Significance
Pre-messenger RNA (pre-mRNA) splicing is a key regulatory step in gene expression. The splicing reaction is mediated by the spliceosome, a dynamic complex comprising five small nuclear ribonucleoproteins (snRNPs), which assembles onto each intron in multiple steps. We present detailed structural analysis and supporting functional data of an important protein–RNA interaction between human U1 and U2 snRNP. Our structure shows that an intrinsically disordered arginine-glycine (RGG/RG)–rich motif of a U2 snRNP subunit forms an RNA-sequence–specific connection with U1 snRNP. This study broadens the functional scope of unstructured RGG/RG-rich motifs in RNA binding proteins and provides a molecular basis of early steps of spliceosome assembly, which may help develop innovative therapeutic strategies against diseases originating from splicing defects.
Keywords: splicing, spliceosome assembly, RGG motif, ubiquitin-like domain
Abstract
In mammals, the structural basis for the interaction between U1 and U2 small nuclear ribonucleoproteins (snRNPs) during the early steps of splicing is still elusive. The binding of the ubiquitin-like (UBL) domain of SF3A1 to the stem-loop 4 of U1 snRNP (U1-SL4) contributes to this interaction. Here, we determined the 3D structure of the complex between the UBL of SF3A1 and U1-SL4 RNA. Our crystallography, NMR spectroscopy, and cross-linking mass spectrometry data show that SF3A1-UBL recognizes, sequence specifically, the GCG/CGC RNA stem and the apical UUCG tetraloop of U1-SL4. In vitro and in vivo mutational analyses support the observed intermolecular contacts and demonstrate that the carboxyl-terminal arginine-glycine-glycine-arginine (RGGR) motif of SF3A1-UBL binds sequence specifically by inserting into the RNA major groove. Thus, the characterization of the SF3A1-UBL/U1-SL4 complex expands the repertoire of RNA binding domains and reveals the capacity of RGG/RG motifs to bind RNA in a sequence-specific manner.
The evolutionarily conserved heterotrimeric complex SF3A, composed of subunits SF3A1 (SF3a120), SF3A2 (SF3a66), and SF3A3 (SF3a60), is essential for pre-messenger RNA (pre-mRNA) splicing (1). Together with the core U2 particle and the SF3B complex, SF3A forms the mature 17S U2 snRNP (small nuclear ribonucleoprotein) (2). During the early stages of the splicing reaction, U2 snRNP interacts with U1 snRNP, while the latter is bound to the pre-mRNA at the 5′-splice site (ss) (3). In mammals, this interaction is mediated via a direct contact between SF3A1 and U1 snRNP stem-loop 4 (U1-SL4) that brings together the 5′- and 3′-ss to form the prespliceosomal A complex (4). Subsequently, the A complex recruits the preformed U4/U6.U5 tri-snRNP to generate the spliceosomal pre-B complex. Thus far, no 3D structures of any mammalian prespliceosomal A complex have been determined, and the cryogenic electron microscopy (cryo-EM) structures of the human pre-B complex do not reveal any contact between U1 and U2 snRNPs, which is possibly only transient and specific for complex A (5, 6). The cryo-EM structure of the analogous yeast A complex identifies two regions of contact between the pre-mRNA bound U1 and U2 snRNPs, involving both protein–protein and protein–RNA interfaces (7). However, none of these contacts have been functionally tested thus far (8). In addition, for these early splicing steps, transfer of structural insights from yeast to human spliceosomes is limited because of significant compositional differences between their respective U1 and U2 snRNPs (2, 9–11). Compared to human U1 snRNP, which consists of three particle-specific proteins, yeast U1 snRNP contains seven additional proteins. Furthermore, the yeast U1 small nuclear RNA (snRNA) is much longer (568 nucleotides) than its human paralog (164 nucleotides); however, it lacks a structure analogous to the human U1-SL4 downstream of the heptameric Sm ring. In total, the human U1 snRNA consists of four SLs, of which SL1 and 2 are bound by U1-specific proteins, while SL3 and 4 are interaction sites for spliceosomal proteins UAP56 and SF3A1, respectively, and also for splicing regulators, including FUS and PTBP1 (4, 12–14). Human U2 snRNP protein SF3A1 contains two tandem suppressor-of-white-apricot domains (SURP1 and SURP2) and a short segment of charged residues at its amino terminus (Fig. 1A). The region harboring the SURP2 domain [amino acids (aa) 145 to 243] mediates binding to SF3A3, while a short motif (aa 269 to 295) directly adjacent to the charged sequence stretch contacts SF3A2. The carboxyl-terminal half of SF3A1 comprises proline-rich stretches and a ubiquitin-like (UBL) domain. PRP21, the yeast ortholog of SF3A1, is homologous to the N terminus of SF3A1 but lacks the carboxyl-terminal half of SF3A1 (15). The recently published 3D cryo-EM structure of the human 17S U2 snRNP contains SF3A1, but only part of the N terminus is visible (aa 160 to 286) (2). Recently, we reported that the carboxyl-terminal UBL domain of SF3A1 is a noncanonical RNA binding domain interacting with U1-SL4 in mammals (16). Our study also indicated an involvement of the positively charged carboxyl-terminal tail of SF3A1-UBL, containing an arginine-glycine-glycine-arginine motif (RGGR) followed by two lysines with U1-SL4 binding. However, atomic details of the SF3A-UBL/U1-SL4 interaction are not known, and the structural and sequence requirements remain poorly understood.
Fig. 1.
