Significance
How bacteria with multipartite genomes organize and segregate their DNA is poorly understood. Here, we investigate a prototypical multipartite genome in the plant pathogen Agrobacterium tumefaciens. We identify previously unappreciated interreplicon interactions: the four replicons cluster through interactions at their centromeres, and the two chromosomes, one circular and one linear, interact along their replication arms. Our data suggest that these interreplicon contacts play critical roles in the organization and maintenance of multipartite genomes.
Keywords: Agrobacterium tumefaciens, SMC, ParB, Hi-C, ChIP-seq
Abstract
Bacterial species from diverse phyla contain multiple replicons, yet how these multipartite genomes are organized and segregated during the cell cycle remains poorly understood. Agrobacterium tumefaciens has a 2.8-Mb circular chromosome (Ch1), a 2.1-Mb linear chromosome (Ch2), and two large plasmids (pAt and pTi). We used this alpha proteobacterium as a model to investigate the global organization and temporal segregation of a multipartite genome. Using chromosome conformation capture assays, we demonstrate that both the circular and the linear chromosomes, but neither of the plasmids, have their left and right arms juxtaposed from their origins to their termini, generating interarm interactions that require the broadly conserved structural maintenance of chromosomes complex. Moreover, our study revealed two types of interreplicon interactions: “ori-ori clustering” in which the replication origins of all four replicons interact, and “Ch1-Ch2 alignment” in which the arms of Ch1 and Ch2 interact linearly along their lengths. We show that the centromeric proteins (ParB1 for Ch1 and RepBCh2 for Ch2) are required for both types of interreplicon contacts. Finally, using fluorescence microscopy, we validated the clustering of the origins and observed their frequent colocalization during segregation. Altogether, our findings provide a high-resolution view of the conformation of a multipartite genome. We hypothesize that intercentromeric contacts promote the organization and maintenance of diverse replicons.
Approximately 10% of sequenced bacterial genomes contain more than one replicon (1). These multipartite genomes are scattered throughout different bacterial phyla, including species of the Borrelia, Burkholderia, Brucella, Rhizobium, and Vibrio genera, which are mostly animal and plant symbionts or pathogens (1, 2). It has been proposed that having multiple replicons allows faster genome duplication and imparts an advantage to these bacteria to quickly adapt when switching hosts or environments (1–3). However, multipartite genomes pose challenges for genome maintenance and management. How multiple replicons are organized inside bacterial cells, how they interact with each other, and whether these interactions help the maintenance or segregation of these multipartite genomes, and if so how, are unanswered questions.
In unichromosomal bacteria, chromosome dynamics and genome-wide DNA interactions have been characterized in several species (4–10). The highly conserved parABS system and structural maintenance of chromosomes (SMC) complex have been found to play critical roles in both chromosome segregation and interactions. The parABS system is composed of centromeric parS sequences that are commonly present in multiple copies in the origin region (11). These sequences are bound by the ParB protein that spreads to adjacent regions, forming large nucleoprotein complexes (12–14). Finally, the ParA ATPase binds nonspecifically throughout the chromosome when bound to adenosine triphosphate (ATP). ParA-ATP interaction with ParB/parS catalyzes ATP hydrolysis and release of ParA from the DNA (14–16). The system is tuned in a manner such that ParA functions like a “burnt bridge” Brownian ratchet that pulls replicated origin regions toward opposite cell poles (17, 18).
The ParB/parS nucleoprotein complex has a second function in chromosome dynamics, in site-specifically loading the SMC complex onto the chromosome (19–21). After loading, the large ring-shaped ATPases of the SMC complex translocate away from their origin-proximal loading site toward the replication terminus, juxtaposing the DNA on the left and right arms (7, 22). This activity is called DNA-loop extrusion and is thought to help topoisomerase IV remove entanglements between the two newly replicated sister chromosomes and thus facilitate chromosome segregation (23). Loop extrusion generates interarm interactions, the most prominent genome-wide long-range interactions observed in bacteria. Interestingly, Escherichia coli and a subset of gamma proteobacteria lack the parABS system and do not have the canonical SMC complex. These bacteria contain a structural analog of the SMC complex called MukBEF (24). E. coli lacks interarm interactions and instead adopts a distinct organization pattern (6). The current view is that MukBEF generates long-range DNA interactions throughout the genome by loading nonspecifically on DNA and extruding loops from these sites (25). Evidence suggests that canonical SMC complexes can also load nonspecifically on DNA and extrude loops in addition to their specific loading at parS sites by ParB (19, 26).
Chromosome dynamics have been investigated in several bacteria with multipartite genomes by visualizing specific genetic loci using fluorescence microscopy (27–30). However, genome-wide interactions have only been examined in Vibrio cholerae (31, 32). V. cholerae has two circular chromosomes (Ch1 and Ch2); Ch2 has interarm interactions that resemble unichromosomal bacteria like Bacillus subtilis, whereas Ch1 is folded similarly to E. coli and lacks interarm interactions. Interestingly, weak interactions were observed between the termini of the two chromosomes and between the origin of Ch2 (ori2) and a sequence element called crtS on Ch1 (31, 32). Replication of crtS triggers the initiation of Ch2 replication, ensuring that replication of the two chromosomes terminates at approximately the same time (32, 33).
V. cholerae, as with E. coli, has MukBEF instead of a canonical SMC complex (34), and it is currently unclear what drives the interarm interactions on Ch2. Here, we investigate the organization of a more complex multipartite genome in the plant pathogen, Agrobacterium tumefaciens. Importantly, this bacterium encodes the canonical SMC complex, suggesting its genome-wide organization is likely to be more prevalent among bacteria with multipartite genomes.
A. tumefaciens C58 has a circular chromosome (Ch1, 2841 kb), a linear chromosome (Ch2, 2,076 kb) and two large plasmids, pAt (542 kb) and pTi (216 kb) (Fig. 1A) (35). Ch1 contains a parABS system, whereas Ch2 and the two plasmids use individual repABC systems, distantly related to parABS. RepAs resemble ParA, RepBs are homologs of ParB, and RepCs are the replication initiator proteins. The centromeric sequences are all called parS sites (36). Cell division proceeds through a polar mechanism in which the daughter cell grows out from a single pole rather than by classic binary fission (37). Here, we combined chromosome conformation capture (Hi-C), chromatin immunoprecipitation sequencing (ChIP-seq), whole-genome sequencing (WGS), and time-lapse fluorescence microscopy to investigate genome organization and segregation in A. tumefaciens C58. We report that both chromosomes, but neither plasmid, have interarm interactions that depend on the SMC complex. In addition, we identify two types of interreplicon contacts: interactions among all four origins that we call ori clustering and interchromosomal interactions along the chromosome arms that we call Ch1-Ch2 alignment. Both types of interreplicon contacts require centromeric ParB/RepB proteins. Finally, we use fluorescence microscopy to track the localization and dynamics of the chromosome and plasmid replication origins during the cell cycle. Collectively, our data support a model in which interreplicon contacts promote organization and maintenance of multipartite genomes during cell division.
