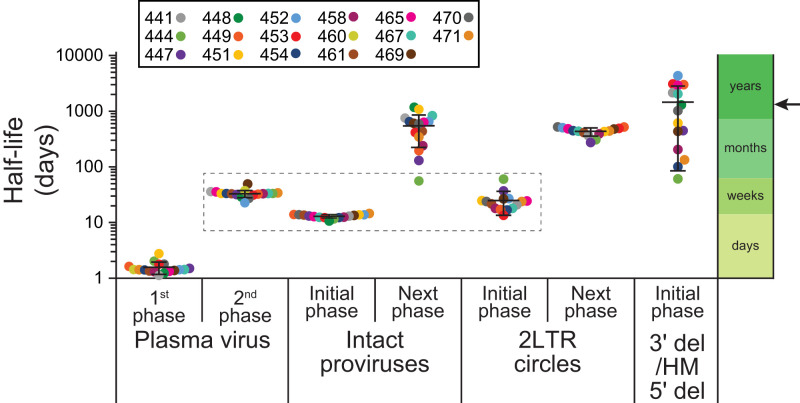
Comparison of the first- and second-phase half-lives for plasma virus, the initial- and next-phase half-lives for intact proviruses and env+ 2LTR circles, and the single-phase half-lives of defective proviruses. Half-lives are plotted on a logarithmic time axis. Bars show mean ± SD of the estimated individual parameter values based on all study participants. Participant 460 had an unusual IPDA dot plot pattern suggestive of multiple expanded clones (SI Appendix, Fig. S2) and was not included in the analysis of proviral decay. The second-phase decay of plasma virus is kinetically similar to the initial decay of cells with intact proviruses and cells with 2LTR circles (dashed box). Arrow indicates the half-life of latently infected CD4+ T cells in PLWH on long-term ART, as measured by the QVOA (12–14).
As shown in Fig. 3, the slope of the decay curve for circulating CD4+ T cells with intact proviruses changes markedly after the first 3 to 4 mo of ART. After the initial decline, there is a subsequent (next) phase, in which intact proviruses decay at a slower rate. The half-life of the next phase was 19.0 mo, with wide 95% CIs (8.2 to 43.7 mo) (SI Appendix, Tables S4 and S5). This decay rate is still faster than the 44-mo value that was reported for latently infected CD4+ T cells in PLWH on long-term ART (12–14, 45). Thus, additional phases of decay may become apparent with long-term follow-up (42, 44, 45, 49). The more rapid initial decay of intact proviruses (t1/2 = 12.9 d) means that accurate measurements of the stable latent reservoir cannot be made during the first 3 to 4 mo of ART. Together, these results delineate at least three distinct populations of infected cells with half-lives measured in days, weeks, and months-to-years.
Monophasic Decay of Cells with Defective Proviruses.
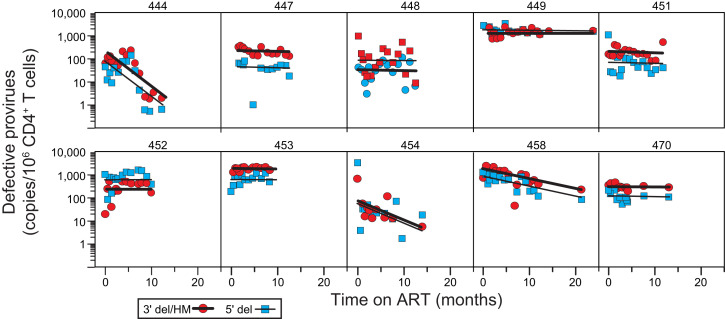
We also used the IPDA to measure decay of cells carrying defective proviruses (Figs. 5 and 6 and SI Appendix, Tables S4 and S6). The IPDA separately quantitates proviruses with deletions in the 5′ end of the genome and proviruses with deletions at the 3′ end of the genome and/or APOBEC3-mediated hypermutation (Fig. 2A) (41). Analysis of the decay of cells with defective proviruses revealed two striking features. First, in contrast to the decay of viremia and of cells with intact proviruses, the decay data for cells with defective proviruses were best fit by a single exponential model (Fig. 6 and SI Appendix, Tables S4 and S6). For most participants, defective proviruses did not show a clear biphasic pattern of decay. Rather, defective proviruses declined slowly in most participants and more rapidly in a subset of participants (see below). A second striking feature is that, within a given donor, proviruses with 5′ deletions and those with 3′ deletions and/or hypermutation behaved similarly. This is surprising, given that these types of defective proviruses may differ in the capacity for viral gene expression. For example, many proviruses with 3′ deletions and/or hypermutation cannot produce the viral transactivator Tat (41). Nevertheless, several recent studies indicate that some defective proviruses can give rise to viral RNA and protein production (50, 51), which could contribute to the more rapid decay observed in a subset of donors. Decay of defective proviruses was observed in 7 of the 15 donors for whom defective provirus measurements were available (Fig. 6 and SI Appendix, Tables S4 and S6). In the subset with observable decay, the estimated half-life was 6.2 mo for proviruses with 3′ deletions and/or hypermutation and 5.17 mo for proviruses with 5′ deletions. Considering all 15 donors for whom defective provirus measurements were available, the estimated half-life was 62 mo for proviruses with 3′ deletions and/or hypermutation and 30 mo for proviruses with 5′ deletions. The unique aspects of the decay of defective proviruses described here highlight the importance of distinguishing intact and defective proviruses in reservoir assays.