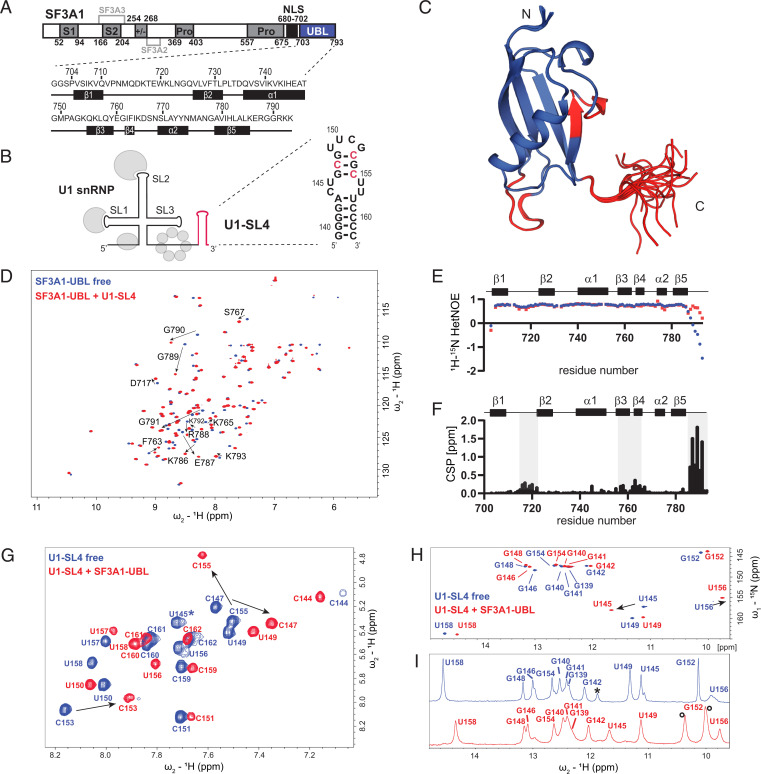
Interaction of SF3A1-UBL with U1-SL4. (A) Domain organization of SF3A1 and primary sequences of the SF3A1-UBL construct used in this study, including the secondary structure elements shown below. S1 and S2, SURP1 and SURP2; Pro, proline-rich sequence; +/−, charged residues; NLS, nuclear localization signal. Interaction sites for SF3A2 and SF3A3 are highlighted in light gray. (B) Schematic representation of U1 snRNP with U1-SL4 shown in red and the predicted secondary structure of U1-SL4 on the right. Nucleotides with the strongest CSP in 2D 1H-1H TOCSY are shown in red. (C) Solution structure of the free SF3A1-UBL. Overlay of the 20 lowest energy structures is shown. Amide CSPs of D and F shown in red. (D) Overlay of 2D 1H-15N heteronuclear single quantum coherence (HSQC) spectra of 15N-labeled SF3A1-UBL in the free (blue) and U1-SL4 RNA-bound (red) 1:1 complex form. CSPs of C-terminal residues are indicated with black arrows. (E) Backbone dynamics data of SF3A1-UBL in the free (blue) and U1-SL4 bound states (red). (F) Plot of the combined chemical shift difference between amide group resonances of the free and bound forms of SF3A1-UBL. (G) Overlay of 2D 1H-1H TOCSY spectra of U1-SL4 free (blue) and bound to SF3A1-UBL (red). Asterisk indicates U145, which was not identified in the bound state in 2D 1H-1H TOCSY. (H and I) Overlay of 2D 1H-15N HSQC and 1D 1H spectra, respectively, of imino signals of U1-SL4 free (blue) and bound to SF3A1-UBL (red). Black asterisk in I indicates an imino signal deriving from a duplex conformation. Black circles in I indicate protein amide signals.
Here, we performed structural, biochemical, and functional analyses of the interaction between the human SF3A1 UBL domain and U1-SL4 (Fig. 1B). The presented crystal structure unravels the structural determinants for U1-SL4 recognition by SF3A1 and expands the repertoire of RNA-binding domains to UBL domains. Surprisingly, sequence specificity is achieved through the RGGR motif of SF3A1 C-terminal to the UBL domain, while the globular ubiquitin fold ensures the shape recognition of the structured UUCG tetraloop. Furthermore, our study provides structural evidence of how human U1 and U2 snRNP interact during the early steps of spliceosome assembly.
Results
Importance of the Carboxyl-terminal Tail of SF3A1-UBL for U1-SL4 Binding.
The previously determined solution structure of SF3A1-UBL by the Structural Genomics Consortium (Protein Data Bank, PDB ID: 1ZKH, aa 704 to 789) demonstrated that the domain adopts a globular UBL fold made of five β-strands that are packed against an α-helix. As the carboxyl-terminal residues including the RGGR motif were not present in this structure, we solved the solution structure of SF3A1-UBL comprising residues 704 to 793 by NMR spectroscopy (Fig. 1C; SI Appendix, Table S1). The protein backbone of the core domain in both structures (residues 704 to 785) is very similar with a backbone root-mean-square deviation (r.m.s.d.) deviation of 1.58 Å (SI Appendix, Fig. S1A). Consistent with the poor chemical shift dispersion of the amide signals (Fig. 1D) and with low and negative values of the {1H}-15N heteronuclear Overhauser effect (hetNOE) (Fig. 1E), the carboxyl-terminal tail of SF3A1-UBL is disordered in our structural ensemble (Fig. 1C). This tail extends the positively charged surface of the core domain by adding five conserved basic residues (SI Appendix, Fig. S1B). To analyze the binding of SF3A1-UBL to U1-SL4, we performed an NMR titration of the 15N-labeled protein with the 24-nucleotide U1-SL4 RNA (Fig. 1B). Upon addition of the RNA, the amide protein resonances experienced large chemical shift perturbations (CSPs) (Fig. 1D; SI Appendix, Fig. S1C). Saturation of the CSP was obtained at equimolar protein and RNA concentrations, consistent with a 1:1 binding stoichiometry. Moderate CSP localized on the loop between β1 and β2 and on β3 and β4, while the strongest changes were observed for the carboxyl-terminal tail of the protein, which contains the RGGR motif (Fig. 1 C and F). Interestingly, backbone dynamics data (hetNOE) indicate that the carboxyl-terminal tail of SF3A1-UBL becomes partially ordered upon RNA binding (Fig. 1E). However, formation of additional secondary structure elements was not observed based on the analysis of backbone chemical shifts (SI Appendix, Fig. S1D). These data confirmed an involvement of the RGGR-containing carboxyl terminus of SF3A1 in U1-SL4 binding. To obtain an initial mapping of the interaction site on U1-SL4, NMR titrations were performed by following the resonances of the RNA. The U1-SL4 helical stem comprises eight Watson-Crick base pairs; an internal, pyrimidine-rich mismatched loop; and a structured UUCG tetraloop (Fig. 1B). Binding of the SF3A1-UBL to U1-SL4 induced global CSP of the RNA resonances (Fig. 1 G–I). Upon addition of SF3A1-UBL, essentially all pyrimidine bases of U1-SL4 underwent CSP of their H5-H6 resonances, and the strongest changes were observed for C147, C153, and C155 that are located in the upper stem (Fig. 1G). In total, 12 imino signals were detected for U1-SL4, corresponding to the Watson-Crick base-paired helix and the closing noncanonical U-G base pair of the UUCG tetraloop (Fig. 1 H and I; SI Appendix, Fig. S1E). Furthermore, two strong imino signals of U145 and U156 of the internal loop of U1-SL4 were observed, indicating base pairing between these nucleotides. An imino signal for U157, which might form a noncanonical base pair with C144 in the RNA helix, was not detected, as observed previously for an SL with an identical internal loop (17). Upon addition of the protein, the imino signals remained detectable and experienced CSP, in particular of the internal loop bases U145 and U156 (Fig. 1 H and I; SI Appendix, Fig. S1F). These results suggest that SF3A1-UBL engages in contact with a large surface of U1-SL4 (from the internal loop to the tetraloop) and that the RNA base pairing is preserved upon complex formation.