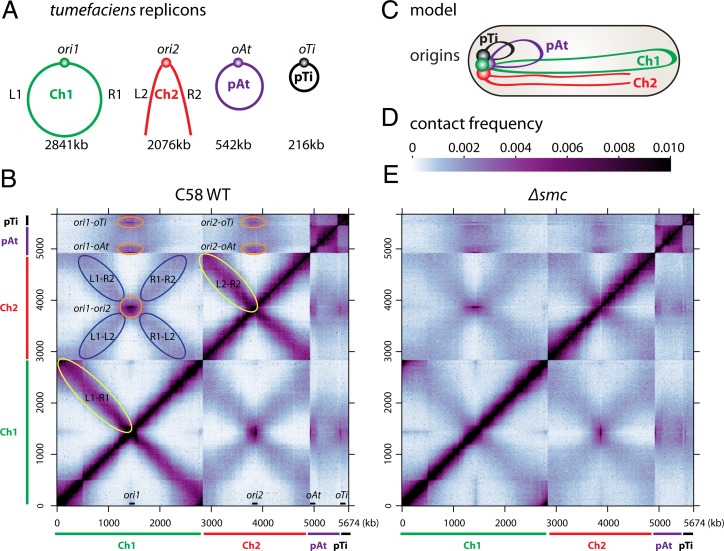
Fig. 1.
Genome-wide organization of A. tumefaciens replicons. (A) A. tumefaciens C58 wild-type cells have a circular chromosome (Ch1), a linear chromosome (Ch2), and two plasmids (pAt and pTi). The replication origins are labeled ori1, ori2, oAt, and oTi. The left and right arms of the chromosomes are labeled L1, R1, L2, and R2. The sizes of these four replicons are indicated. (B) Normalized Hi-C contact map displaying contact frequencies for pairs of 10-kb bins across the genome of A. tumefaciens C58 wild-type (WT). The x and y axes indicate genome positions. To better visualize contacts in the origin region of Ch1, the reference genome of Ch1 is rearranged with the origin (ori1) at the center and the two arms on either side. Ch1, Ch2, pAt, and pTi are indicated by green, red, purple, and black bars, respectively. The positions of the four origins are indicated on the x axis. Interarm interactions on both Ch1 and Ch2 are circled in yellow. Interactions between origins are circled in orange. The interactions between the arms of Ch1 and the arms of Ch2 are circled in blue. (C) Schematic model of genome organization in a newborn cell. The four origins are clustered together at the old cell pole. Ch1 and Ch2 are aligned along the cell length. (D) The scale depicts Hi-C interaction scores (contact frequency) for all contact maps presented in this study. (E) Normalized Hi-C contact map of Δsmc (AtWX108). Interarm interactions are absent in Ch1 and Ch2.
Results
Global Conformation of the A. tumefaciens Genome.
To study the global folding pattern of the A. tumefaciens genome, we performed Hi-C (38) on C58 wild-type (39) cells during exponential growth in minimal medium, ATGN (doubling time = 202 min ± 7 min, mean ± SD) (40) (Fig. 1B). To better visualize the chromosome origin regions on the Hi-C interaction matrix, we oriented the reference sequence such that the origins were at the center of each chromosome. Consistent with previous studies on unichromosomal bacteria that contain an SMC complex (4, 5, 7–9), we found that the two replication arms of the circular Ch1 interacted along their lengths (Fig. 1B, yellow ovals). Interestingly, the linear Ch2 also had interarm interactions, whereas the plasmids, pAt and pTi, did not (Fig. 1B and SI Appendix, Fig. S1). In addition to these intrareplicon contacts, the Hi-C matrix revealed two types of interreplicon contacts. First, the origins of all four replicons interacted with each other (Fig. 1B, orange circles), which we refer as “ori-ori clustering.” This interaction pattern is consistent with their polar localization that has been reported previously (41). Second, the arms of Ch1 interacted with the arms of Ch2 along their lengths. This latter interaction, which we call “Ch1-Ch2 alignment,” can be seen on the Hi-C map as an X-shaped pattern (Fig. 1 B, blue ovals, and C).
To investigate whether the interaction patterns we observed were specific to strain C58, we tested a phylogenetically distinct wild-type strain, A. tumefaciens 15955 (42). This strain has four replicons but differs significantly from C58 in gene content and sequence. As can be seen in SI Appendix, Fig. S2, the overall interaction patterns were similar in these two strains. Moreover, the interactions remained in all growth media tested, including virulence-inducing growth conditions (40, 43), minimal medium ATGN, and rich LB medium. Finally, in strains that are missing the pTi plasmid or a large region of the pAt plasmid (43), chromosome interactions were not altered (SI Appendix, Fig. S2 C, D, G, and H).
SMC Is Required for Interarm Interactions on Both Chromosomes.
To investigate whether SMC is required for the interarm interactions observed for the circular and linear chromosomes, we generated a markerless deletion of the smc gene (Atu0801/ATU_RS03945) using allelic replacement. Analysis of chromosome interactions by Hi-C in Δsmc mutant revealed that the interarm interactions on both chromosomes were lost (Fig. 1E and SI Appendix, Fig. S3A). Interestingly, despite the absence of these long-range interactions, cells lacking SMC had no growth defects, and replicon segregation was only mildly impaired (Fig. 2F and SI Appendix, Fig. S3B) (Discussion). In addition, the two types of interchromosomal contacts (ori-ori clustering and Ch1-Ch2 alignment) were largely unchanged in the Δsmc mutant (Fig. 1E and SI Appendix, Fig. S3A) and thus were independent of SMC. Therefore, as observed previously in unichromosomal bacteria, the SMC complexes in A. tumefaciens act in cis to disentangle the sister chromatids, juxtaposing the left and right arms of the circular and linear chromosomes, presumably by loop extrusion. Furthermore, SMC complexes do not function in bridging between replicons, nor do they function in isolating the replicons from each other.
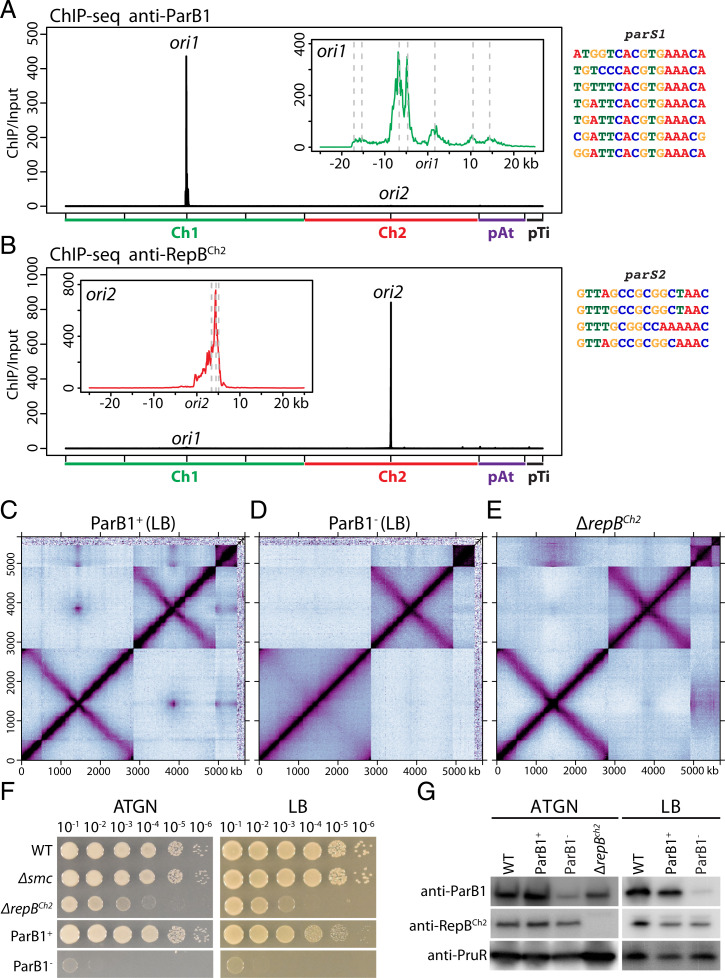
Fig. 2.