Fig. 6.
Decay of defective proviruses in circulating CD4+ T cells. Proviruses with 3′ deletions and/or APOBEC3-mediated hypermutation (red circles, thick black line) and proviruses with 5′ deletions (blue squares, thin black line) were separately quantitated as single positive droplets using the IPDA (Fig. 2A). Lines show the best-fit single-exponential decay model for each participant. Data are shown for participants for whom samples were obtained throughout the first year of ART. Decay parameters for all participants are given in SI Appendix, Table S6. Samples for early time points for participants 441 and 447 were not available. Defective proviruses could not be accurately measured for participant 461 over the background level of single positive events generated by DNA shearing. Baseline measurements of 3′ defective proviruses for participants 449 and 453 could not be made for the same reason. Participant 460 had an unusual IPDA dot plot pattern suggestive of multiple expanded clones (SI Appendix, Fig. S2) and was not included in the analysis of proviral decay.
Kinetics of 2LTR Circles.
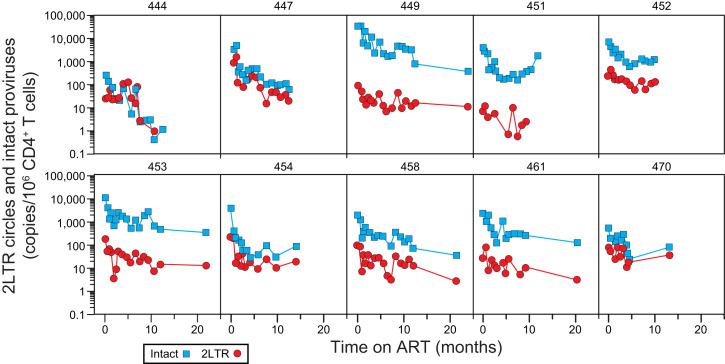
We also measured the decay of 2LTR circles following initiation of ART in the same donors. As previously described for simian immunodeficiency virus (SIV) (52), some 2LTR circles have deletions and/or hypermutation. We quantitated 2LTR circles by digital droplet PCR using the IPDA env amplicon (41) and an amplicon spanning the 2LTR junction (53), allowing us to separately quantitate env+ and env− circles (SI Appendix, Fig. S3 A and B). Prior to ART, frequency of cells with 2LTR circles was substantially lower than the frequency of cells with intact proviruses (on average, 32-fold; SI Appendix, Fig. S3C). The ratio of intact proviruses to 2LTR circles at the time of ART initiation tended to be higher in donors with higher starting viral loads (SI Appendix, Fig. S3C). Following initiation of ART, there was a striking parallel in the decay of intact proviruses and 2LTR circles (Fig. 7 and SI Appendix, Tables S4 and S7). In both cases, a biexponential decay model provided the best fit for the data. Population-level estimates for the half-lives of the initial and next phases of decay of 2LTR circles were 0.789 mo (24 d) and 14.8 mo, respectively (SI Appendix, Table S4). These values are similar to the half-lives of the initial and next phases of decay of intact proviruses (12.9 d and 19 mo, respectively; Fig. 5 and SI Appendix, Table S4). The similar biexponential decay of intact proviruses and 2LTR circles illustrated in Fig. 5 may reflect similar processes affecting cells with these different forms of HIV-1 DNA. Importantly, the 2LTR circles are diluted by cell proliferation (29, 30), which may account for the fact that their long-term decay is more rapid than that of defective proviruses.