Structure of SF3A1-UBL Bound to U1-SL4.
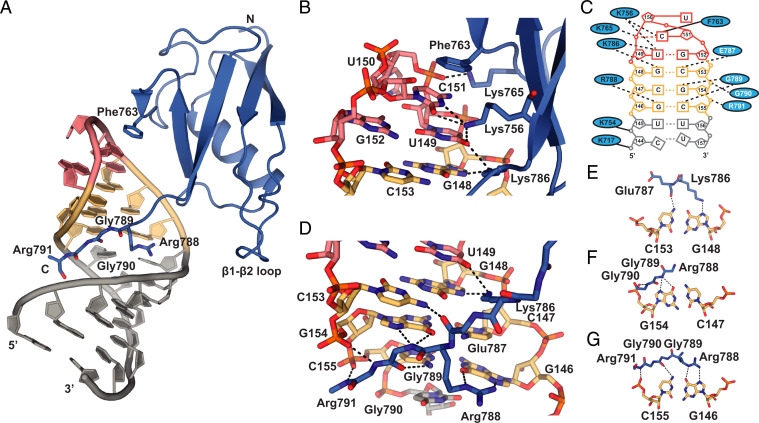
In order to understand the structural basis of RNA recognition by SF3A1-UBL, we crystallized the complex of SF3A1-UBL and U1-SL4 (Table 1; SI Appendix, Fig. S2A). Crystals obtained belonged to space group I222 and diffracted to 1.56 Å resolution. The structure was solved by a combination of molecular replacement using the structures of SF3A1-UBL and a UUCG tetraloop and native single-wavelength anomalous dispersion (SAD). The crystals contained one protein–RNA complex per asymmetric unit, and all nucleotides of U1-SL4 (nucleotides 139 to 162), as well as most SF3A1-UBL residues (aa 704 to 790 and main chain of R791), were visible in the electron density map (SI Appendix, Fig. S2 B and C). In complex with U1-SL4 (Fig. 2A), the core of SF3A1-UBL retains essentially the same structure as in its free form (r.m.s.d. of 1.08 Å for 82 Cα atoms of residues 704 to 785; SI Appendix, Fig. S2D). Consistent with our NMR data, SF3A1-UBL establishes extensive contacts with the upper part of the SL4 RNA, and the carboxyl-terminal tail adopts a rigid conformation (Fig. 2D). The carboxyl-terminal residues insert into the RNA major groove, thereby enabling direct contacts with the three G-C base pairs located between the UUCG tetraloop and pyrimidine-rich internal loop. The base identity of G146 just upstream of the internal loop is specifically recognized by hydrogen bonds involving the side chain of Arg788 (Fig. 2G). The guanidinium group of Arg788 aligns with the Hoogsteen edge of G146 to form hydrogen bonds with O6 and N7 atoms. Importantly, the main chain of Gly789 forms additional base-specific intermolecular hydrogen bonds to the major-groove edges of C155 (N4) via the carbonyl oxygen and of G154 (O6, N7) via the amide proton (Fig. 2 F and G). Furthermore, the main chain atoms of Gly790 and Arg791 interact with the phosphate oxygens of G154 and C155, respectively (Fig. 2 F and G). Thus, both arginines and both glycines of the RGGR motif mediate sequence-specific recognition of the GCG/CGC upper stem of U1-SL4. Additional sequence-specific contacts to the G148–C153 base pair are mediated by the side chain of Lys786 and the main chain oxygen of Glu787, which precede the RGGR motif (Fig. 2E). The involvement of the carboxyl-terminal residues in direct sequence readout is consistent with the large CSP observed in NMR titrations (Fig. 1E). Since the SF3A1-UBL tail and the bound section of U1-SL4 are not involved in crystal packing, the observed contacts are unlikely to result from crystallization artifacts (SI Appendix, Fig. S2E). The SF3A1-UBL folded core contacts the apical UUCG tetraloop through aromatic stacking and hydrogen-bonding interactions enabling shape- and sequence-specific readout (Fig. 2B). The aromatic ring of Phe763 stacks on the base of C151 within the UUCG tetraloop. This agrees well with strong intermolecular NOE cross-peaks observed between the side chain of Phe763 (Hδ) and the H1′, H5, and H6 of C151 observed in the 2D 1H-1H nuclear Overhauser effect spectroscopy (NOESY) spectrum and the chemical shift changes of C151 seen in the 2D 1H-1H total correlation spectroscopy (TOCSY) spectrum (SI Appendix, Fig. S2F and Fig. 1G). Additionally, Lys765 forms a salt bridge with the phosphate backbone of U150. The amide group of the Lys765 main chain is stabilized by a hydrogen bond with the highly conserved Tyr773. This likely explains the fivefold decrease in affinity and reduced U1 snRNP pull-down efficiency previously observed for a SF3A1-UBL Tyr773Cys mutant (16). Furthermore, Lys756 and Lys786 side chains contribute to the recognition of the UUCG tetraloop. Lys756 of β3 forms hydrogen bonds with the bases of U149 and C151. Lys786 forms hydrogen bonds with U149 of the UUCG tetraloop and G148 of the stem (Fig. 2B). Finally, the lower stem of U1-SL4 is also contacted by SF3A1-UBL via two salt bridges formed by the side chains of Lys717 and Lys754 to the RNA phosphate backbone of C144 and U145, respectively (SI Appendix, Fig. S2G). Sequence alignment of SF3A1-UBL of various species shows that the residues involved in RNA recognition are all conserved (SI Appendix, Fig. S1G). The SF3A1-UBL/U1-SL4 binding interface seen in the crystal structure is consistent with the CSP observed upon RNA binding in solution (SI Appendix, Fig. S2H). Overall, SF3A1-UBL RNA recognition is both shape- and sequence-specific, with protein–RNA contacts involving the RNA bases and the sugar–phosphate backbone of U1-SL4 (Fig. 2C). Interestingly, compared to the structure of SL4 from U1 snRNP (6, 9), the overall geometry of U1-SL4 bound to SF3A1-UBL is different with a smaller major-groove width, as determined by the distance between the phosphate atoms of G141 and the cross-strand i + 6 base pair (G152) (9.2 Å SF3A1-UBL bound and 12.9 Å free U1-SL4) (SI Appendix, Fig. S2I) (18). This conformational difference could be explained by the presence of the tandem noncanonical 5′-CU-3′/5′-UU-3′ internal loop that allows the insertion of the carboxyl-terminal tail of SF3A1-UBL in the major groove. The internal loop in the SF3A1-UBL bound state adopts base-pairing configurations that differ from those found in the unbound state (SI Appendix, Fig. S2J). Similar base-pairing configurations have been reported before for such internal loops (17, 19). Consistently, NMR data of the imino RNA resonances around the internal loop indicated altered base stacking, and the conformational change of U1-SL4 could explain the CSP observed in the β1–β2 loop of SF3A1-UBL upon RNA binding (Fig. 1 D, E, H, and I).