SMC loaders on Ch1 and Ch2. (A) Left: ParB1 enrichment in wild-type cells. To match the presentation of Hi-C, the reference genome of Ch1 is rearranged with the origin (ori1) at the center and the two arms on either side. Sequencing reads from ChIP and input samples were normalized to the total number of reads. The x axis shows genome positions, and the y axis indicates ChIP enrichment (ChIP/input) in1-kb bins. Inset: High-resolution plots of a 50-kb region encompassing ori1. Putative parS1 sites are indicated by gray dotted lines. Data are plotted in 100-bp bins. Right: Individual parS1 sequences are aligned. (B) Left: RepBCh2 enrichment in wild-type cells. Inset: High-resolution plots of a 50-kb region encompassing ori2. Putative parS2 sites are indicated by gray dotted lines. Right: Individual parS2 sequences are aligned. (C and D) Normalized Hi-C contact maps for ParB1 depletion strain (AtWX192) grown in LB with (ParB1+) or without (ParB1−) AHL and theophylline. A different genetic isolate is shown in SI Appendix, Fig. S4 K and L. 30-h depletion was used for this experiment. An earlier time point (12-h depletion) is shown in SI Appendix, Fig. S4N. The same strains growing in ATGN were shown in SI Appendix, Fig. S4 E–H. When ParB1 was depleted, interarm interactions are absent on Ch1 but unchanged on Ch2. Quantification of the interactions can be found in SI Appendix, Fig. S5. We observed the loss of pTi in this strain in two genetic isolates (SI Appendix, Fig. S4 E–L), likely during strain construction. (E) Normalized Hi-C contact maps for ΔrepBCh2 (AtWX089). When repBCh2 was deleted, interarm interactions on both chromosomes were unchanged. (F) Tenfold serial dilutions of the indicated strains spotted on ATGN plate (left) or LB plate (right). ParB1+ plates contain 1 μM AHL and 2 mM theophylline, and ParB1− plates lack these additives. (G) Immunoblot analysis showing levels of ParB1, RepBCh2, and PruR, a loading control. Wild-type, ParB1 depletion strain (AtWX192), and ΔrepBCh2 (AtWX089) were grown in LB or ATGN as indicated. After 30-h depletion, ParB1 has ∼3% remaining in LB (also SI Appendix, Fig. S4M) and 21.0% in ATGN.
SMC Loaders on Ch1 and Ch2.
The requirement of SMC for interarm interactions on both Ch1 and Ch2 prompted us to investigate the role of ParB1 (Atu2828/ATU_RS13770) and RepBCh2 (Atu3923/ATU_RS18280) in loading the SMC complex. As a first step in our characterization of these proteins, we experimentally determined their binding sites on the genome. We purified recombinant ParB1 and RepBCh2 and generated polyclonal antibodies against them to perform chromatin immunoprecipitation sequencing (ChIP-seq). Our analysis revealed that ParB1 was enriched over an ∼40-kb region surrounding ori1 (Fig. 2A), whereas RepBCh2 occupied an ∼8-kb region encompassing ori2 (Fig. 2B). Guided by previously characterized binding motifs of ParB and RepB proteins (11, 36), we identified seven parS1 sites on Ch1 (Fig. 2A) and four parS2 sites on Ch2 (Fig. 2B), which are within the ChIP-seq enrichment regions.
To test whether ParB1 is required for interarm interactions along Ch1, we sought to generate a conditional allele for the essential parB1 gene. Our attempts to deplete ParB1 using IPTG-regulated promoters [Plac and Ptac (44)] were unsuccessful due to leaky transcription in the absence of inducer. Instead, we combined a tightly controlled inducible promoter [PtraI (45)] with a riboswitch that controls translation (46, 47). The PtraI promoter is activated by TraR in the presence of the acylhomoserine lactone (AHL) produced by the TraI AHL synthase. To enable exogenous control of the PtraI promoter, we deleted the traI gene that encodes the AHL synthase (48). The riboswitch is regulated by the small molecule theophylline (46, 47). In the absence of theophylline, transcripts containing this riboswitch are poorly translated. We inserted a PtraI-riboswitch-parB1 traR cassette at the tetRA locus on Ch2 (40) and deleted the native parB1 gene in the presence of AHL and theophylline. In the presence of the two inducers, the strain grew similarly to the wild type; in the absence of inducers, the cells failed to form colonies, supporting the essentiality of parB1 and its successful depletion using this system (Fig. 2F).
We used the ParB1 depletion strain to compare the genome conformation in the presence and absence of ParB1. For these experiments we used our standard ATGN medium (doubling time = 202 min ± 7 min, mean ± SD) and LB (doubling time = 89 min ± 2 min, mean ± SD). We grew the strain in the presence of AHL and theophylline and collected ParB1+ samples. We then washed the cells, diluted them in media lacking the inducers, and we collected ParB1-depleted samples (ParB1−) at various time points (SI Methods). To prevent the cells from entering stationary phase, we diluted the cultures when their optical density at 600 nm (OD600) reached 0.6. As shown in Fig. 2G and SI Appendix, Fig. S4M, ParB1 depletion in ATGN for 30 h or in LB for 12 h was incomplete (∼79% or ∼83% depleted, respectively), and interarm interactions on Ch1 were reduced but still detectable (SI Appendix, Fig. S4 E–H and N). However, after depletion in LB for 30 h (Fig. 2G and SI Appendix, Fig. S4M), ParB1 depletion was more pronounced (∼97% depleted). Under these conditions, interarm interactions on Ch1 were barely above the background (Fig. 2 C and D and SI Appendix, Figs. S4 I–L and S5 A and B). Importantly, the interarm interactions on Ch2 were unchanged. Therefore, ParB1 is required for interarm interactions on Ch1 and not Ch2, supporting the idea the SMC complexes are loaded onto Ch1 by ParB1 bound to parS1 sites in ori1.
In the absence of ParB1, interarm interactions on Ch2 were unchanged (Fig. 2 C and D and SI Appendix, Figs. S4 I–L and S5 A and B), suggesting that RepBCh2 was responsible for loading SMC complexes onto this linear chromosome. To investigate this possibility, we generated an in-frame deletion of repBCh2. The mutant strain had a severe growth defect but was viable (Fig. 2F), enabling us to investigate the genome conformation by Hi-C. Strikingly, interarm interactions on Ch2 and Ch1 were unaffected in this mutant (Fig. 2E and SI Appendix, Fig. S5). These data indicate that RepBCh2 is not required for SMC-dependent interarm interactions on Ch2 and suggest that loading SMC onto the origin of Ch2 is mediated by a currently unknown factor.
ParB1 and RepBCh2 Are Required for Interchromosomal Contacts.