Fig. 7.
Decay of 2LTR circles in circulating CD4+ T cells. The 2LTR circles (red circles) were measured by duplex digital droplet PCR as droplets with fluorescent signal for the both the circle junction and env amplicons (SI Appendix, Fig. S3 A and B). Intact provirus measurements (blue squares) for the same samples are shown for comparison. Data are show for participants for whom samples were obtained throughout the first year of ART. Circles could not be measured for participant 448 due to circle junction polymorphisms.
Discussion
Analysis of viral decay processes has contributed valuable insights into mechanisms of HIV-1 pathogenesis and persistence. Classic studies defined the rapid biphasic decay of viremia following initiation of ART and attributed it to the elimination of two populations of cells with half-lives measured in hours and in weeks (1–4). Defining the nature of these populations has proven challenging because accurate measurement of the decay of infected cells is more difficult than measuring the decay of viremia. Some infected cells carry intact, replication-competent proviruses, but many others contain replication-defective forms of the viral genome, including 2LTR circles and proviruses with large deletions and/or APOBEC3-mediated hypermutation (34, 37–39, 41, 54). These defective forms are captured in many PCR-based reservoir assays and confound the analysis of decay. In contrast to the rapid decay of viremia is the extremely slow decay of latently infected resting CD4+ T cells carrying intact, replication-competent proviruses. Studies of PLWH on long-term ART have established that this decay occurs on a time scale of years (12–14). Between the extremely rapid initial decay of viremia and the very slow decay of the latent reservoir are decay processes that occur over intermediate time scales. These processes have been difficult to define due to the assay issues described above. Nevertheless, the intermediate decay processes are important because infected cells that survive these processes can potentially enter the stable latent reservoir that is the major barrier to cure.
To better understand viral decay processes that influence the composition of the reservoir, we used the novel IPDA (41) to measure the frequency of intact and defective proviruses in participants initiating ART after a recent diagnosis or reinitiating ART after a long interruption. We found that intact proviruses in circulating CD4+ T cells decay in a biphasic pattern, with a more rapid decline (t1/2 = 12.9 d) in the first 3 mo of ART followed by a slower decay (t1/2 = 19.0 mo). In contrast, circulating CD4+ T cells carrying defective provirus showed a generally slower, monophasic decay that within a given participant was similar for proviruses with different types of defects. However, there was considerable variation from donor to donor in the rate of this decay. Surprisingly, 2LTR circles showed a decay pattern that closely resembled that of intact proviruses. Together, these observations define the intermediate decay processes occurring in blood and tissues that may shape the latent reservoir and provide important insights into the correct use of reservoir assays. Findings from this work are summarized below.
Prior to ART, circulating CD4+ T cells with intact proviruses outnumber cells with defective proviruses and are present at frequencies that are over 40-fold higher than those observed in the PLWH on long-term ART. These findings reflect active replication occurring in the absence of ART and illustrate why accurate measurements of the stable latent reservoir cannot be made in untreated individuals. Although latently infected cells are certainly present prior to ART, infected cell populations and early on-ART dynamics are dominated by productively infected cells.