Table 1.
Data collection and refinement statistics
| Data set | SF3A1-UBL U1-SL4 (native) | SF3A1-UBL U1-SL4 (native SAD) |
| Space group | I222 | I222 |
| Unit cell | ||
| dimensions (a, b, c) (Å) | 42.993, 62.722, 138.28 | 43.24, 62.78, 139.09 |
| angles (α, β, γ) (°) | 90, 90, 90 | 90, 90, 90 |
| Data collection* | ||
| Wavelength (Å) | 0.999987 | 2.07505 |
| Resolution range (Å) | 41.06–1.56 (1.66–1.56) | 41.05–2.00 (2.09–2.00) |
| Rmeas, % | 4.3 (335.7) | 5.9 (44.4) |
| Rpim, %† | 1.2 (96.1) | 0.6 (8.3) |
| Completeness, % | 99.6 (98.8) | 99.58 (96.57) |
| Mean I/σ(I) | 21.74 (0.75) | 59.7 (6.0) |
| Multiplicity | 13.3 (13.3) | 85.6 (24.1) |
| CC 1/2 | 100 (66.6) | 100 (98.7) |
| Wilson B | 42.83 | 46.07 |
| Refinement | ||
| Data range (Å) | 41.06–1.56 | |
| Rcryst, % | 21.79 | |
| Rfree, % | 25.79 | |
| No. of atoms per asymmetric unit | ||
| all atoms | 1,270 | |
| protein | 699 | |
| RNA | 506 | |
| ligand | 17 | |
| water | 48 | |
| Average B-factor (Å2) | ||
| all atoms | 75.97 | |
| protein | 54.97 | |
| RNA | 107.45 | |
| ligand | 63.18 | |
| water | 54.48 | |
| Ramachandran plot | ||
| favored regions, % | 98.86 | |
| disallowed regions, % | 0 | |
| Rmsd from ideal geometry | ||
| bond lengths (Å) | 0.007 | |
| bond angles (°) | 0.861 |
Values in parentheses are for highest-resolution shell.
†Rpim gives the precision of averaged intensities and is a better indicator for data quality in highly redundant datasets than Rmerge, which penalizes redundancy (55).
Fig. 2.
Molecular basis of the interaction between SF3A1-UBL and U1-SL4. (A) Overall view of the crystal structure of SF3A1-UBL (residues 704 to 791) (blue) and U1-SL4 (UUCG tetraloop in red, GCG base pairs in yellow, and the rest of the SL in gray). (B) Close-up views of the contacts to the UUCG tetraloop of U1-SL4. Putative hydrogen bonds are shown as dashed lines. Solid lines indicate electrostatic interactions. (C) Schematic representation of the intermolecular interactions; side chain–mediated contacts are written in black, while amino acids using the main chain are written in white. (D) Specific recognition of RNA by carboxyl-terminal residues Lys786, Glu787, Arg788, Gly789, Gly790, and Arg791. (E–G) Base-specific recognition of the RNA base pairs in the upper part of the RNA duplex by carboxyl-terminal residues of SF3A1-UBL.
Mutational Analysis of the SF3A1-UBL/U1-SL4 Interaction.
To quantify the thermodynamic importance of the individual contacts between SF3A1-UBL and U1-SL4, we performed several mutational analyses and determined the binding affinities of several protein mutants by surface plasmon resonance (SPR) spectroscopy. Using this method, we found that SF3A1-UBL binds U1-SL4 with a dissociation constant (Kd) of 330 nM. We introduced changes in aa residues that mediated protein–RNA contacts in the crystal structure and exhibited NMR CSP upon RNA binding (SI Appendix, Fig. S3 A and B). Replacing the RGG motif with three alanine residues led to the strongest reduction in affinity (Kd higher than 50 μM, which was the highest concentration tested and a more than 167-fold increase in Kd compared to wild-type) (Table 2; SI Appendix, Fig. S3 C and D). A similarly strong effect was observed by mutating only Arg788 of the RGG motif, which forms two hydrogen bonds to guanine, to alanine (Kd higher than 25 μM and a more than 83-fold increase in Kd). Mutation of Arg788 to lysine, which has the same charge as arginine but can only form one hydrogen bond, also resulted in a more than 83-fold increase in Kd compared to wild-type (WT). However, the SPR sensograms for this mutant (SI Appendix, Fig. S3C) showed stronger responses at the same concentrations than for R788A, indicating higher affinity than the alanine mutant (R788A). These data demonstrate the important role of the two arginine-mediated hydrogen bonds of Arg788 for high-affinity binding of SF3A1-UBL to U1-SL4. Mutating the individual glycine residues of the RGG motif to bulky isoleucines also reduced affinity (130- and 44-fold increase in Kd for G789I and G790I, respectively). These larger side chains likely induced steric clashes with the RNA, thereby highlighting the importance of having glycine residues that allow the main chain to mediate sequence-specific contacts with the RNA major groove. Mutation of Arg791 to alanine reduced the affinity to a similar extent as mutating Lys765, which is in contact with the phosphate oxygens at the apical loop (19- and 25-fold increase in Kd, respectively). Interestingly, Lys792 and Lys793, which are not ordered in our crystal structure, showed a strong reduction in affinity when mutated to alanine (more than 83-fold increase in Kd). Although potentially too dynamic to be observed in the crystal structure, these two lysine residues seem to contribute to the interaction with U1-SL4. In agreement with this, shortening of U1-SL4 by removal of the last G-C base pair induced altered amide CSP of these lysine residues compared to the longer U1-SL4 used in our study (SI Appendix, Fig. S4 A–D), indicating a potential interaction with the G-C base pairs of the lower stem of U1-SL4. Alanine mutation of Phe763, which stacks on C151 of the UUCG tetraloop, had only a mild effect on RNA binding (ninefold increase in Kd). Similarly, Lys717, which forms a salt bridge with the phosphate backbone, showed a moderate decrease in affinity when mutated to alanine (fivefold increase in Kd). Overall, the protein mutation experiments are all in agreement with the intermolecular contacts identified in the crystal structure of SF3A1-UBL/U1-SL4.