Strikingly, when ParB1 was depleted, all interreplicon contacts (ori-ori clustering and Ch1-Ch2 alignment) were diminished (Fig. 2D and SI Appendix, Fig. S4 E–L). We note that even the interreplicon contacts among ori2, pAt, and pTi were absent. When RepBCh2 was deleted, interactions between Ch2 and the other replicons were absent. However, interactions between Ch1 and the plasmids were unchanged (Fig. 2E). These results and the Hi-C analysis of the Δsmc mutant (Fig. 1E and SI Appendix, Fig. S3A) indicate that ori-ori clustering and Ch1-Ch2 alignment are mediated by the centromeric ParB/RepB proteins, but not through the loading the SMC complex; ParB1 is the central component for interactions among all four origins and a prerequisite for ori-ori contacts between secondary replicons.
We also noticed that pTi was lost in two independent isolates of the ParB1 depletion strain during the strain building procedure. However, pTi was not lost in any other mutants analyzed by Hi-C (Fig. 2 C and D and SI Appendix, Fig. S4 E–L). The exact reason for pTi loss was uncertain. We speculate that ParB1 is important for the stable maintenance of pTi, and ori-ori clustering promotes the stable maintenance of secondary replicons of multipartite genomes.
Hierarchical Localization and Replication of the Four Replicons.
Our Hi-C assays identified a genome-wide interaction pattern in A. tumefaciens. To complement this population-averaged assay, we used single cell–based methods to investigate if and how interreplicon contacts impact genome segregation and maintenance for multipartite genomes. To directly visualize the localization and dynamics of the four replicons, we used two independent approaches. In the first approach, we generated fluorescent fusions to ParB1 and the three RepB proteins and expressed them from a low-copy-number plasmid, pSRKKm (49) (Fig. 3A). As a validation to this approach, we performed ChIP-seq experiments and found that GFP-ParB1 (SI Appendix, Fig. S6A) and GFP-RepBCh2 (SI Appendix, Fig. S6B) had the same enrichment profiles as the untagged proteins (Fig. 2 A and B), and mYpet-RepBpAt (SI Appendix, Fig. S6C) and mYpet-RepBpTi(parS*) (SI Appendix, Fig. S6D) (also see below) were only enriched at the origins of their cognate plasmids. Fluorescence imaging of exponentially growing cells expressing GFP-ParB1, GFP-RepBCh2, mYpet-RepBpAt, or mYpet-RepBpTi(parS*) revealed that each fusion protein had unipolar or bipolar foci (Fig. 3A). These data suggest that the centromeres (ParB1 and RepBs bound to their parS sites) at the replication origins from all four replicons localize at the cell poles.
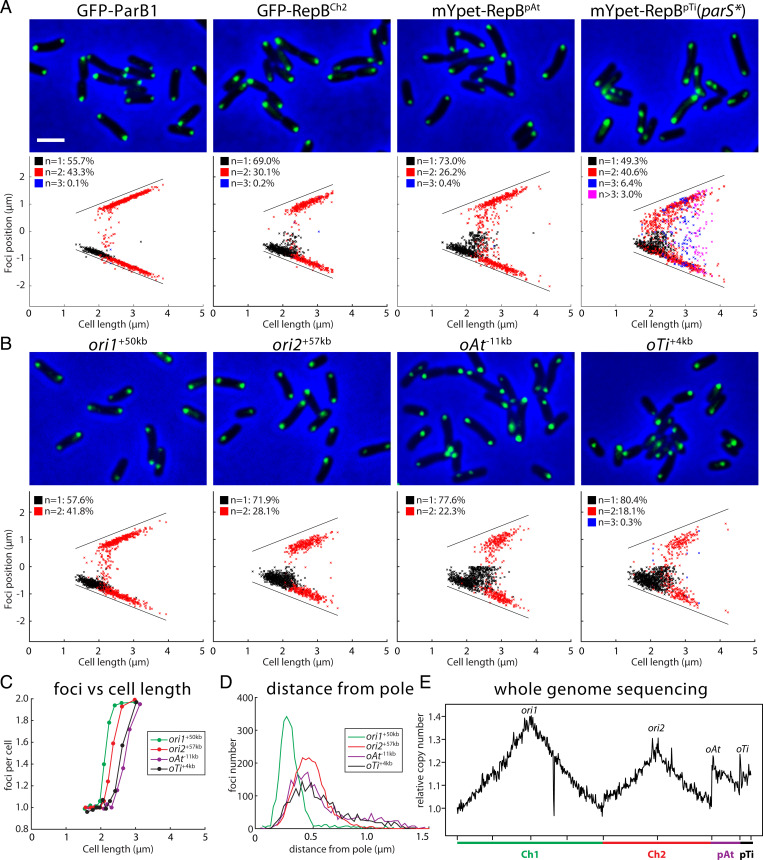
Fig. 3.
Visualization of the four origins. (A) Localization of the four centromeric regions visualized using fluorescent protein fused to ParB1/RepBs expressed from pSRKKm-based plasmids. Top panels are cropped microscopy images of GFP-ParB1 (AtWX226), GFP-RepBCh2 (AtWX228), mYpet-RepBpAt (AtWX179), and mYpet-RepBpTi(parS*) (AtWX372). Fluorescent fusions were expressed by adding 0.25 mM IPTG for 4 h. Bottom panels are plots showing relative position of the foci. One-thousand cells were analyzed for each strain. The solid black lines indicate the positions of two poles. The same population of cells were used to quantify the distance of the foci from pole (SI Appendix, Fig. S8A). (Scale bar, 2 μm.) (B) Localization of the four origin regions visualized using the ParBpMT1-parSpMT1 system (AtWX278, AtWX295, AtWX359, AtWX351). Top panels are microscopy images and Bottom panels are plots of foci position. (C) Average number of ParBpMT1-parSpMT1 labeled origins per cell. Cells are divided into 100-cell bins. The x axis indicates the cell length, and the y axis shows foci number per cell in the bin. (D) Distance of the ParBpMT1-parSpMT1–labeled origins from the nearest pole. The x axis shows the distance from the pole, and the y axis indicates the number of foci at that length. (E) Maker frequency analysis using WGS data from the wild type. The x axis shows the genome position, and the y axis shows relative copy number. The reference genome of Ch1 is arranged the same way as in Hi-C and ChIP-seq plots.
In a second approach, we localized the four origins by inserting a cassette containing gfp-parBpMT1-parSpMT1 near the origin regions of the four replicons (Fig. 3B). This ParB/parS system is derived from pMT1 plasmid and is commonly used to visualize chromosomal loci (50). Importantly, the parSpMT1 sequence is distinct from the parS sites on the four replicons in A. tumefaciens. Using WGS and Hi-C experiments, we found that labeling the chromosome origins using this cassette did not perturb DNA replication (SI Appendix, Fig. S7A) or genome organization (SI Appendix, Fig. S7B). Consistent with the experiments described above, the origins of all four replicons localized as one or two polar foci. These data are consistent with the ori-ori clustering interactions observed by Hi-C (Fig. 1B).
