Significance
Microbial cells organized on solid surfaces are the most ancient form of biological communities. Yet how single cells interact with surfaces and integrate a variety of signals to establish a sessile lifestyle is poorly understood. We developed and used sensitive biosensors to determine the kinetics of second messengers’ responses to surface attachment. This allowed us to examine cell-by-cell variability of the initial signaling events and establish that some of these events depend on flagellar motor function while others do not. Environmentally determined factors, like the energetic status of the cell, can modulate all signaling events. The complex interplay between the surface interaction inputs and external conditions can now be studied using our system.
Keywords: cyclic-di-GMP, surface sensing, pH, biofilm, cAMP
Abstract
Bacterial cells interact with solid surfaces and change their lifestyle from single free-swimming cells to sessile communal structures (biofilms). Cyclic di-guanosine monophosphate (c-di-GMP) is central to this process, yet we lack tools for direct dynamic visualization of c-di-GMP in single cells. Here, we developed a fluorescent protein–based c-di-GMP–sensing system for Escherichia coli that allowed us to visualize initial signaling events and assess the role played by the flagellar motor. The sensor was pH sensitive, and the events that appeared on a seconds’ timescale were alkaline spikes in the intracellular pH. These spikes were not apparent when signals from different cells were averaged. Instead, a signal appeared on a minutes’ timescale that proved to be due to an increase in intracellular c-di-GMP. This increase, but not the alkaline spikes, depended upon a functional flagellar motor. The kinetics and the amplitude of both the pH and c-di-GMP responses displayed cell-to-cell variability indicative of the distinct ways the cells approached and interacted with the surface. The energetic status of a cell can modulate these events. In particular, the alkaline spikes displayed an oscillatory behavior and the c-di-GMP increase was modest in the presence of glucose.
Free-swimming bacterial cells attach to solid supports and initiate a transcriptional program essential for the transition to a sessile lifestyle (1). The trigger event, surface sensing, is not well understood. Many cell surface proteins and cellular appendages have been shown to mediate a response to surface attachment (2). Ideally, one would want to link each such “surface detector” to an initial intracellular signal and then further link that signal to an established long-term response. It is challenging to study how single cells respond dynamically to the surface encounter, mainly because not all initial signals can be identified by genetics and suspected secondary messengers’ dynamics are difficult to detect with high temporal resolution (2).
Single-cell imaging experiments indicate that cyclic di-guanosine monophosphate (c-di-GMP), an established mediator of the planktonic-to-sessile lifestyle transition (3, 4), is also an initial signal to surface attachment (5, 6). The signaling networks that respond to surface attachment are species specific, yet the connection between surface attachment, c-di-GMP increase, and biofilm formation has been demonstrated in several bacteria (3, 4). For Pseudomonas aeruginosa, one of the most studied models of biofilm formation, the signaling networks appear to generate a hierarchical cascade of secondary messengers (7) and result in distinct developmental states within a bacterial population (5, 6, 8, 9). The ability to visually track c-di-GMP concentration changes in individual cells is essential to understanding how various inputs increase the c-di-GMP concentration in different bacteria.
Here, we developed a fluorescent protein–based c-di-GMP–sensing system that allowed us to visualize initial signals to surface attachment in single cells of Escherichia coli and assess the role of the flagellar motor input. E. coli possesses specific challenges to dynamic c-di-GMP detection. In particular, the rapid cell doubling requires fast folding proteins as part of a sensing system, and the low c-di-GMP concentration requires sensing in the 100 to 700 nM range (10, 11). Key to achieving the required sensitivity is a biosensor bright enough to be detected at low intracellular concentrations.
Results
Development of a c-di-GMP–Sensing System for E. coli.
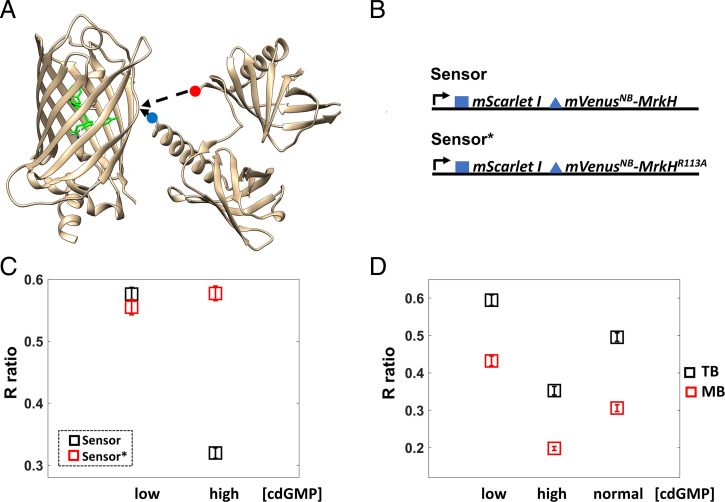
Biosensors based on fluorescence resonance energy transfer (FRET) were previously used to detect changes in c-di-GMP concentration (12, 13). They require overnight incubation at 4 °C for fluorescent protein maturation in E. coli and therefore do not allow for measurements in unperturbed, free-swimming cells. Here, we developed a single fluorescent protein biosensor using mVenusNB, the fastest-folding yellow fluorescent protein (YFP) (14), and MrkH [Klebsiella pneumoniae transcription factor with 100 to 200 nM affinity for c-di-GMP (15, 16)] inserted between Y145 and N146 of mVenusNB (Fig. 1A and Methods). This location mediates YFP fluorescence emission changes in response to conformational changes of inserted protein domains (17), and MrkH does undergo a conformational change upon c-di-GMP binding. Specifically, the MrkH amino-terminal domain rotates 138° relative to its carboxyl-terminal domain (15); this rotation should disrupt the mVenusNB chromophore environment, resulting in reduced fluorescence of the c-di-GMP–bound biosensor.