Table 2.
Affinities and splicing activity of SF3A1-UBL mutants measured by SPR spectroscopy and in vivo reporter assays, respectively
| SF3A1-UBL construct | Kd [μM] | Fold increase of Kd compared to WT | Average exon 2 inclusion (%) | ΔPSI |
| WT | 0.332 ± 0.03 | 1.0 | 57.46 | 0.00 |
| K717A | 1.586 ± 0.27 | 4.9 | 50.05 | 7.41 |
| K754A | 16.733 ± 4.38 | 52.0 | 46.78 | 10.68 |
| F763A | 2.764 ± 0.3 | 8.6 | 49.25 | 8.21 |
| K765A | 7.889 ± 0.71 | 24.5 | 50.19 | 7.27 |
| K786A | >50 | >167 | 44.73 | 12.73 |
| R788A | >25 | >83 | 42.97 | 14.49 |
| R788K | >25 | >83 | n.d. | n.d. |
| RGG2AAA | >50 | >167 | 50.52 | 6.94 |
| G789I | 41.9 ± 5.7 | 130.1 | 46.12 | 11.34 |
| G790I | 14.1 ± 3.12 | 43.8 | 49.65 | 7.81 |
| R791A | 5.97 ± 0.5 | 18.5 | 48.98 | 8.48 |
| KK2AA | >25 | >83 | 51.76 | 6.38 |
| RKK2AAA | n.d. | n.d. | 51.76 | 5.7 |
| K754A/RGG2AAA | n.d. | n.d. | 42.33 | 15.13 |
| K765/RGG2AAA | n.d. | n.d. | 42.18 | 15.28 |
See SI Appendix, Fig. S3 for the sensograms of the SPR measurements. See Fig. 5 for primer extension analysis that yield the exon 2 inclusions. n.d., not determined (55).
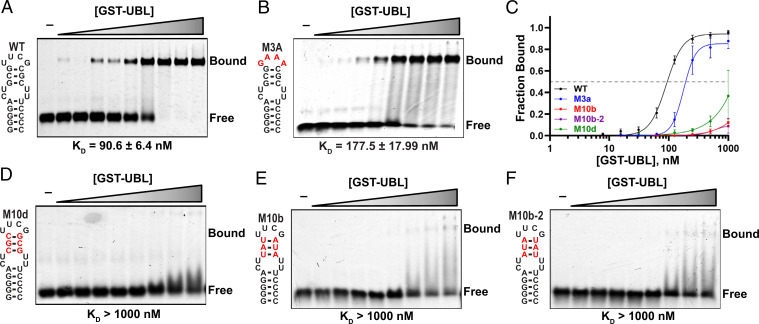
We next studied the contribution of individual nucleotides in U1-SL4 to complex formation by electrophoretic mobility shift assays (EMSAs). Replacing the UUCG tetraloop with a GNRA (N, any nucleotide; R, purine) type tetraloop (GAAA) or mutating U149 of the UUCG tetraloop to a C only mildly decreased RNA binding (Fig. 3 A and B; SI Appendix, Fig. S4 E–I). Similarly, introducing perfect Watson-Crick base pairing in the mismatched internal loop by replacing U156 and U157 with 5′-AG-3′ to introduce a regular A-form helix also had only a mild effect on RNA binding (SI Appendix, Fig. S4F). In contrast, when we swapped the G-C base pairs in the upper part of U1-SL4 or replaced them with A-U base pairs (Fig. 3 D–F), all mutants strongly reduced binding because of unfavorable placement of the bases' functional groups that disrupt crucial hydrogen bonds. Previous splicing assays performed with U1-SL4 mutants highlighted the importance of G-C base pairs of U1-SL4 for SF3A1 binding and are in agreement with these binding assays (4). These data clearly demonstrate that the sequence-specific contacts mediated by the carboxyl terminus of SF3A1 to the GCG/CGC stem are crucial for the SF3A1-UBL/U1-SL4 interaction.
Fig. 3.
Mutational analysis of the U1-SL4 RNA. (A) EMSA experiments performed with SF3A1-UBL and U1-SL4. (B) Loop mutant of U1-SL4 probed for binding to SF3A1-UBL. Bases different from WT U1-SL4 are shown in red. (C) Binding curves of the indicated U1-SL4 variants. (D–F) Mutants of the upper helical part of U1-SL4. Respective Kd values were derived from curve fitting to the relative bound fraction per lane. GST, glutathione-S-transferase.
Validation of the SF3A1-UBL/U1-SL4 Interface in the Context of U1 snRNP.
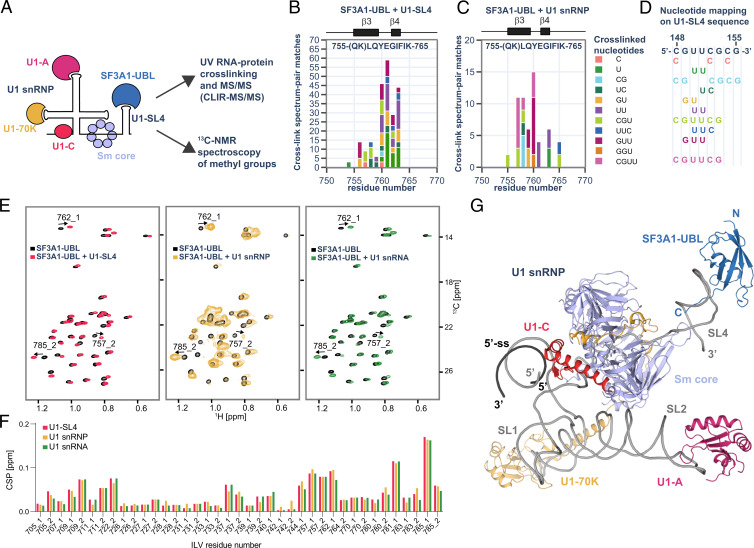
Pull-down experiments from HeLa cell nuclear extracts previously showed that SF3A1-UBL binds U1-SL4 in the context of U1 snRNP (16). To examine the interaction of SF3A1-UBL and U1-SL4 in the context of U1 snRNP, we analyzed an in vitro reconstituted U1 snRNP/SF3A1-UBL complex using the cross-linking of isotope-labeled RNA coupled with tandem mass spectrometry (CLIR-MS/MS) method (20) (Fig. 4A). U1 snRNP was reconstituted using recombinant components (21), and SF3A1-UBL was added before ultraviolet (UV) irradiation to induce protein–RNA cross-linking. The detection of all U1 snRNP proteins and protein–RNA cross-links for U1-70K and SmD2 fit well with the U1 snRNP structure and confirmed the correct reconstitution of the particle (SI Appendix, Fig. S5A). For SF3A1-UBL, most cross-links localized around β3 and β4, containing Lys756, Phe763, and Lys765 that contact the UUCG tetraloop of U1-SL4. Consistently, the cross-linked nucleotides could also be mapped to the UUCG tetraloop sequence (Fig. 4 C and E). This cross-linking site was also detected in a 1:1 complex of SF3A1-UBL/U1-SL4 and was in agreement with the crystal structure (Fig. 4B; SI Appendix, Fig. S5B), highlighting that the interaction is identical in the context of both the isolated U1-SL4 and the U1 snRNP particle.