Taking advantage of the correlation between cell length and cell cycle progression, we compared the relative segregation timing of each replicon. We ordered the cells with increasing cell length, divided them into 100-cell bins and analyzed the average number of foci per cell in each bin. We found that ori1 segregated first, followed by ori2 and then oAt and oTi (Fig. 3C). These data are consistent with experiments reported previously (41, 51). The staggered segregation patterns suggested that the initiation of replication of the four replicons was not synchronous. To investigate this more directly, we performed genome-wide marker frequency analysis. The ratios of relative copy number of the four origins were 1:0.87:0.73:0.75 (ori1:ori2:oAt:oTi) (Fig. 3E), consistent with the idea that the delay in segregation by microscopy was due to a delay in the initiation of replication (Fig. 3 A–C). We conclude that A. tumefaciens cells are born with a single copy of each replicon with the replication origins localized at the old cell pole. At some time after septation, ori1 initiates replication and segregation; this is followed by ori2 replication initiation and segregation, and then the plasmids.
Finally, we compared the polar localization of the four origins by analyzing the distance between the cell pole and each focus. We found that ParB1 was closer to the cell poles than RepBCh2, RepBpAt, or RepBpTi(parS*) (SI Appendix, Fig. S8A). Similar results were obtained using the ParBpMT1-parSpMT1 labeled origins (Fig. 3D). Thus, the origins not only have a hierarchy of replication initiation and segregation timing but a similar hierarchy of polar localization. Both hierarchies were previously observed in A. tumefaciens (41) and other species with multiple replicons (27–29). We note that the ParBpMT1-parSpMT1–labeled origins were further away from the cell pole than their respective centromeres (SI Appendix, Fig. S8B). A similar phenomenon was observed in Caulobacter crescentus (52). These observations are consistent with the idea that the centromeres are sites of force exertion for origin anchoring and origin segregation and that the nearby regions can be stretched (52).
pTi Can Be Cured due to a Cryptic parS in repBpTi, and This Confounds Copy-Number Measurements.
Although consistent with one earlier study (41), our data that pTi had one or two copies localized to the cell poles contradicted a recent report by Robalino-Espinosa and coworkers showing that pTi had one to six copies and did not occupy specific regions of the cell (51). We note that in our initial experiments, we visualized mYpet-RepBpTi expressed from the pSRKKm plasmid and observed three to ten foci distributed throughout the cell (SI Appendix, Fig. S9C). The mYpet-RepBpTi fluorescent foci were also fainter and appeared fuzzy (SI Appendix, Fig. S9C), similar to what was observed by Robalino-Espinosa et al. using eGFP-RepBpTi (51). ChIP-seq on the strain expressing mYpet-RepBpTi revealed that there is a parSpTi site within the repBpTi gene (SI Appendix, Fig. S9 A and C), and the presence of the pSRKKm plasmid expressing mYpet-RepBpTi led to the loss of the pTi plasmid (SI Appendix, Fig. S9 A and B). Thus, our mYpet-RepBpTi (and likely eGFP-RepBpTi of Robalino-Espinosa et al., also expressed from a pSRK derivative) is reporting on the copy number and subcellular position of the pSRK plasmid and not those of pTi. To circumvent this problem, we generated an mYpet-RepBpTi(parS*) fusion in which the parSpTi sequence within the fusion was mutated without altering the protein sequence (SI Appendix, Fig. S9C). Fluorescence imaging of a strain harboring this fusion revealed one or two polar foci (SI Appendix, Fig. S9C), similar to the other three replicons.
ori-ori Clustering during Segregation.
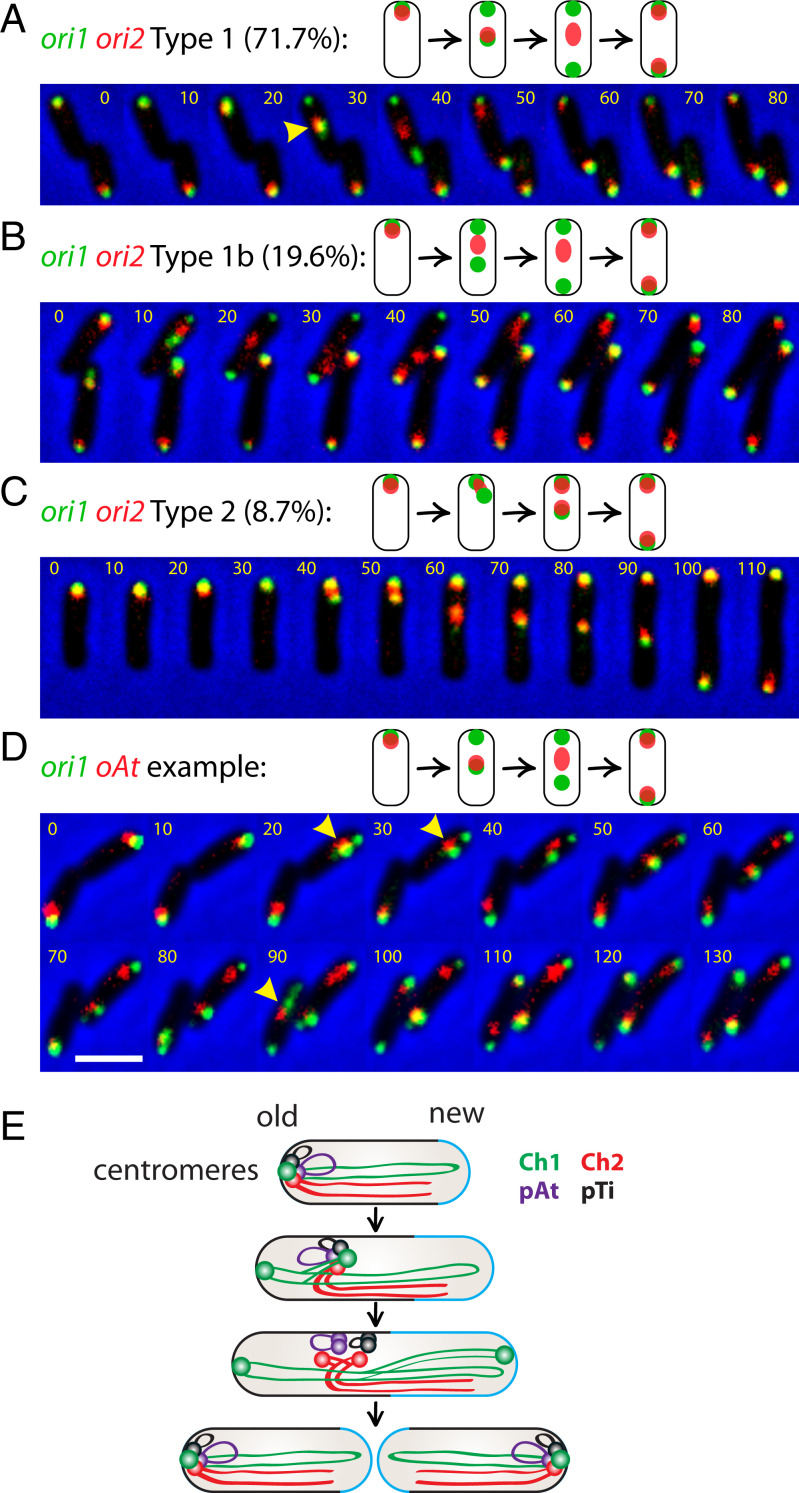
After visualizing individual replication origins in snapshots, we further investigated the dynamic behavior of the origins of each replicon and covisualized ori1 (eGFP-ParB1) and ori2 (RFP- RepBCh2) or oAt (RFP- RepBpAt) using time-lapse microscopy (Fig. 4). This analysis revealed that ori1 and ori2 colocalize at the poles during most of the cell cycle, consistent with the ori-ori clustering interactions observed in Hi-C (Fig. 1B). However, upon initiation of replication and segregation, not all cells behaved the same. In the vast majority of cells (71.7%; n = 45) called type 1 (Fig. 4A and SI Appendix, Fig. S10), ori1 duplicated and segregated before ori2; then, one of the ori1 foci remained at the old pole, and the other moved to the new pole. Interestingly, the single ori2 focus initially was colocalized with the translocating ori1 focus. Upon initiation of ori2 replication, as indicated by a smeary fluorescent focus, the ori2 smear no longer colocalized with the migrating ori1 focus. It then split into two foci and segregated to opposite poles, colocalizing with the two ori1 foci. In type 1b (19.6%) (Fig. 4B), the fluorescent foci followed the same above steps as in type 1, except for the colocalization step during ori1 translocation. It is possible that ori2 and the moving ori1 colocalized for less than 10 min, which was the time interval of image acquisition, and was missed in our time-lapse imaging. In type 2 (8.7%) (Fig. 4C), we could not resolve the timing of ori1 and ori2 duplication, and one pair of ori1-ori2 foci stayed at the old pole, and the other pair of foci segregated to the new pole at the same time. The time-lapse movies of ori1 and oAt were similar to the ori1-ori2 dynamics, in which the single oAt focus initially colocalized with the translocating ori1 focus during segregation (Fig. 4D). Thus, the time-lapse experiments are consistent with the single-color snapshots in regard to origin localization and segregation timing (Fig. 3 and SI Appendix, Fig. S8A) and further revealed that before duplication, ori2 and oAt were colocalized with the segregating ori1, likely through the interactions between the centromeres (Fig. 2 D and E). These data suggest that ori1/ParB1 holds the ori-ori clustering at the cell pole, and upon duplication of this region, the unreplicated origins specifically associate with the nonanchored Ch1 centromere.