Following initiation of ART, we observed rapid, biphasic decay in viremia, confirming that the decay dynamics described in early studies (1–4) apply also to PLWH taking modern InSTI-based ART regimens, despite the additional complexities arising from where InSTIs act in the virus life cycle (26, 46–48). We show that the initial decay of viremia is at least 10-fold faster than the initial decay of circulating CD4+ T cells with intact proviruses. Thus, the circulating population of CD4+ T cells with intact proviruses includes few of the rapidly decaying cells that produce most of the plasma virus. There is considerable variation in the amount of viral RNA in individual productively infected cells (55), and we cannot exclude the possibility that a small fraction of circulating CD4+ T cells carrying intact proviruses make a significant contribution to the plasma virus. However, our results show that very few of the circulating CD4+ T cells with intact provirus decay with first-phase kinetics. This finding is consistent with the conclusion that most of the plasma virus is produced by cells residing in the secondary lymphoid organs where CD4+ T cells encounter antigen, leading to cellular activation, up-regulation of adhesion molecules, and increased susceptibility to productive infection. Subsequently, these cells produce virus and die within days from viral cytopathic effects or host immune surveillance without entering the circulation. Although infected cells carrying identical proviruses can be detected in blood and lymph node (56–58), recently activated, productively infected CD4+ T cells may be retained in the lymph node until adhesion molecules are down-regulated. The rapid death of the tissue CD4+ T cells that produce most of the plasma virus may contribute to CD4+ T cell depletion, but other mechanisms are likely also involved (59). Higher levels of T cell activation and CCR5 expression in the gut-associated lymphoid tissue may account for the depletion of gut CD4+ T cells seen during acute infection (60–63).
The initial decay of circulating CD4+ T cells carrying intact proviruses more closely resembles the second-phase decay of viremia. Both processes occur over the course of several weeks. Our results do not establish that circulating CD4+ T cells with intact proviruses are producing virus during the second phase, and we observed that their decay rate is slightly faster than the second-phase decay of viremia in the same individuals. However, it is possible that circulating CD4+ T cells with intact proviruses are part of a much larger population of cells that produce virus during the second phase. Their entry into the circulation may reflect a decreasing activation state, exit from the cell cycle, and down-regulation of adhesion molecules. In the circulation, these cells may be susceptible to antibody-dependent clearance mechanisms. These factors might account for the fact that these cells decay at a slightly faster rate than most of the cells producing virus during the second phase. Importantly, the decreasing frequency of cells with intact provirus indicates that this phase of decay is primarily due to cell death or preferential exit from the circulation rather than transition to latency, as transition to a state of nonproductive infection would not account for the observed loss of intact proviral DNA. Understanding if some CD4+ T cells with intact proviruses survive this phase of decay would provide valuable insights into reservoir formation.
Multiple mechanisms may account for the initial decay of circulating CD4+ T cells with intact proviruses. CD8+ cytolytic T lymphocytes (CTLs) exert potent selective pressure on replicating virus (64–67), as evidenced by the early accumulation of CTL escape mutations (68–70). Viruses in the latent reservoir often contain escape mutations in immunodominant, but not subdominant, CTL epitopes (71, 72), and it is possible that the selective elimination of cells with wild-type epitopes occurs over the course of several weeks during second-phase decay. However, CD8+ T cell depletion did not alter decay kinetics in SIV-infected macaques started on ART (73, 74). Productively infected cells could also be eliminated by viral cytopathic effects. During the initial decay of circulating CD4+ T cells with intact proviruses, there could thus be selection for cells that are resistant to cell-death pathways (75, 76). The initial decay of circulating CD4+ T cells carrying intact proviruses could simply reflect virus-independent, contraction-phase elimination of recently activated CD4+ T cells (33). Understanding the mechanism of decay could provide insights into the composition of the latent reservoir, assuming that infected CD4+ T cells surviving these decay processes can down-regulate HIV-1 gene expression and enter the reservoir.
Following the initial decay of circulating CD4+ T cells with intact proviruses during the first 3 mo of ART, we observed a change in the decay slope. The half-life for the next phase of decay is 19 mo. This is still faster than the decay of latently infected cells in PLWH on long-term ART (t1/2 = 44 mo) (12–14). It is not clear that this phase of decay of cells with intact proviruses represents decay of a distinct population or rather decay of a complex population that cannot be defined by a single decay rate due to ongoing changes in the composition and properties of the population. For example, over the long term, the proliferation of infected cells continuously alters the composition of the population of latently infected cells and likely prevents any clinically meaningful decay (35, 77–83).