Fig. 4.
Analysis of SF3A1-UBL binding to U1 snRNP. (A) Schematic representation of the SF3A1-UBL/U1 snRNP complex analysis. (B) Protein–RNA cross-links identified for 1:1 complex of SF3A1-UBL and U1-SL4 by CLIR-MS/MS plotted on the sequence of SF3A1-UBL. The bar colors represent the nucleotide composition of the RNA adducts. Protein–RNA cross-links are shown as counts of cross-link spectrum matches. (C) Cross-links identified for SF3A1-UBL bound to in vitro–reconstituted U1 snRNP. (D) Mapping of nucleotides cross-linked to SF3A1-UBL on the sequence of U1-SL4. (E) Overlay of the 2D 1H-13C HMQC spectra of the free SF3A1-UBL protein (black) and in complex with U1-SL4 (red), U1 snRNP (yellow), and U1 snRNA (green). (F) Plot showing the CSPs of the methyl groups of isoleucine, leucine, and valine (ILV) of SF3A1-UBL observed upon addition of U1-SL4 (red), U1 snRNP (yellow), or U1 snRNA (green). Methyl groups are labeled according to the residue number; 1 or 2 stands for HD1/CD1 and HD2/CD2 in the case of leucine or HG1/CG1 and HG2/CG2 in the case of valine. (G) Structural model of SF3A1-UBL bound to U1 snRNP.
To further validate these results, chemical shifts of 13C-labeled methyl groups of isoleucine, leucine, and valine of SF3A1-UBL were monitored upon addition of in vitro reconstituted U1 snRNP using NMR spectroscopy. 2D 1H-13C heteronuclear multiple quantum coherence (HMQC) spectra of SF3A1-UBL showed identical chemical shifts for the methyl groups when bound to U1 snRNP and U1-SL4, respectively (Fig. 4 E and F). Consistent with the crystal structure and the initial NMR titrations, CSP localized on the loop between β1 and β2, on β3 and β4, while the strongest CSP was found at Leu785 of the carboxyl terminus (SI Appendix, Fig. S5C). This confirmed that the contacts made by SF3A1-UBL with U1-SL4 in the context of U1 snRNP are identical to those observed in the crystal structure, and structural modeling further showed that the conformation of SF3A1-UBL bound to U1-SL4 is compatible with the remainder of the U1 snRNP structure (Fig. 4G).
Functional Splicing Data Support the SF3A1-UBL/U1-SL4 Structure.
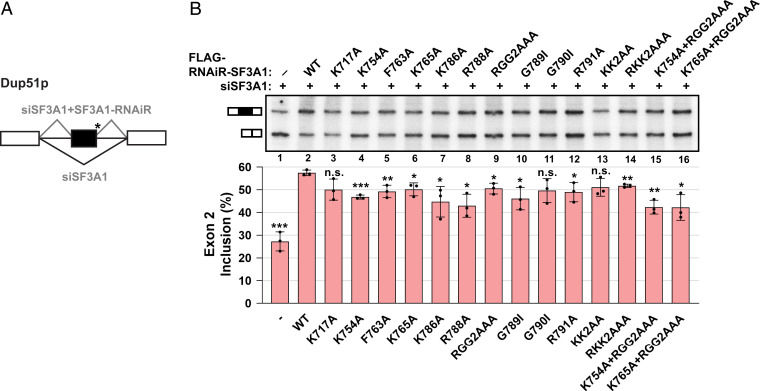
To assess the functional importance of the SF3A1-UBL/U1-SL4 interface, we performed in vivo splicing assays using a mini-gene reporter in HeLa cells (Fig. 5A). This reporter contains three exons with a mutated 5′-ss downstream of the second exon, which impairs base pairing of the endogenous U1 snRNA and therefore requires coexpression of a complementary U1 snRNA from a plasmid (U1-5a) for efficient exon 2 inclusion (4). Endogenous SF3A1 was silenced by treatment with SF3A1-targeting small interfering RNA (siRNA, siSF3A1), and exon 2 inclusion of the reporter was rescued by cotransfection with RNA interference (RNAi)-resistant SF3A1 constructs (SF3A1-RNAiR) (SI Appendix, Fig. S6 A and C). Previously, we have shown that splicing of the reporter transcript is affected by mutations in SL3 and SL4 of the U1 snRNA, and the effect of SF3A1-UBL mutations could be masked by other spliceosomal components that can interact with SL3 to support splice site pairing (see Discussion in Ref. 14). Therefore, a U1-5a/SL3-M1d variant (SI Appendix, Fig. S6B) was used in all splicing assays, which prevents the UAP56/U1-SL3 interaction. Under these conditions, knockdown of endogenous SF3A1 drastically reduced exon 2 inclusion in the reporter transcript (SI Appendix, Fig. S6C, lanes 1 and 2). Exon 2 inclusion could be efficiently rescued by WT SF3A1-RNAiR in cotransfections with U1-5a snRNA harboring SL4-WT, but not with the SL4 mutant M10r (SI Appendix, Fig. S6C, lanes 3 and 4); we have previously shown that U1-5a/SL3-M1d/SL4-M10r mutant expresses efficiently in HeLa cells and localizes to the nucleus bound to U1 snRNP–specific proteins (14). Seven of the nine point mutations that were tested significantly impaired the capacity of SF3A1-RNAiR to rescue exon 2 inclusion, although the magnitude of the effect was surprisingly small (change in percent spliced in, ΔPSI > ∼6 to 15% compared to WT) (Fig. 5B; Table 2). Importantly, mutant and WT proteins accumulated to a similar extent and incorporated into U2 snRNP particles with similar efficiencies (SI Appendix, Fig. S6 D–G). Two of the mutations, Lys717Ala (K717A) and Gly790Ile (G790I) that had only a mild reduction of binding to U1-SL4 as determined by SPR (Table 2), were also found to not significantly reduce SF3A1 splicing activity. Although the Lys792Ala-Lys793Ala (KK2AA) double mutant was also not significantly changed in splicing ratios, the SF3A1 mutant harboring changes to the full RKK motif (Arg791Ala-Lys792Ala-Lys793Ala, RKK2AAA) exhibited a significant reduction in exon 2 inclusion compared to WT (ΔPSI = 5.7%). Additionally, combining mutations of residues that interact with the tetraloop and stem of U1-SL4 such as Lys765Ala (K765A, ΔPSI = 7.3%) and Arg788Ala-Gly789Ala-Gly790Ala (RGG2AAA, ΔPSI = 7.0%), respectively, had an additive effect on SF3A1 splicing activity, as in the mutant K765A/RGG2AAA (ΔPSI = 15.3%). Thus, mutations of SF3A1 residues involved in binding to U1-SL4 led to impaired splicing in cells.