Fig. 4.
Covisualization of replicons. (A–C) Three types of segregation patterns of ori1 and ori2 (AtWX263). GFP-ParB1 and RFP-RepBCh2 are expressed from a single pSRKKm-based plasmid. A preculture was grown for overnight in the absence of IPTG. The cells were subcultured to medium containing 0.2 mM IPTG for 4 h before setting up time lapse. Time-lapse progression (10-min intervals) of ori1 (green) and ori2 (red) dynamics monitored in cells grown on agarose pad containing ATGN and 0.2 mM IPTG. Two biological experiments were performed. The percentages were calculated from a total of 45 cells, in which we captured the entire replication and segregation cycle. Yellow carets show where the red focus colocalizes/overlaps with the moving green focus. (D) A representative of time-lapse progression of ori1 (GFP-ParB1 green) and oAt (RFP-RepBpAt) (AtWX319). (Scale bar, 2 μm.) (E) Schematic model for the organization and segregation of A. tumefaciens genome. The four origins/centromeres are clustered together at the old pole in a newborn cell. Ch1 initiates its replication and segregation first. One copy of ori1 stays anchored at the old pole, whereas the other ori1 moves toward the new pole. ori2/oAt/oTi travels with the moving ori1. ori2 and then oAt/oTi disassociate from ori1 upon their replication. After duplication, the two copies of ori2, oAt, and oTi translocate to the two opposite poles and tether with ori1 again before cytokinesis. The old and new cell poles are indicated. The newly added cell wall materials are labeled in blue.
Discussion
In this study, we use A. tumefaciens as a model to investigate the global interactions of a multipartite genome and how these interactions might function in the maintenance and segregation of the multiple replicons during the cell cycle. Within each chromosome, we observed interarm interactions on both the circular Ch1 and the linear Ch2 and these interactions required the SMC complex (Fig. 1E). These Hi-C patterns can most easily be understood by specific loading of SMC complexes adjacent to the replication origins followed by loop extrusion of these complexes as they translocate to the replication terminus. Our data showed that ParB1 is required for interarm interactions on Ch1 and therefore is likely to load the SMC complex at origin-proximal parS1 sites as has been reported in several bacteria (4, 5, 7, 8, 10). However, our data indicate that RepBCh2 is not required for the interarm interactions on Ch2 and therefore is unlikely to function in SMC loading (Fig. 3). These results suggest that A. tumefaciens encodes an unknown protein that binds to unique sequences adjacent to the origin of Ch2 and facilitates the site-specific loading of SMC complexes. Identification and characterization of this factor may reveal a broadly conserved mechanism for SMC loading onto secondary replicons.
It is noteworthy that despite the complete loss of interarm interactions in the absence of SMC, cells lacking this protein exhibited no observable growth defect (Fig. 2F) and only a mild defect in chromosome segregation (SI Appendix, Fig. S3B). A similar phenomenon has been reported for a related alpha proteobacterium, C. crescentus (8). Interestingly, in both organisms, the ParABS system is essential. By contrast, SMC and MukBEF in B. subtilis and E. coli are essential during rapid growth, whereas the ParABS system is either dispensable or completely absent (53–55). Thus, different bacteria have evolved to rely more heavily on the partitioning system or the SMC family of proteins for chromosome segregation.
We discovered that the origins of the four replicons are clustered at the cell pole by their centromeres (Figs. 1 and 2), and the two chromosomes are linearly aligned along their lengths. This organization is reminiscent of the “bouquet” arrangement in meiotic prophase I cells, in which the telomeres are clustered to help homologous chromosomes pairing and removing entanglement between different chromatids (56). In bacteria with multiple replicons, the clustered origins and the linear alignment of the large replicons could reduce chromosome entanglement and help genome organization. This arrangement could also locate genes with related functions on different chromosomes in close proximity (57). Moreover, a recent study preformed Hi-C on 24 species across the eukaryotic domain of life and proposed that centromeric clustering is conserved since the last eukaryotic common ancestor (58). Because we show that the origins of the four replicons of A. tumefaciens cluster through their centromeres, it will be exciting to investigate whether centromere clustering in eukaryotes and in bacteria was inherited from a common ancestor or has evolved independently.
We found that the primary chromosome’s centromere (ParB1/parS1) plays a key role in ori-ori clustering. Moreover, disruption of ParB1 might have caused the loss of pTi (Fig. 2 C and D and SI Appendix, Fig. S4 E–L). Based on these observations, we propose that centromeric clustering could be a general solution for maintaining multipartite genomes. By clustering centromeres, the secondary replicons are attached to the primary chromosome, ensuring their maintenance even under conditions in which there is no selective advantage for their inheritance. This clustering may also serve as a counting mechanism to ensure one copy of each replicon segregates with each primary chromosome. Our cytological analyses reveal that the ori-ori clustering is lost shortly after the replication initiation of the secondary replicons. We hypothesize that replication through the parS sequences disrupts the centromeric nucleoprotein complex and the clustering. Reformation of the replicated centromeric complexes initiates their segregation to opposite poles and re-establishes the origin clustering.