Fig. 1.
Biosensor development and validation. (A) The MrkH insertion into the mVenusNB β-barrel is depicted. The image was generated in UCSF Chimera (49) using 1HUY.pdb for mVenusNB and 5KEC.pdb for MrkH. The red dot marks the amino terminus, and the blue dot marks the carboxyl terminus of MrkH. The amino acids that make up the chromophore are highlighted in green. (B) A diagram of the expression system is shown; the blue square represents the T7 phage ribosome-binding site, and the blue triangle represents the less efficient ribosome-binding site (GGAACAGAC sequence). Both mScarletI and the mVenusNB–MrkH fusion are expressed using the same promoter (proC, black arrow). The Sensor* platform contains the MrkH R113A mutation that abolishes c-di-GMP binding. Thus, only the Sensor platform is expected to be c-di-GMP responsive. (C) The Sensor platform is validated in cells grown exponentially in TB and added directly to channel slides. The average ratio (R) of mVenusNB to mScarletI fluorescence emissions is plotted for cell populations expressing the Sensor (black squares) or the Sensor* platform (red squares) overexpressing either the phosphodiesterase PdeH (low intracellular c-di-GMP) or the diguanylate cyclase WspR:D70E (high intracellular c-di-GMP). Each square represents the average measurement for 179 cells, and brackets represent 95% CIs. Notice that the Sensor platform R ratio is sensitive to changes in c-di-GMP concentration, in contrast to the Sensor*’s R ratio. A decrease in the Sensor R value corresponds to an increase in c-di-GMP concentration. (D) Cells in TB have higher internal pH than cells in motility buffer (MB), and the Sensor’s R values vary accordingly. Black squares represent the average measurements in TB and red squares the average measurements in motility buffer. More than 126 cells were measured for each average, and the brackets represent 95% CIs.
We figured that the key to achieving the required sensitivity [100 to 700 nM range (10, 11)] is a biosensor that can itself be detected at low concentrations. We therefore constitutively expressed the mVenusNB-MrkH fusion using a low copy plasmid developed by Davis et al. (18) and further characterized by Balleza et al. (14). For a ratiometric measurement, we coexpressed mScarletI, a bright, rapidly folding red fluorescent protein (19) (Fig. 1B). As a control, we introduced the R113A mutation in MrkH (numbering as in ref. 15) previously shown to eliminate its c-di-GMP–binding ability (15, 16). We thus produced two systems, a biosensor (Sensor) and a mutant biosensor (Sensor*) (Fig. 1B), with the expectation that the former is sensitive to c-di-GMP while the latter is not and thus serves as a control for fluorescence changes unrelated to c-di-GMP concentration changes.
The Biosensor Responds to c-di-GMP Concentration Changes in Motile E. coli.
We created cell populations with low c-di-GMP concentrations (expressing the phosphodiesterase PdeH) and high c-di-GMP concentrations [expressing a constitutively active form of the diguanylate cyclase WspR (20)] and measured the ratio (R) of mVenusNB to mScarletI fluorescence emission for individual cells (Fig. 1C). We found that the average R value for cells that express the Sensor is ∼2-fold higher in the low c-di-GMP than in the high c-di-GMP regimen. This is consistent with our expectation that the c-di-GMP–bound biosensor has reduced mVenusNB fluorescence. In contrast, cells that express the mutant biosensor (Sensor*) have similar average R values between the two c-di-GMP regimens (low and high c-di-GMP concentrations, Fig. 1C and SI Appendix, Fig. S1A). This indicates that Sensor, but not Sensor*, responds to changes in c-di-GMP concentration and that the dynamic range of our sensing system is at least twofold.
Note that E. coli with unperturbed levels of c-di-GMP registers between the low and high c-di-GMP regimens (Fig. 1D). In addition, cells that lack stators (ΔmotAmotB) and are therefore not motile have lower than wild-type intracellular c-di-GMP (higher average R ratio, SI Appendix, Fig. S1B). This suggests that both increases and decreases in the c-di-GMP concentration could be detected with our system.
The Biosensor and the Mutant Biosensor Are pH Sensitive.
YFPs with insertions suffer from pH sensitivity in the physiological pH range (17). In contrast, mScarletI fluorescence, as well as MrkH c-di-GMP–binding ability, are not sensitive to pH in that range (13, 19, 21) (SI Appendix, Fig. S3). We collapsed E. coli internal pH with the uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and found that both biosensors (Sensor and Sensor*) are pH sensitive in the 7 to 8 pH range (SI Appendix, Fig. S2A, green and gray symbols, respectively). Therefore, fluorescence changes that occur for both biosensors likely reflect changes in internal pH, while changes that occur only in the Sensor’s fluorescence correspond to changes in c-di-GMP concentration. The pH sensitivity is in line with previous reports, such as refs. 17 and 22. However, the pKa of our sensors (∼7.7) is close to E. coli’s internal pH (∼7.6) (23–26), making them sensitive to physiological pH changes. Note that the Sensor platform exhibits similar pH sensitivity for both the low c-di-GMP and the high c-di-GMP regimens (SI Appendix, Fig. S2A, red and black symbols, respectively), possibly indicating that the two states (c-di-GMP bound and c-di-GMP unbound) have similar pH sensitivities.