For defective proviruses, we observed monophasic decays that were generally slower than the initial decay of intact proviruses. Surprisingly, for each participant, the decay was similar for proviruses with 5′ deletions and proviruses with 3′ deletions and/or hypermutation. The IPDA aggregates a wide variety of defective proviruses into just two categories, and our results indicate that the aggregate behavior of these two categories is similar. The generally slower decay of defective proviruses relative to intact proviruses (Fig. 5) may reflect defects in viral gene expression and the absence of gene products targeted by CTL. In long-term studies, it is clear that cells with defective proviruses decay more slowly than cells with intact proviruses (41, 44, 45). This finding has been interpreted as reflecting differential susceptibility to immune surveillance, leading to more rapid loss of cells with intact proviruses. Similarly, selection against proviruses integrated into regions that are permissive for transcription has been documented in elite controllers (84). On the other hand, several recent studies have documented viral gene expression from some defective proviruses (38, 50, 51). Thus, the mechanisms underlying the different early dynamics of cells with intact and defective proviruses remain unclear.
Finally, we examined the decay of 2LTR circles in circulating CD4+ T cells following the initiation of ART. These defective DNA forms have generated great interest as a potential marker of ongoing viral replication (27, 28, 31, 85, 86). However, there has been controversy regarding their stability (29–31). Our results help to resolve this controversy. We observed a biphasic decay pattern for 2LTR circles that closely parallels that of intact proviruses. The initial phase has a t1/2 of 24 d. It is not clear whether this represents degradation of the circles, virus-independent T cell turnover, or virus-dependent elimination of cells carrying circles. Although 2LTR circles are replication-defective, gene expression from 2LTR circles has been detected and could allow recognition by CTL (87, 88). The next phase of decay of 2LTR circles has a t1/2 of ∼15 mo. Unlike intact and defective proviruses, circles are not copied when cells proliferate and may thus decay more quickly. Nevertheless, our results demonstrate that 2LTR circles can persist for long periods of time in the setting of ART, and therefore simply detecting 2LTR circles cannot be used as evidence for ongoing cycles of replication.
Our study has several limitations. Because we measured decay of free virus and infected cells in the blood, we can only make inferences about decay processes occurring in tissues. We have measured net changes in proviral frequency, which could reflect more rapid decay partially offset by proliferation of infected cells. The IPDA does not provide sequence information that would be needed to demonstrate proliferation. As the IPDA was developed using clade B sequences, we have not examined decay for other clades, and our cohort consists only of male participants. Further studies are needed to establish the generality of these findings.
Our analysis of viral decay following initiation of ART has revealed decay processes occurring on time scales of days, weeks, and months-to-years. It is important to note that the decay processes described here are taking place continuously in PLWH who are not on treatment. These ongoing decay processes are simply revealed when new infection events are blocked by ART. With regard to reservoir measurements, differences in the initial decay kinetics of intact and defective proviruses emphasize the importance of using reservoir assays that are selective for intact or replication-competent proviruses. The relatively rapid initial decay of circulating CD4+ T cells with intact proviruses (t1/2 = 12.9 d) also means that the reservoir cannot be accurately measured in the first months of ART. Understanding the mechanisms responsible for the decay could provide important insights into HIV-1 pathogenesis and persistence.
Methods
Study Participants.
We evaluated viral decay in a longitudinal cohort of participants enrolled through the Baltimore Rapid HIV Treatment Initiation and Treatment Reinitiation programs. All participants were newly diagnosed with HIV-1 and initiating ART or reinitiating ART after a long interruption (>6 mo). Peripheral blood mononuclear cell (PBMC) samples were collected before the initiation of ART, every 2 wk for the first 3 mo, and once a month thereafter for up to a year. Plasma HIV-1 RNA levels were measured by using the clinical Cobas assay for every time point. Characteristics of study participants are given in SI Appendix, Table S1.
CD4+ T Cell and Plasma Isolation.
PBMCs and plasma were isolated by density gradient centrifugation using Ficoll–Paque PLUS (GE Healthcare Life Sciences) per the manufacturer’s instructions. Untouched total CD4+ T cells were then enriched from PBMCs by using negative immunomagnetic selection with the EasySep Human CD4+ T-Cell Enrichment Kit (StemCell Technologies).
IPDA and 2LTR Circle Analysis.
We performed IPDA as described (41) to separately measure genetically intact and defective (3′ deleted/hypermutated and 5′ deleted) proviral DNA. The 2LTR circles were measured by using a validated droplet digital PCR assay as described (53). Custom primers and/or probes for the 2LTR circle and the IPDA were designed when amplification failed due to mismatches as described (43, 89).
Mixed-Effect Modeling.