Our single-cell localization data indicate that the four replicons have a hierarchical organization. The primary chromosome is located closest to the cell pole, followed by secondary replicons; the primary chromosome replicates and segregates first, followed by the secondary replicons (Fig. 3). Similar phenomena have been observed in other species (27–29). We hypothesize that ori-ori clustering plays a role in regulating the initiation of replication and in facilitating the segregation of secondary replicons. In V. cholerae, the replication of Ch2 is triggered by the replication of crtS on Ch1 (32, 33). It is unclear how A. tumefaciens coordinates the timing of replication of the four replicons. Our data suggesting that the unreplicated origins migrate with the replicated ori1 away from the pole raise the possibility that displacement of the origins of the secondary replicons from a replication inhibitor at the cell pole could trigger replication initiation of these origins. It is also possible that timing of Ch1 replication and the delay in secondary replicons are mediated by ParA and RepA proteins, since ParA is implicated in replication control in B. subtilis (59, 60).
It is unclear whether the clustering of the origins is through direct interactions between the centromeres or bridged by other proteins. Previous work in C. crescentus, B. subtilis, and V. cholerae identified polarly localized proteins that interact with sequence-specific DNA-binding proteins and anchor the origin regions at the cell poles (61, 62). The ori-ori clustering interactions observed here could be mediated by such polar proteins. A. tumefaciens has a unipolar growth mode (37) in which the cell envelope is only synthesized at the new cell pole (also called a growth pole). So far, three pole-organizing proteins have been characterized: PopZ, GPR, and PodJ. PopZ and GPR are growth-pole proteins required for unipolar growth, cell shape, and normal cell division (46, 63–66); PodJ localizes to the old pole and plays an essential role in the growth-pole to old-pole transition (66, 67). Future studies will be directed at testing whether any of these polar proteins anchor the origin regions and serve as mediators of the interreplicon contacts.
In summary, our study reveals the global interaction pattern of a divided bacterial genome. We postulate that clustering of the centromeres facilitates the organization, segregation, and maintenance of multipartite bacterial genomes.
Materials and Methods
A. tumefaciens strains were derived from the strain C58 (39) or 15955 (42). Cells were grown as specified in defined minimal medium (40) (ATGN), virulence induction broth (43), or LB broth at 30 °C with aeration. The doubling time of wild-type is 202 min ± 7 min (mean ± SD) in ATGN (n = 3) and 89 min ± 2 min (mean ± SD) in LB (n = 3).
In liquid media, when appropriate, the following antibiotics or supplements were added at the indicated concentrations: kanamycin (IBI, IB02120) 150 μg/mL, carbenicillin (GoldBio, C-103-5) 25 μg/mL, gentamicin (ACROS Organics, AC613980010) 150 μg/mL, IPTG (Dot Scientific, DS102125) 0.2 or 0.25 mM as indicated, theophylline (Sigma, T1633-100G) 2 mM, and AHL (N-3-oxooctanoyl-L-homoserine lactone) (Sigma, O1764-10MG) 1 μM. Antibiotics were doubled when applied on solid media. Virulence induction broth (43) is a modified ATGN medium, replacing AT buffer with 50 mM phosphate buffer (NaH2PO4 and Na2HPO4, pH 5.7) with 0.02 M 2-N-[morpholino] ethanesulfonate, and supplementing with 200 μM acetosyringone.
For Hi-C, ChIP-seq, WGS, or microscopy experiments, cells were streaked on ATGN plates. Single colonies were inoculated into 5 mL ATGN medium and rolled overnight. In the next morning, the cultures were diluted into 30 mL ATGN liquid with a starting OD600 of 0.15. Cultures were grown in a shaking water bath for 6 h to reach an OD600 of 0.5–0.6 before harvest. For ParB1+, AtWX192/193 were supplied with inducers (1 μM AHL and 2 mM theophylline) when growing on solid media or in liquid media. For ParB1−, AtW192/193 were grown in ATGN or LB solid medium or liquid media without inducers for indicated amount of time. Cultures were diluted before their OD600 reached 0.8 to prevent cells from entering stationary phase. The best depletion was achieved in LB after 30 h (SI Appendix, Fig. S4M).
Detailed procedures of Hi-C, ChIP-seq, WGS, sequence analysis, fluorescence microscopy, image analysis, immunoblot analysis, antibody generation, and strain and plasmid construction can be found in SI Appendix, Materials and Methods. Lists of strains, plasmids, oligonucleotides, and next-generation sequencing samples can be found in SI Appendix, Tables S1–S4.
Data are deposited in the Gene Expression Omnibus repository (accession no. GSE182881). Further information and requests for resources, reagents, and analytical scripts should be directed to and will be fulfilled by the corresponding author. Plasmids and strains generated in this study are available from the corresponding author with a completed Materials Transfer Agreement.
Supplementary Material
Acknowledgments
We thank the Wang and Fuqua labs for discussions and support, Kathy Zhang and Jordan Winn for initial observations, and the Indiana University Center for Genomics and Bioinformatics for assistance with high-throughput sequencing. We thank Ivan Surovtsev, Paula Montero Llopis, and Ellen Quardokus for advice on microscopy and analysis; Pam Brown, Ankur Dalia, Patricia Zambryski, and John Zupan for strains and plasmids; and Dan Kearns and David Rudner for critical reading of the manuscript. Support for this work comes from NIH Grants R01GM141242 (X.W.) and R01GM120337 (C.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115854119/-/DCSupplemental.
Data Availability
Next-generation sequencing data have been deposited in the Gene Expression Omnibus repository (accession no. GSE182881). Plasmids and strains generated in this study are available from the corresponding author with a completed Materials Transfer Agreement.