Using Sensor* as a pH indicator, we found that internal pH decreased when the cells were transferred from tryptone broth (TB) to motility buffer (Methods and SI Appendix, Fig. S2B). The Sensor’s R values varied accordingly between TB and motility buffer for both the low c-di-GMP and the high c-di-GMP regimens (Fig. 1D).
The c-di-GMP Concentration Increases after E. coli Surface Attachment.
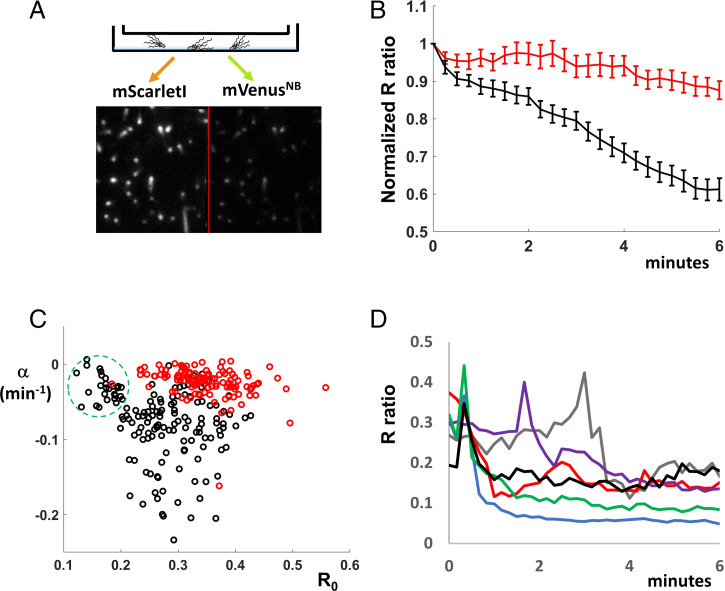
To detect changes in the intracellular c-di-GMP concentration following surface attachment, we imaged cells attached to the coverslips of premade channel slides (from Ibidi, see Methods). We acquired mVenusNB and mScarletI fluorescence using our total internal reflection fluorescence (TIRF) microscope setup for cells resuspended in motility buffer with 10 mM lactate as an external energy source (Fig. 2A). Following rapid attachment (less than 1 min), images were acquired every 15 s. Fig. 2B shows the average R ratios for cells expressing either Sensor (black symbols) or Sensor* (red symbols) as a function of time. The decrease observed for Sensor-expressing cells, but not for Sensor*-expressing cells, is consistent with an increase in c-di-GMP within the first 5 min of surface attachment. Interestingly, the c-di-GMP increase did not occur in the presence of 40 mM potassium benzoate and 40 mM methylamine (SI Appendix, Fig. S4), which dissipate the transmembrane pH gradient. Thus, the c-di-GMP increase depends on the presence of this proton concentration gradient. Note that the modest R decrease for Sensor*-expressing cells (Fig. 2B, red symbols) and for both Sensor- and Sensor*-expressing cells in SI Appendix, Fig. S4 is consistent with mVenusNB bleaching at a faster rate than mScarletI during the experiment (SI Appendix, Fig. S5A).
Fig. 2.
Single-cell imaging reveals an increase in c-di-GMP following attachment. (A) A diagram of our imaging system is shown. Cells are added to channel slides, and they attach to poly-L-lysine–treated coverslips. They are imaged on an inverted Nikon microscope in TIRF mode using a 514-nm laser and a split electron multiplying CCD camera that allows simultaneous recording of mScarletI (Left) and mVenusNB (Right) fluorescence. For each cell within the image, the corresponding mScarletI and mVenusNB intensity values were measured in ImageJ and the ratio R of mVenusNB to mScarletI fluorescence was computed. (B) E. coli cells in motility buffer with lactate were added to channel sides. Cells attached for 45 s, excess motility buffer was added to the channels, and image acquisition (every 15 s) was started 1 min after cell addition. The average R ratios (individual traces normalized to initial values) are plotted as a function of time for Sensor-expressing cells (146 cells averaged, black line) and Sensor*-expressing cells (120 cells averaged, red line). The 95% CIs are shown on the plot for each time point. The Sensor's R value decrease corresponds to an increase in c-di-GMP concentration. (C) The cell-to-cell variable kinetics of the c-di-GMP increase is displayed by fitting the individual traces averaged in Fig. 2B to a single term exponential R0eαt and plotting the rate constant α (min−1) as a function of R0; Sensor, black circles; Sensor*, red circles. The green dashed circle outlines a cell population with low R0 (high c-di-GMP) and little or no c-di-GMP increase. (D) Sensor-expressing cells in lactate motility buffer were imaged every 10 s as they attached to coverslips. The ratios R for individual cells (differently colored lines) were plotted as a function of time. Some cells decrease R (increase c-di-GMP) abruptly within the first minute of attachment.