We used a mixed-effect modeling approach to fit either a single-phase or two-phase decay model to the plasma HIV-1 RNA, intact proviruses as measured by IPDA, defective proviruses, and 2LTR circles. The full model is given by
where y is the variable of interest (plasma HIV-1 RNA, intact proviruses, 2LTR circles, or defective proviruses), y0 is its baseline value, A is the fraction of y that decays in the first phase at rate λ1, and (1 − A) is the fraction that decays in the second phase at rate λ2. We can test if a biphasic or a single-phase decay is better by setting A = 1 in the expression above, which then causes y to decay as a single exponential, and no estimation of A or λ2. We compare the fits using the Akaike information criteria (AIC) and the log-likelihood ratio test since we typically compare nested models. The AIC was used for model selection, with the lowest AIC indicating the preferred model (SI Appendix, Table S2). The AIC takes into consideration the number of parameters used to fit the data and penalizes models with more parameters.
To fit the model to the data, we used population fitting, using Monolix 2020R1 (Lixoft SAS, 2020) (90), in which the data (for each variable of interest) from all participants were fitted simultaneously on a log10 scale. In this modeling approach, λ1 and λ2 are assumed to follow a log-normal distribution in the population, and the parameter value for an individual i can be expressed as , where θ is the median value of the population distribution and ηi is the individual random affect, assumed to be normally distributed as N(0, ω2), accounting for variability between individuals. The coefficient A is restricted to lying between zero and one and hence is assumed to be logit-normally distributed. The first data point below the limit of detection of the experimental assay was handled as censored data (91).
Statistics.
Descriptive statistics, tests for normality, Spearman’s correlation, two-tailed Student’s t test, and one-way ANOVA were used to determine statistical significance with GraphPad Prism 8.0 (GraphPad Software). A P value of less than 0.05 was considered significant, unless otherwise stated.
Study Approval.
The Johns Hopkins Institutional Review Board approved this study. All participants provided written informed consent.
Supplementary Material
Acknowledgments
This work was supported by NIH Martin Delaney Collaboratories for HIV Cure Research Grant Awards, Immunotherapy for Cure (I4C) 2.0 (UM1AI164556), Delaney Collaboratory to Cure HIV-1 Infection by Combination Immunotherapy (BEAT-HIV) (UM1AI164570), and Delaney AIDS Research Enterprise (DARE) (UM1AI164560); the Johns Hopkins Center for AIDS Research (P30AI094189); the Bill and Melinda Gates Foundation (OPP1115715); and the Howard Hughes Medical Institute. Portions of this work were performed under the auspices of the U.S. Department of Energy through Los Alamos National Laboratory, which is operated by Triad National Security, LLC, for the National Nuclear Security Administration of the US Department of Energy (Contract 89233218CNA000001). Support was also provided by NIH Grants R01-AI028433, R01-OD011095 (to A.S.P.), and R01-AI15270301 (to R Ke); and Fundacao para a Ciencia e Tecnologia, Portugal Grant PTDC/MAT-APL/31602/2017 (to R.M.R.).
Footnotes
Reviewers: J.V.G., University of North Carolina at Chapel Hill; O.S., Aarhus Universitetshospital.