Fig. 5.
Mutations in SF3A1-UBL interfere with splicing rescue of the Dup51p minigene reporter under SF3A1 knockdown conditions. (A) Schematic representation of three-exon/two-intron Dup51p reporters depicting the splicing pattern upon siRNA-mediated knockdown and rescue with an siRNA-resistant construct. The asterisk indicates a mutant 5′-ss. (B) Primer extension analysis monitors the inclusion of exon 2 in RNA isoforms of the Dup51p minigene reporter. The mRNA products are shown schematically to the left of the gel image. All cells were transfected with siSF3A1 and plasmid harboring WT or mutant FLAG-RNAiR (RNA interference-resistant)-SF3A1. In the absence of the RNAi-resistant clone, exon 2 inclusion is inhibited (lane 1). Cotransfection with the WT RNAi-resistant SF3A1 clone rescues exon 2 inclusion under siSF3A1 treatment (lane 2), which is reduced if splicing rescue is performed using mutant RNAi-resistant SF3A1 (lanes 3 to 16). Percent exon 2 inclusion (n =3; *P < 0.05, **P < 0.01, ***P < 0.001) is plotted below the gel.
Discussion
The UBL Domain of SF3A1 Is a Sequence-Specific RNA Binding Domain.
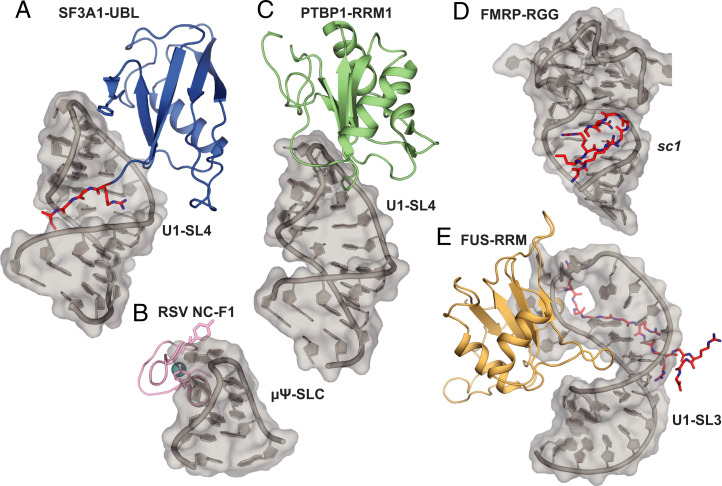
In this study, we solved the crystal structure of the SF3A1-UBL in complex with U1-SL4 RNA and uncovered the molecular details of this recognition. In contrast to previously characterized ubiquitin-like domains, SF3A1 UBL is capable of binding RNA in a sequence-specific manner. This study might provide insights into nucleic acid recognition by other UBL domains, like the N-terminal domain of TDP-43, which was also shown to be capable of binding nucleic acids (22, 23). Additionally, we identified another type of UNCG (N, any nucleotide) tetraloop recognition by an RNA binding protein. The only other structure of a protein bound to this abundant family of tetraloops is the RSV nucleocapsid protein in complex with the μΨ RNA packaging signal. This protein engages in a very different mode of binding, as two tyrosine residues of a CCHC-type zinc knuckle motif sandwich a solvent-exposed guanosine in the minor groove side of a UGCG tetraloop (Fig. 6B) (24, 25). In addition, based on our recent structural investigation of the RNA binding properties of the N-terminal RRM (RNA recognition motif) of PTBP1 bound to a pyrimidine-rich RNA pentaloop, we proposed that PTBP1-RRM1 would bind the UUCG tetraloop of U1-SL4 from the major-groove side (Fig. 6C) (26), similar to what we observed here for SF3A1-UBL. Since PTBP1 was previously shown to bind U1-SL4 to inhibit splicing (13), our structure reveals that the binding of PTBP1 and SF3A1 could be mutually exclusive, and therefore, competition between the two proteins may regulate splicing (see Discussion). PTBP1-RRM1 could prevent the contacts of SF3A1 to the UUCG tetraloop and the correct positioning of the UBL core and the carboxyl-terminal tail. Additionally, PTBP1-RRM2, which has also been shown to bind U1-SL4, could increase the affinity of PTBP1 for U1-SL4 and might contribute to the steric hindrance of SF3A1-UBL binding (13, 21, 27).
Fig. 6.
Structural comparison of UNCG tetraloop recognition and RNA binding RGG/RG motifs. (A) SF3A1-UBL bound to U1-SL4. (B) Solution structure of RSV nucleocapsid protein (NC) zinc knuckle motif (F1) bound to μΨ RNA packaging signal containing a UNCG-type tetraloop. Zinc atom shown in cyan (PDB ID: 2IHX). (C) Structural model of PTBP1-RRM1 bound to U1-SL4 based on Ref. 26. (D) Crystal structure of RGG motif of FMRP bound to sc1 RNA (PDB ID: 5DEA). (E) Solution structure of FUS-RRM bound to U1-SL3 (PDB ID: 6SNJ). RGG/RG motifs are highlighted in red.
RNA Sequence Readout by RGG Motif.