Among individual cells, the signal descent (c-di-GMP increase) is heterogeneous, with some cells descending rapidly (within 1 min), some gradually, and some not at all. To display this heterogeneity and to directly compare individual traces of Sensor- and Sensor*-expressing cells, we fitted each trace to an exponential function (R0eαt) and plotted the descent constant α (min−1) versus the initial R0 value. Notice that decent constants for most Sensor*-expressing cells (Fig. 2C, red circles) cluster around zero, while decent constants for Sensor-expressing cells (Fig. 2C, black circles) are heterogeneous. The Sensor-expressing population with low R0 values (high c-di-GMP) and small decent constants (Fig. 2C, green dashed circle) suggested the possibility that some cells increase c-di-GMP within the first minute of surface attachment. We imaged Sensor-expressing cells every 10 s as they attached to the coverslip (Fig. 2D) and found, indeed, that some cells decreased their R value (increased their c-di-GMP concentration) within the first minute of surface attachment (Fig. 2D), in contrast to Sensor*-expressing cells (SI Appendix, Fig. S6). Interestingly, some of the Sensor-expressing cells that decreased their R values abruptly also “spiked” just before the decrease (Fig. 2D). Cells expressing Sensor* also spiked during the experiment (SI Appendix, Fig. S6), but they lacked the subsequent rapid downward decrease of the Sensor-expressing cells (Fig. 2D).
The Intracellular pH Spikes after E. coli Surface Attachment.
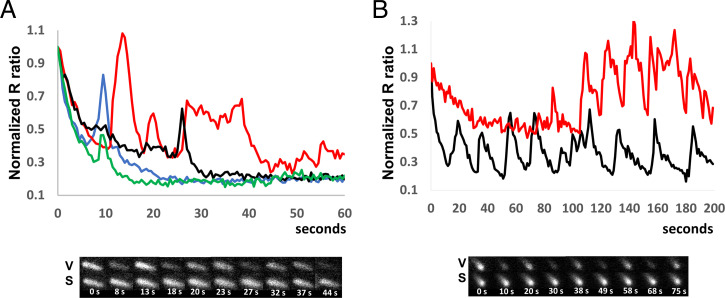
To further evaluate these spikes, we acquired images with high temporal resolution (every 0.5 and/or 1 s) for both Sensor- and Sensor*-expressing cells. During the first 5 min of surface attachment, we detected spikes for both cell types. This suggests that the spikes represent pH changes, specifically, pH increases (alkaline spikes). The spikes’ duration, amplitude, and position vary widely from cell to cell. Some cells display well-defined narrow spikes, while others exhibit broad and/or low-amplitude events. Fig. 3A is an example of “spectacular” spiking, with data acquired every half second following cell attachment. Spikes with full width at half maximum (FWHM) under 2 s (blue trace), along with a spike with FWHM of ∼15 s (red trace), can be seen. Fine structure can also be observed in many of the larger spikes (such as the red trace), suggesting that the larger spikes might result from temporal superposition of several more basic events. Note that at this acquisition rate (every 0.5 s), the main reason for the global decrease in R values is mVenusNB bleaching faster than mScarletI (SI Appendix, Fig. S5B).
Fig. 3.
The intracellular pH spikes after E. coli surface attachment. (A) Sensor*-expressing cells in motility buffer with lactate were imaged every 0.5 s as they attached to coverslips. The ratio R for individual cells (normalized to the initial value) was plotted as a function of time. The R value spikes represent transient pH increases (alkaline spikes), since they occur for both Sensor- and Sensor*-expressing cells. Note the variable amplitude, position, and width of the spikes. Differently colored lines correspond to different cells. Below, images from the red trace cell are shown, with mVenusNB fluorescent images labeled with V and the mScarletI images labeled with S. Time (s) is shown below each pair of images. (B) Sensor-expressing cells in motility buffer with glucose were imaged every 1 s. The ratio R for individual cells (normalized to the initial value) was plotted as a function of time. Many traces, such as the two shown, display a burst of periodic spikes in contrast to the spikes generated in motility buffer with lactate. Below, images from the black trace cell are shown, labeled as in Fig. 3A.
Approximately 50% of the cells spike in the first 3 min of attachment, and only around 10% of cells spike after they have been attached for 5 min (SI Appendix, Table). This suggests that alkaline spiking is a transient event that occurs in the initial stages of surface encounters.
When 40 mM potassium benzoate and 40 mM methylamine were added to the motility buffer, no alkaline spikes were observed—as expected, since potassium benzoate and methylamine clamp the cytoplasmic pH to the external pH (SI Appendix, Fig. S7).
The Alkaline Spikes Display Periodicity and the c-di-GMP Increase Is Modest in Motility Buffer with Glucose.
When glucose, rather than lactate, was used as the external energy source, elaborate, persistent spiking events were observed (Fig. 3B). Many spiking events were periodic with a ∼20-s period (Fig. 3B), resembling the glycolytic pH oscillations observed in Saccharomyces cerevisiae (27). “Glucose spiking” is more persistent than “lactate spiking,” since many cells still display periodic spiking events 5 min after surface encounter (SI Appendix, Table). Like the lactate spikes, the glucose spikes were not observed when benzoate and methylamine were added to motility buffer (SI Appendix, Fig. S8). Thus, both spiking events are blocked by a pH clamp.