Competing interest statement: Aspects of the IPDA are the subject of patent application PCT/US16/28822 filed by Johns Hopkins University with R.F.S. as an inventor and licensed to AccelevirDx. R.F.S. holds no equity interest in AccelevirDx.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120326119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Ho D. D., et al. , Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Wei X., et al. , Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373, 117–122 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Perelson A. S., Neumann A. U., Markowitz M., Leonard J. M., Ho D. D., HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Perelson A. S., et al. , Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387, 188–191 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Hammer S. M., et al. , A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 337, 725–733 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Gulick R. M., et al. , Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337, 734–739 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Chun T. W., et al. , In vivo fate of HIV-1-infected T cells: Quantitative analysis of the transition to stable latency. Nat. Med. 1, 1284–1290 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Chun T. W., et al. , Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Finzi D., et al. , Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Chun T. W., et al. , Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong J. K., et al. , Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Finzi D., et al. , Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5, 512–517 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Siliciano J. D., et al. , Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Crooks A. M., et al. , Precise quantitation of the latent HIV-1 reservoir: Implications for eradication strategies. J. Infect. Dis. 212, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archin N. M., et al. , Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen T. A., et al. , Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: A phase 1/2, single group, clinical trial. Lancet HIV 1, e13–e21 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Nixon C. C., et al. , Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 578, 160–165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honeycutt J. B., et al. , Macrophages sustain HIV replication in vivo independently of T cells. J. Clin. Invest. 126, 1353–1366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrade V. M., et al. , A minor population of macrophage-tropic HIV-1 variants is identified in recrudescing viremia following analytic treatment interruption. Proc. Natl. Acad. Sci. U.S.A. 117, 9981–9990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veenhuis R. T., Abreu C. M., Shirk E. N., Gama L., Clements J. E., HIV replication and latency in monocytes and macrophages. Semin. Immunol. 51, 101472 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blankson J. N., et al. , Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J. Infect. Dis. 182, 1636–1642 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Zack J. A., et al. , HIV-1 entry into quiescent primary lymphocytes: Molecular analysis reveals a labile, latent viral structure. Cell 61, 213–222 (1990). [DOI] [PubMed] [Google Scholar]
- 23.Bukrinsky M. I., Stanwick T. L., Dempsey M. P., Stevenson M., Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254, 423–427 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson T. C., et al. , Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76, 8518–8531 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y., Zhang H., Siliciano J. D., Siliciano R. F., Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J. Virol. 79, 2199–2210 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardozo E. F., et al. , Treatment with integrase inhibitor suggests a new interpretation of HIV RNA decay curves that reveals a subset of cells with slow integration. PLoS Pathog. 13, e1006478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Picado J., Zurakowski R., Buzón M. J., Stevenson M., Episomal HIV-1 DNA and its relationship to other markers of HIV-1 persistence. Retrovirology 15, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharkey M. E., et al. , Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6, 76–81 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierson T. C., et al. , Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76, 4138–4144 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler S. L., Johnson E. P., Bushman F. D., Human immunodeficiency virus cDNA metabolism: Notable stability of two-long terminal repeat circles. J. Virol. 76, 3739–3747 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharkey M., Triques K., Kuritzkes D. R., Stevenson M., In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J. Virol. 79, 5203–5210 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Boer R. J., Homann D., Perelson A. S., Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J. Immunol. 171, 3928–3935 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Zhan Y., Carrington E. M., Zhang Y., Heinzel S., Lew A. M., Life and death of activated T cells: How are they different from naive T cells? Front. Immunol. 8, 1809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho Y. C., et al. , Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosmane N. N., et al. , Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon K. J., et al. , Different human resting memory CD4+ T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci. Transl. Med. 12, eaax6795 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruner K. M., et al. , Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imamichi H., et al. , Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 113, 8783–8788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiener B., et al. , Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep. 21, 813–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksson S., et al. , Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9, e1003174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruner K. M., et al. , A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Besson G. J., et al. , HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin. Infect. Dis. 59, 1312–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simonetti F. R., et al. , Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc. Natl. Acad. Sci. U.S.A. 117, 18692–18700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gandhi R. T., et al. ; AIDS Clinical Trials Group A5321 Team, Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J. Infect. Dis. 223, 225–233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peluso M. J., et al. , Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight 5, 132997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sedaghat A. R., Dinoso J. B., Shen L., Wilke C. O., Siliciano R. F., Decay dynamics of HIV-1 depend on the inhibited stages of the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 105, 4832–4837 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrade A., et al. ; AIDS Clinical Trials Group A5248 Team, Three distinct phases of HIV-1 RNA decay in treatment-naive patients receiving raltegravir-based antiretroviral therapy: ACTG A5248. J. Infect. Dis. 208, 884–891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrade A., et al. ; ACTG A5249s Protocol Team, Early HIV RNA decay during raltegravir-containing regimens exhibits two distinct subphases (1a and 1b). AIDS 29, 2419–2426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer S., et al. , Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 105, 3879–3884 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollack R. A., et al. , Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe 21, 494–506.