References
- 1.diCenzo G. C., Finan T. M., The divided bacterial genome: Structure, function, and evolution. Microbiol. Mol. Biol. Rev. 81, e00019-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misra H. S., Maurya G. K., Kota S., Charaka V. K., Maintenance of multipartite genome system and its functional significance in bacteria. J. Genet. 97, 1013–1038 (2018). [PubMed] [Google Scholar]
- 3.Jha J. K., Baek J. H., Venkova-Canova T., Chattoraj D. K., Chromosome dynamics in multichromosome bacteria. Biochim. Biophys. Acta 1819, 826–829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lioy V. S., Junier I., Lagage V., Vallet I., Boccard F., Distinct activities of bacterial condensins for chromosome management in Pseudomonas aeruginosa. Cell Rep. 33, 108344 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Böhm K., et al. , Chromosome organization by a conserved condensin-ParB system in the actinobacterium Corynebacterium glutamicum. Nat. Commun. 11, 1485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lioy V. S., et al. , Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172, 771–783.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Wang X., et al. , Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29, 1661–1675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le T. B., Imakaev M. V., Mirny L. A., Laub M. T., High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szafran M. J., et al. , Spatial rearrangement of the Streptomyces venezuelae linear chromosome during sporogenic development. Nat. Commun. 12, 5222 (2021). [DOI] [PMC free article] [PubMed]
- 10.Marbouty M., et al. , Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution Imaging. Mol. Cell 59, 588–602 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Livny J., Yamaichi Y., Waldor M. K., Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J. Bacteriol. 189, 8693–8703 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breier A. M., Grossman A. D., Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64, 703–718 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Murray H., Ferreira H., Errington J., The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 61, 1352–1361 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Fogel M. A., Waldor M. K., A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20, 3269–3282 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang L. C., et al. , ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 32, 1238–1249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ptacin J. L., et al. , A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12, 791–798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu L., Vecchiarelli A. G., Mizuuchi K., Neuman K. C., Liu J., Brownian Ratchet mechanism for faithful segregation of low-copy-number plasmids. Biophys. J. 112, 1489–1502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surovtsev I. V., Campos M., Jacobs-Wagner C., DNA-relay mechanism is sufficient to explain ParA-dependent intracellular transport and patterning of single and multiple cargos. Proc. Natl. Acad. Sci. U.S.A. 113, E7268–E7276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhelm L., et al. , SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4, e06659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan N. L., Marquis K. A., Rudner D. Z., Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137, 697–707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruber S., Errington J., Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137, 685–696 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Tran N. T., Laub M. T., Le T. B. K., SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 20, 2057–2071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlandini E., Marenduzzo D., Michieletto D., Synergy of topoisomerase and structural-maintenance-of-chromosomes proteins creates a universal pathway to simplify genome topology. Proc. Natl. Acad. Sci. U.S.A. 116, 8149–8154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rybenkov V. V., Herrera V., Petrushenko Z. M., Zhao H., MukBEF, a chromosomal organizer. J. Mol. Microbiol. Biotechnol. 24, 371–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mäkelä J., Sherratt D. J., Organization of the Escherichia coli chromosome by a MukBEF axial core. Mol. Cell 78, 250–260.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandão H. B., Ren Z., Karaboja X., Mirny L. A., Wang X., DNA-loop-extruding SMC complexes can traverse one another in vivo. Nat. Struct. Mol. Biol. 28, 642–651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubarry N., Willis C. R., Ball G., Lesterlin C., Armitage J. P., In vivo imaging of the segregation of the 2 chromosomes and the cell division proteins of Rhodobacter sphaeroides reveals an unexpected role for MipZ. MBio 10, e02515-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frage B., et al. , Spatiotemporal choreography of chromosome and megaplasmids in the Sinorhizobium meliloti cell cycle. Mol. Microbiol. 100, 808–823 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Deghelt M., et al. , G1-arrested newborn cells are the predominant infectious form of the pathogen Brucella abortus. Nat. Commun. 5, 4366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David A., et al. , The two Cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLoS Genet. 10, e1004448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cockram C., Thierry A., Gorlas A., Lestini R., Koszul R., Euryarchaeal genomes are folded into SMC-dependent loops and domains, but lack transcription-mediated compartmentalization. Mol. Cell 81, 459–472.e10 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Val M. E., et al. , A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci. Adv. 2, e1501914 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek J. H., Chattoraj D. K., Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet. 10, e1004184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heidelberg J. F., et al. , DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood D. W., et al. , The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Pinto U. M., Pappas K. M., Winans S. C., The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 10, 755–765 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Brown P. J., et al. , Polar growth in the Alphaproteobacterial order Rhizobiales. Proc. Natl. Acad. Sci. U.S.A. 109, 1697–1701 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieberman-Aiden E., et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson B., Currier T. C., Gordon M. P., Chilton M. D., Nester E. W., Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 123, 255–264 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morton E. R., Fuqua C., Laboratory maintenance of Agrobacterium. Curr. Protoc. Microbiol. Chapter 1, Unit3D.1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahng L. S., Shapiro L., Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J. Bacteriol. 185, 3384–3391 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dessaux Y., Tempé J., Farrand S. K., Genetic analysis of mannityl opine catabolism in octopine-type Agrobacterium tumefaciens strain 15955. Mol. Gen. Genet. 208, 301–308 (1987). [DOI] [PubMed] [Google Scholar]
- 43.Barton I. S., Platt T. G., Rusch D. B., Fuqua C., Destabilization of the tumor-inducing plasmid from an octopine-type Agrobacterium tumefaciens lineage drives a large deletion in the co-resident at megaplasmid. G3 (Bethesda) 9, 3489–3500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Figueroa-Cuilan W., Daniel J. J., Howell M., Sulaiman A., Brown P. J., Mini-Tn7 Insertion in an artificial attTn7 site enables depletion of the essential master regulator CtrA in the phytopathogen Agrobacterium tumefaciens. Appl. Environ. Microbiol. 82, 5015–5025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danhorn T., Hentzer M., Givskov M., Parsek M. R., Fuqua C., Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186, 4492–4501 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zupan J. R., Grangeon R., Robalino-Espinosa J. S., Garnica N., Zambryski P., GROWTH POLE RING protein forms a 200-nm-diameter ring structure essential for polar growth and rod shape in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 116, 10962–10967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Topp S., et al. , Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 76, 7881–7884 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwang I., et al. , TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. U.S.A. 91, 4639–4643 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan S. R., Gaines J., Roop R. M. II, Farrand S. K., Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74, 5053–5062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen H. J., Ottesen J. R., Youngren B., Austin S. J., Hansen F. G., The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62, 331–338 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Robalino-Espinosa J. S., Zupan J. R., Chavez-Arroyo A., Zambryski P., Segregation of four Agrobacterium tumefaciens replicons during polar growth: PopZ and PodJ control segregation of essential replicons. Proc. Natl. Acad. Sci. U.S.A. 117, 26366–26373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toro E., Hong S. H., McAdams H. H., Shapiro L., Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 105, 15435–15440 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Badrinarayanan A., Lesterlin C., Reyes-Lamothe R., Sherratt D., The Escherichia coli SMC complex, MukBEF, shapes nucleoid organization independently of DNA replication. J. Bacteriol. 194, 4669–4676 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gruber S., et al. , Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr. Biol. 24, 293–298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Tang O. W., Riley E. P., Rudner D. Z., The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr. Biol. 24, 287–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleckner N., et al. , A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. U.S.A. 101, 12592–12597 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright M. A., Kharchenko P., Church G. M., Segrè D., Chromosomal periodicity of evolutionarily conserved gene pairs. Proc. Natl. Acad. Sci. U.S.A. 104, 10559–10564 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoencamp C., et al. , 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372, 984–989 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray H., Errington J., Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135, 74–84 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Scholefield G., Errington J., Murray H., Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31, 1542–1555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X., Montero Llopis P., Rudner D. Z., Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14, 191–203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Badrinarayanan A., Le T. B., Laub M. T., Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 31, 171–199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howell M., et al. , Absence of the polar organizing protein popz results in reduced and asymmetric cell division in Agrobacterium tumefaciens. J. Bacteriol. 199, e00101-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grangeon R., Zupan J., Jeon Y., Zambryski P. C., Loss of PopZ At activity in Agrobacterium tumefaciens by deletion or depletion leads to multiple growth poles, minicells, and growth defects. MBio 8, e01881-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ehrle H. M., et al. , Polar organizing protein PopZ is required for chromosome segregation in Agrobacterium tumefaciens. J. Bacteriol. 199, e00111-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grangeon R., Zupan J. R., Anderson-Furgeson J., Zambryski P. C., PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 112, 11666–11671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson-Furgeson J. C., Zupan J. R., Grangeon R., Zambryski P. C., Loss of PodJ in Agrobacterium tumefaciens leads to ectopic polar growth, branching, and reduced cell division. J. Bacteriol. 198, 1883–1891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data have been deposited in the Gene Expression Omnibus repository (accession no. GSE182881). Plasmids and strains generated in this study are available from the corresponding author with a completed Materials Transfer Agreement.