Cells attaching in lactate motility buffer increased the intracellular c-di-GMP concentration within 5 min of finding the surface (Fig. 2B). In contrast, we detected only a modest increase in intracellular c-di-GMP for cells attaching in glucose motility buffer. Specifically, the average R ratio decreased for both Sensor- and Sensor*-expressing cells when attaching in glucose buffer (SI Appendix, Fig. S9A), consistent with a decrease in intracellular pH. However, the R ratio decrease was more pronounced for Sensor- than for Sensor*-expressing cells, suggesting a modest increase in intracellular c-di-GMP in addition to the decrease in internal pH. To assay this differently and eliminate the impact of mVenusNB bleaching during the time course of the experiment, we measured R values for cell populations that have just found the surface and compared them to R values of cell populations that have been attached for 5 to 6 min at opposite ends of the channel slide. We found that Sensor* cells have lower R values after 5 to 6 min attachment but similarly shaped distributions (SI Appendix, Fig. S9 B and C), consistent with a decrease in internal pH. The average R value for Sensor-expressing cells also decreased (SI Appendix, Fig. S9B). However, the distribution of R values tightened, suggesting that the initial population (broad distribution and diverse c-di-GMP concentrations) became more homogenous as it modestly increased the c-di-GMP concentration and decreased the cytosolic pH (SI Appendix, Fig. S9C, black and gray circles).
Note that individual traces show a wide range of behavior, with R values abruptly decreasing, R values spiking and gradually decreasing, and R values spiking with little downward trend (SI Appendix, Fig. S9D).
Flagellar Motor Function Is Required for c-di-GMP Increase in Lactate Buffer.
Genetic analysis in E. coli revealed that motility is critical to normal biofilm formation (28). In addition, flagellar motor activity was shown to be essential for the c-di-GMP response in Caulobacter crescentus and P. aeruginosa (6, 29).
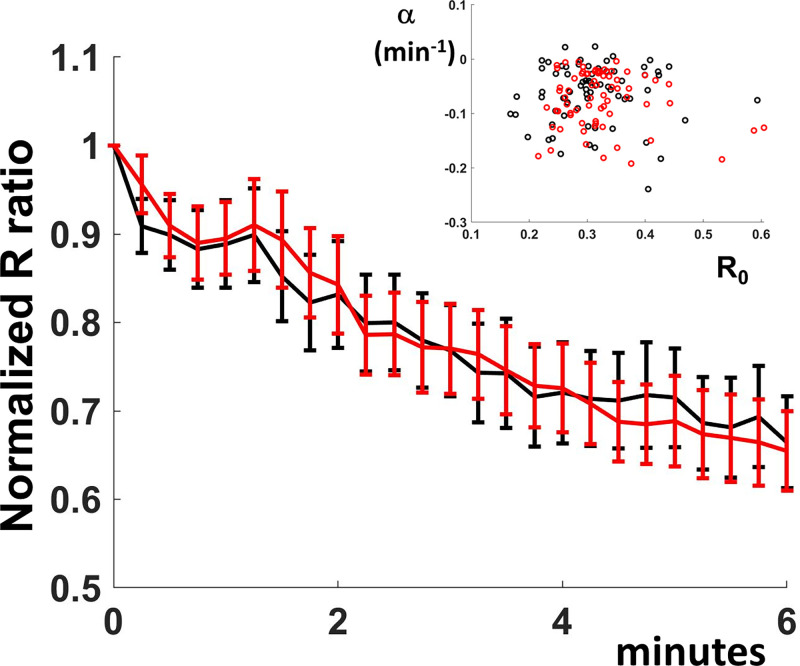
To address the role of the flagellar motor function in the initial surface-sensing events, we imaged cells that lacked stator-unit function (cells with genomic deletions of either motB or both motA and motB). Flagella assemble in these cells, but they do not rotate, so the cells are not motile. We found that the average R value decreased for both the Sensor- and the Sensor*-expressing cells, consistent with a decrease in pH following attachment (Fig. 4). There was no significant difference between the two average traces (Fig. 4), consistent with a lack of c-di-GMP increase within the attached cell population. In addition, as shown in Fig. 4, Inset, individual cell behavior is similar between the Sensor- and the Sensor*-expressing cells. Motor function is thus required for both maintaining intracellular pH and increasing the c-di-GMP concentration following attachment.
Fig. 4.
Flagellar motor function is required for c-di-GMP increase. ΔmotB E. coli cells in motility buffer with lactate were added to channel sides. Cells were attached for 45 s, excess motility buffer was added to the channels, and image acquisition (every 15 s) was started 1 min after cell addition. Average R values (individual traces normalized to initial values) are plotted as a function of time for Sensor- (black line) and Sensor*-expressing cells (red line). The 95% CIs are shown on the plot for each time point. The individual traces were fitted to a single term exponential R0eαt. The inset shows the rate constant α (min−1) as a function of the initial ratio value R0 for both Sensor- (black circles) and Sensor*- (red circles) expressing cells.
Images acquired every 1 s show that ΔmotAmotB cells display alkaline spikes when attached to glass coverslips (SI Appendix, Fig. S10). Therefore, E. coli does not require functional flagellar motors for the generation of alkaline spikes. However, ΔmotAmotB cells did not display clear, periodic events in glucose motility buffer (SI Appendix, Fig. S11), suggesting that the motor’s function is important for the complex spiking behavior of wild-type cells in glucose buffer.
Discussion
When bacterial cells attach to surfaces, a variety of “surface detectors” generate intracellular signals that inform the altered motility state (2). c-di-GMP stands out as an initial signal to surface encounter as well as a longer-term surface-sensing signal integrator. Indeed, its concentration increases within minutes of P. aeruginosa and C. crescentus surface attachment (6, 29, 30). In addition, c-di-GMP is essential to both forming and maintaining a biofilm structure in various bacteria (4).