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sannier G., et al. , Combined single-cell transcriptional, translational, and genomic profiling reveals HIV-1 reservoir diversity. Cell Rep. 36, 109643 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Bender A. M., et al. , The landscape of persistent viral genomes in ART-treated SIV, SHIV, and HIV-2 infections. Cell Host Microbe 26, 73–85.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massanella M., Gianella S., Lada S. M., Richman D. D., Strain M. C., Quantification of total and 2-LTR (long terminal repeat) HIV DNA, HIV RNA and herpesvirus DNA in PBMCs. Bio Protoc. 5, e1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee G. Q., et al. , Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Invest. 127, 2689–2696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiegand A., et al. , Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc. Natl. Acad. Sci. U.S.A. 114, E3659–E3668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kearney M. F., et al. , Well-mixed plasma and tissue viral populations in RT-SHIV-infected macaques implies a lack of viral replication in the tissues during antiretroviral therapy. Retrovirology 12, 93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McManus W. R., et al. , HIV-1 in lymph nodes is maintained by cellular proliferation during antiretroviral therapy. J. Clin. Invest. 129, 4629–4642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin A. R., et al. ; HOPE in Action Study Team, Similar frequency and inducibility of intact human immunodeficiency virus-1 proviruses in blood and lymph nodes. J. Infect. Dis. 224, 258–268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doitsh G., et al. , Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veazey R. S., et al. , Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280, 427–431 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Smit-McBride Z., Mattapallil J. J., McChesney M., Ferrick D., Dandekar S., Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J. Virol. 72, 6646–6656 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehandru S., et al. , Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200, 761–770 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brenchley J. M., et al. , CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200, 749–759 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker B. D., et al. , HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature 328, 345–348 (1987). [DOI] [PubMed] [Google Scholar]
- 65.Koup R. A., et al. , Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68, 4650–4655 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmitz J. E., et al. , Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283, 857–860 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Deeks S. G., Walker B. D., Human immunodeficiency virus controllers: Mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27, 406–416 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Borrow P., et al. , Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3, 205–211 (1997). [DOI] [PubMed] [Google Scholar]
- 69.Jones N. A., et al. , Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 200, 1243–1256 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salazar-Gonzalez J. F., et al. , Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206, 1273–1289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng K., et al. , Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warren J. A., et al. , The HIV-1 latent reservoir is largely sensitive to circulating T cells. eLife 9, e57246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klatt N. R., et al. , CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 6, e1000747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wong J. K., et al. , In vivo CD8+ T-cell suppression of SIV viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog. 6, e1000748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang S. H., et al. , Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J. Clin. Invest. 128, 876–889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ren Y., et al. , BCL-2 antagonism sensitizes cytotoxic T cell-resistant HIV reservoirs to elimination ex vivo. J. Clin. Invest. 130, 2542–2559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tobin N. H., et al. , Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: Expression of archival virus and replication of virus. J. Virol. 79, 9625–9634 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bailey J. R., et al. , Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80, 6441–6457 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maldarelli F., et al. , HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner T. A., et al. , HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simonetti F. R., et al. , Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 1883–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bui J. K., et al. , Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13, e1006283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z., et al. , Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. U.S.A. 115, E2575–E2584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang C., et al. , Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature 585, 261–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buzón M. J., et al. , HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat. Med. 16, 460–465 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Hatano H., et al. , Increase in 2-long terminal repeat circles and decrease in D-dimer after raltegravir intensification in patients with treated HIV infection: A randomized, placebo-controlled trial. J. Infect. Dis. 208, 1436–1442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonczkowski P., et al. , Protein expression from unintegrated HIV-1 DNA introduces bias in primary in vitro post-integration latency models. Sci. Rep. 6, 38329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Policicchio B. B., et al. , Dynamics of simian immunodeficiency virus two-long-terminal-repeat circles in the presence and absence of CD8(+) cells. J. Virol. 92, e02100-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kinloch N. N., et al. , HIV-1 diversity considerations in the application of the Intact Proviral DNA Assay (IPDA). Nat. Commun. 12, 165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chow S.-C., Wang H., Shao J., “Sample size calculations in clinical research,” in Chapman & Hall/CRC Biostatistics Series (Chapman & Hall/CRC, New York, NY, 2014) [Google Scholar]
- 91.Samson A., Lavielle M., Mentre F., Extension of the SAEM algorithm to left-censored data in nonlinear mixed-effects model: Application to HIV dynamics model. Comput. Stat. Data Anal. 51, 1562–1574 (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.