Here, we developed a bright fluorescent protein–based biosensor to determine the kinetics of c-di-GMP concentration changes that follow surface attachment in single E. coli. Our sensing system, which consists of c-di-GMP–responsive and c-di-GMP–unresponsive platforms (Fig. 1B), informs us of changes in intracellular c-di-GMP as well as intracellular pH. We detected an increase in the c-di-GMP concentration within minutes of E. coli attachment to coverslips (Fig. 2A), consistent with reports from other species (6). In addition, transient pH increases (alkaline spikes) were observed within seconds of the surface encounter. These spikes can be immediately followed by an increase in c-di-GMP (Fig. 2D), which suggests that the alkaline spikes may trigger this increase. Indeed, DgcB, the C. crescentus enzyme responsible for c-di-GMP increase following attachment (29), has a pH-dependent activity that increases at alkaline pH (31). However, in our experimental setup, alkaline spikes per se are not sufficient to trigger the c-di-GMP rise, since cells lacking stator units do spike (SI Appendix, Fig. S10), yet they do not increase intracellular c-di-GMP (Fig. 4). As previously suggested (29), functional flagellar motors may be distinctively required to generate a higher-amplitude, local increase in pH (see next paragraph, SI Appendix, Supplementary Note) necessary to activate a motor-associated diguanylate cyclase.
How are the pH spikes generated? Similar pH transients have been observed in S. cerevisiae, in which they appear to be linked to glycolytic oscillations (27), in the mitochondrion as “mitoflashes” (32, 33), and in plants (34, 35). While the mechanism is not well understood, the spikes are thought to be generated by imbalances in the energy metabolism that trigger a pH increase followed by a compensatory mechanism that returns intracellular pH to its homeostatic value (36). How could surface attachment trigger an increase in intracellular pH? One possibility is that an extracellular pH increase at the attachment interface (37) triggers an increase in intracellular pH (26), which is then brought back to the set pH (generating a spike) by the metabolic enzymes involved in pH homeostasis. Such spikes could be generated in both motile and nonmotile cells and would depend on the nature of the attachment surface (37). An intracellular pH spike could be also generated by stalling the flagellar motor through flagellar interactions with the surface, followed by a similar compensatory mechanism. Indeed, at 100 Hz rotation, a flagellar motor translocates around 50,000 protons per second (38, 39). Even though this is a small contributor to the total proton influx (38), the stall can produce measurable pH spikes, particularly if there is a delay in the proton homeostasis feedback mechanism (SI Appendix, Supplementary Note). A spike generated by the second mechanism would require functional flagellar motors and could display a higher amplitude in the vicinity of the motor.
We detected only a modest increase in intracellular c-di-GMP concentration for cells attaching in glucose motility buffer (SI Appendix, Fig. S9). Glucose inhibits E. coli biofilm formation (40, 41), and this effect is mediated in part by the decreased intracellular cyclic AMP (cAMP) that results from glucose utilization as a carbon source (40, 41). Interestingly, for P. aeruginosa, a cAMP increase is thought to precede and trigger the c-di-GMP increase (7, 42). P. aeruginosa initial attachment events do occur on a longer timescale than our attachment experiment (hours versus minutes) (7). However, the same hierarchical second messenger cascade (cAMP, then c-di-GMP) may occur for E. coli surface attachment, too. Alternatively, cAMP may play a permissive, carbon source–informative role in E. coli biofilm formation (43).
The periodic alkaline spikes observed in the glucose motility buffer (Fig. 3B) are striking and reminiscent of the pH oscillations observed in S. cerevisiae during glycolysis (27). Distinctly, though, E. coli need not be starved to trigger such oscillations, since we immediately transferred the cells from growth media to the buffer containing glucose. Why, then, would the internal pH oscillate upon attachment? Glycolytic enzymes do oscillate together with the internal pH (27, 44), and the key enzyme phosphofructokinase is pH sensitive (27). An initial transient increase in pH following surface attachment could trigger an oscillatory behavior of the glycolytic enzymes and of the internal pH.
Both the pH spiking behavior as well as the c-di-GMP increase display a lot of cell-to-cell variability (Figs. 2 C and D and 3). The variability might be due, in part, to the heterogeneity of the E. coli–surface interactions, since some cells approach and interact with the surface over their entire length, while other cells interact closely only at the poles (as seen in our TIRF images, Fig. 2A). It would be interesting to establish if this heterogeneity persists to create a low c-di-GMP population and a high c-di-GMP population, as for P. aeruginosa (5).
Methods
Strains and Plasmids.
The motile MG1655 strain was used as a parent strain for all the experiments. motB, motA and motB deletions were made using the Datsenko and Wanner method and the pKD3 plasmid (45). Following site-directed recombination to eliminate the chloramphenicol resistance gene, an 85-bp scar was left in place of the disrupted gene. Deletions were verified using PCR and Sanger sequencing of the PCR products.
pdeH and wspRD70E were amplified using PCR either from MG1655 cells or, respectively, from the pCOLA-T7-wspR:D70E plasmid (Addgene No. 79164) (20). The resulting fragments were ligated into a pTrc99A backbone (46) for expression from the inducible hybrid trp/lac promoter.
mrkH (codon optimized, as in ref. 15) was amplified from a plasmid provided by Maria Schumacher (Duke University School of Medicine, Durham, NC); mVenusNB and mScarletI fragments were amplified from the pEB1-mVenusNB, and pEB2-mScarletI plasmids (Addgene Nos. 103986 and 104007, respectively) given to us by Enrique Balleza and Philippe Cluzel (Harvard University, Cambridge MA). Fragments with various linkers and truncations were assembled using the NEBuilder HiFi assembly method. The brightest variant contained a three-amino-acid linker at the MrkH amino terminus and an eight-amino-acid truncation at the MrkH carboxyl terminus. The amino acids at the mVenusNB–MrkH junctions were
 GSG
GSG
 and
and

 (green letters represent mVenusNB amino acids, blue letters represent MrkH amino acids, and GSG is the linker sequence). The ribosome-binding sites were the T7 phage ribosome binding site (for mScarletI expression) and the GGAACAGAC nucleotide sequence between the stop codon (TAA) of mScarletI and the start codon (ATG) of mVenusNB-MrkH for biosensor expression. The R113A mutation was introduced by PCR followed by the NEBuilder HiFi assembly method.
(green letters represent mVenusNB amino acids, blue letters represent MrkH amino acids, and GSG is the linker sequence). The ribosome-binding sites were the T7 phage ribosome binding site (for mScarletI expression) and the GGAACAGAC nucleotide sequence between the stop codon (TAA) of mScarletI and the start codon (ATG) of mVenusNB-MrkH for biosensor expression. The R113A mutation was introduced by PCR followed by the NEBuilder HiFi assembly method.
Cell Preparation and Experimental Setup.
Cells for microscopy were grown in tryptone broth (TB, 10 g/L Bacto-Tryptone, 5 g/L sodium chloride) at 30 °C from single colonies and inoculated into fresh TB media with necessary antibiotics at 1:1,000 dilution. The cultures were grown at 28 to 29 °C for 7 to 8 h until the optical density at 600 nm (OD600) was between 0.5 and 0.55. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at 20 μM when required.
Channel slides from Ibidi (µ-Slide VI 0.5 Glass Bottom) were used for microscopy. The channels (40 μL volume) were coated with 0.1% poly-L-lysine solution (Sigma) and washed with at least 2 mL water. Cells were added either directly in growth media or after being washed two times and resuspended in motility buffer at OD600 0.5. Motility buffer is 10 mM potassium phosphate, 100 μM ethylenediaminetetraacetic acid (EDTA), and 10 mM lactate (or 0.2% glucose) pH 6.9.
A total of 200 μL cells were added to the channel reservoir, and image acquisition was started immediately (for imaging cells as they attached). Alternatively, 35 μL cells were added to the channel slide, cells were attached for ∼45 s, and then 200 μL buffer was added to the reservoir to dilute the unattached population. Image acquisition was started ∼1 min after cell addition to the channel slide. The second approach was preferred because signal to noise was better under that scheme (cells were already attached when image acquisition was started and the background was spatially uniform).
CCCP was used at 100 μM in 20 mM potassium phosphate and 100 μM EDTA buffers of various pHs. Potassium benzoate and methylamine were used together, each at a concentration of 40 mM.
Microscope Setup and Imaging Assay.
We used a Nikon Eclipse Ti-U microscope in TIRF mode with a 514-nm laser and an Apo 60× oil objective (numerical aperture 1.49). The emission was split and projected on two halves of an Andor iXon camera. Exposure settings were the same for all the acquired data. Laser power was set at 10 mW, the camera exposure at 100 ms, and the camera gain at 200. Images were acquired every 0.5 s or every 1 s for better resolution of the alkaline spikes. Note that in this regimen, bleaching is the dominant reason for the R ratio decrease, since mVenusNB bleaches faster than mScarletI. To minimize bleaching and assess longer-term changes in the R ratio, we acquired images every 10 or 15 s.
Image Analysis.
The acquired images were processed in ImageJ (47). The background was subtracted from each stack; alignment was performed using linear stack alignment with Scale Invariant Feature Transform (SIFT) (48). Corresponding mScarletI and mVenusNB signals were measured and the measurements were processed and analyzed in Excel and MATLAB.
Supplementary Material
Acknowledgments
We thank Maria Schumacher for sending us the codon-optimized MrkH DNA. We thank Enrique Balleza and Phillipe Cluzel for sharing optimized fluorescent protein DNA before it was available at Addgene. We thank Gabriel Hosu for discussions and help with the TIRF microscope setup. We thank the Harvard University Bauer Core scientists for help with the flow cytometry instrumentation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116830119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.O’Toole G., Kaplan H. B., Kolter R., Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Kimkes T. E. P., Heinemann M., How bacteria recognise and respond to surface contact. FEMS Microbiol. Rev. 44, 106–122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Toole G. A., Wong G. C., Sensational biofilms: Surface sensing in bacteria. Curr. Opin. Microbiol. 30, 139–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenal U., Reinders A., Lori C., Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Armbruster C. R., et al. , Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations. eLife 8, e45084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laventie B. J., et al. , A surface-induced asymmetric program promotes tissue colonization by Pseudomonas aeruginosa. Cell Host Microbe 25, 140–152.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Luo Y., et al. , A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6, e02456-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C. K., et al. , Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc. Natl. Acad. Sci. U.S.A. 115, 4471–4476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webster S. S., Lee C. K., Schmidt W. C., Wong G. C. L., O’Toole G. A., Interaction between the type 4 pili machinery and a diguanylate cyclase fine-tune c-di-GMP levels during early biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 118, e2105566118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simm R., Morr M., Remminghorst U., Andersson M., Römling U., Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Biochem. 386, 53–58 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Spangler C., Böhm A., Jenal U., Seifert R., Kaever V., A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81, 226–231 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Christen M., et al. , Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328, 1295–1297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho C. L., et al. , Visualizing the perturbation of cellular cyclic di-GMP levels in bacterial cells. J. Am. Chem. Soc. 135, 566–569 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Balleza E., Kim J. M., Cluzel P., Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 15, 47–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumacher M. A., Zeng W., Structures of the activator of K. pneumonia biofilm formation, MrkH, indicates PilZ domains involved in c-di-GMP and DNA binding. Proc. Natl. Acad. Sci. U.S.A. 113, 10067–10072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F., et al. , The PilZ domain of MrkH represents a novel DNA binding motif. Protein Cell 7, 766–772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baird G. S., Zacharias D. A., Tsien R. Y., Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 11241–11246 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis J. H., Rubin A. J., Sauer R. T., Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bindels D. S., et al. , mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Wang X. C., Wilson S. C., Hammond M. C., Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Res. 44, e139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botman D., de Groot D. H., Schmidt P., Goedhart J., Teusink B., In vivo characterisation of fluorescent proteins in budding yeast. Sci. Rep. 9, 2234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruni G. N., Weekley R. A., Dodd B. J. T., Kralj J. M., Voltage-gated calcium flux mediates Escherichia coli mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 114, 9445–9450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slonczewski J. L., Rosen B. P., Alger J. R., Macnab R. M., pH homeostasis in Escherichia coli: Measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. U.S.A. 78, 6271–6275 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmond C. V., Kroll R. G., Booth I. R., The effect of food preservatives on pH homeostasis in Escherichia coli. J. Gen. Microbiol. 130, 2845–2850 (1984). [DOI] [PubMed] [Google Scholar]
- 25.Zilberstein D., Agmon V., Schuldiner S., Padan E., Escherichia coli intracellular pH, membrane potential, and cell growth. J. Bacteriol. 158, 246–252 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilks J. C., Slonczewski J. L., pH of the cytoplasm and periplasm of Escherichia coli: Rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189, 5601–5607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodd B. J. T., Kralj J. M., Live cell imaging reveals pH oscillations in Saccharomyces cerevisiae during metabolic transitions. Sci. Rep. 7, 13922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratt L. A., Kolter R., Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Hug I., Deshpande S., Sprecher K. S., Pfohl T., Jenal U., Second messenger-mediated tactile response by a bacterial rotary motor. Science 358, 531–534 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Ellison C. K., et al. , Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358, 535–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abel S., et al. , Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell 43, 550–560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., et al. , Superoxide flashes in single mitochondria. Cell 134, 279–290 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarzländer M., et al. , Mitochondrial ‘flashes’: A radical concept repHined. Trends Cell Biol. 22, 503–508 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Lovy-Wheeler A., Kunkel J. G., Allwood E. G., Hussey P. J., Hepler P. K., Oscillatory increases in alkalinity anticipate growth and may regulate actin dynamics in pollen tubes of lily. Plant Cell 18, 2182–2193 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behera S., et al. , Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30, 2704–2719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., et al. , Protons trigger mitochondrial flashes. Biophys. J. 111, 386–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong Y., Brown D. G., Alteration of bacterial surface electrostatic potential and pH upon adhesion to a solid surface and impacts to cellular bioenergetics. Biotechnol. Bioeng. 105, 965–972 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Meister M., Lowe G., Berg H. C., The proton flux through the bacterial flagellar motor. Cell 49, 643–650 (1987). [DOI] [PubMed] [Google Scholar]
- 39.Blair D. F., Flagellar movement driven by proton translocation. FEBS Lett. 545, 86–95 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Jackson D. W., Simecka J. W., Romeo T., Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184, 3406–3410 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hufnagel D. A., et al. , The catabolite repressor protein-cyclic AMP complex regulates csgD and biofilm formation in uropathogenic Escherichia coli. J. Bacteriol. 198, 3329–3334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schniederberend M., et al. , Modulation of flagellar rotation in surface-attached bacteria: A pathway for rapid surface-sensing after flagellar attachment. PLoS Pathog. 15, e1008149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C., Sun D., Zhu J., Liu J., Liu W., The regulation of bacterial biofilm formation by cAMP-CRP: A mini-review. Front. Microbiol. 11, 802 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hess B., Boiteux A., Krüger J., Cooperation of glycolytic enzymes. Adv. Enzyme Regul. 7, 149–167 (1969). [DOI] [PubMed] [Google Scholar]
- 45.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amann E., Ochs B., Abel K. J., Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69, 301–315 (1988). [DOI] [PubMed] [Google Scholar]
- 47.Abramoff M. D., Magalhaes P. J., Ram S. J., Image Processing with ImageJ. Biophoton. Int. 11, 36–42 (2004). [Google Scholar]
- 48.Lowe D., Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60, 91–110 (2004). [Google Scholar]
- 49.Pettersen E. F., et al. , UCSF Chimera -- A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.