Our structure highlights the importance of the carboxyl-terminal tail of SF3A1 for RNA binding. We found that this tail rigidifies in the complex with U1-SL4 and that the RGGR motif makes sequence-specific contacts in the major groove of the upper helical part of U1-SL4. RGG/RG motifs are frequently found in RNA binding proteins and contribute to RNA binding; however, structural details of these interactions are limited (28, 29). The first arginine (Arg788) of the RGGR motif in SF3A1 mediates a specific readout of the major groove edge of guanine by a widespread type of interaction that has been previously described as the “arginine fork” (30, 31). This critical intermolecular contact of Arg788 to a guanine of U1-SL4 is followed by two glycine residues. The glycines are not acting as passive spacers between arginines but also provide base specificity through hydrogen bonds involving their main chain oxygens and amides. Overall, the RGGR motif of SF3A1 allows the sequence-specific readout of a GCG/CGC stem mediated by six intermolecular hydrogen bonds (Fig. 2D). Consistent with its important role in U1-SL4 binding, mutations of the carboxyl terminus, particularly the RGGR motif, lead to the strongest reductions in affinity. However, the shape-specific contacts of the SF3A1-UBL core to the U1-SL4 helix and its UUCG tetraloop help positioning of the carboxyl terminus inside the major groove, which enables the formation of the network of sequence-specific hydrogen bonds by the RGGR motif, illustrating the complementarity of the interaction sites. Although the overall arrangement and recognition of the RNA conformation are different, the molecular details of RNA binding by the RGGR motif bear parallels to the structure of a peptide of fragile X mental retardation protein (FMRP) bound to a systematic evolution of ligands by exponential enrichment (SELEX)-derived, high-affinity G-quadruplex ligand (32, 33) (Fig. 6D). The FMRP peptide contacts mainly G-C base pairs at the duplex–quadruplex junction, including recognition of guanines by arginine forks. Interestingly, the RGGGGGR motif of FMRP folds into a type I β-turn upon RNA interaction, which is stabilized by hydrogen bonds within this motif and with surrounding glycine residues. Similar secondary structure formation was also suggested for the RGG box of nucleolin (34, 35). However, the shorter RGGR motif in the C terminus of SF3A1 does not form new secondary structure elements upon U1-SL4 binding. This shows that protein secondary structure formation is not a prerequisite for base-specific RNA binding of RGG/RG motifs.
The sequence-specific modes of binding of RGG/RG motifs seen here for SF3A1 and earlier for FMRP are probably only two of the many ways RGG/RG motifs may contribute to RNA binding and protein function. RGG/RG motifs have been shown to exhibit a preference for G-C rich sequences, however, with a certain degree of flexibility for RNA sequence and structure (28, 36, 37). We recently found that the RGG/RG motif of FUS binds to the minor groove of several SL RNAs mainly via unspecific contacts and without adopting a rigid conformation (Fig. 6E) (12, 38). RGG/RG motifs also play a key role in mediating liquid–liquid phase separation of RNA binding proteins with and without RNA (28). Overall, these examples illustrate the great versatility of this small domain for several different tasks depending on the interaction partners present in the cell.
SF3A1-UBL Binding to U1-SL4 Contributes to Spliceosomal Assembly.
The structure of SF3A1-UBL bound to SL4 of U1 snRNP also provides insights into the process of spliceosomal assembly. Previous studies have established the importance of U1-SL4 for splicing and alternative splicing regulation (4, 13, 39). Mutations in U1-SL4 do not completely abolish U1 function, suggesting a redundancy in the interface between U1 and U2 snRNP in the human A complex (3, 29). Similarly to yeast, two or more interfaces could mediate the contact of U1 and U2 snRNP in the A complex. These additional interfaces between U1 and U2 snRNP could explain the comparably mild effects on splicing activity that we observed when SF3A1-UBL was mutated. Furthermore, they could explain the absence of a clear correlation between the loss of affinity for U1-SL4 binding in vitro and the relative splicing activity of the SF3A1-UBL mutants. The potential contacts between other spliceosomal components might be strengthened in the absence of the interaction between SF3A1 and U1-SL4 in order to compensate for the altered interactions. We recently identified an interaction of U1-SL3 with the RNA helicase UAP56 that enhances complex A formation (14), and there are likely other protein–protein or protein–RNA contacts involved during complex A formation. Splicing regulatory factors were found to associate with prespliceosomal complex A and could contribute to the pairing between the ss (40). An interaction between U1-70K and the 3′-ss–bound protein U2AF mediated by SR proteins has been reported in mammals (41). In fission yeast, the SR-like protein Rsd1 and Prp5 were reported to form a bridge between U1A and the U2 snRNP–specific protein SF3B1 (42, 43). The human homolog of Rsd1, RBM39, was shown to bind SF3b155 (another component of U2 snRNP) and was found in prespliceosomal A complexes (40, 44), suggesting alternative contacts that help bridging U1 and U2 snRNP in mammals. It is possible that other proteins can bind to U1-SL4, thereby compensating for the SF3A1-UBL mutations and leading to the differences in exon inclusion observed in this study that do not fully correlate with the SF3A1-UBL/U1-SL4 binding affinities.
Importantly, the structure of the SF3A1-UBL/U1-SL4 complex suggests potential mechanisms of alternative splicing regulation. Arginine residues within RGG/RG motifs are the preferred substrate for protein arginine methyltransferases (45). Therefore, it is conceivable that posttranslational modifications of the carboxyl terminus of SF3A1 could regulate splicing. Additionally, PTBP1 and other heterogeneous nuclear ribonucleoproteins (hnRNPs) were shown to bind U1-SL4 (4, 13). Future studies will investigate whether alternative splicing factors such as PTBP1 could compete with SF3A1-UBL for binding to U1-SL4.
Materials and Methods
Protein Preparation.
SF3A1-UBL (residues 704 to 793) was cloned into pET24b (Novagen) in fusion with an N-terminal GB1 solubility tag and a 6×His tag cleavable by tobacco etch virus (TEV) protease (pET24-GB1-TEV-UBL). Mutants were generated by site-directed mutagenesis using the quick-change protocol and specific primers listed in SI Appendix, Table S3. All plasmids were sequenced and transformed into Escherichia coli BL21‐Codon Plus (DE3)‐RIL cells (Agilent Technologies) for protein expression (detailed expression protocol can be found in SI Appendix). The protein was purified by Ni-affinity chromatography and size exclusion chromatography in the NMR buffer [10 mM sodium phosphate (pH 6), 50 mM NaCl] (see SI Appendix for a detailed purification protocol). Final protein purity was checked by sodium dodecyl sulfate gels and analyzed for nucleic acid contamination using A260nm/A280nm. Protein concentration was estimated using A280nm by calculating with the theoretical extinction coefficient of 9,970 M−1 cm−1 and stored at −80 °C. All point mutants were produced using the same protocol, and their correct folding was assessed by recording a 1D 1H NMR spectrum (SI Appendix, Fig. S3). GST-SF3A1-UBL for EMSA experiments was prepared as before (16). U1 snRNP in vitro reconstitution was performed as described previously (21).
RNA Preparation.
RNA constructs of human U1-SL4 (RNA sequence: 5′-GGGGACUGCGUUCGCGCUUUCCCC-3′) were produced by in vitro runoff transcription with T7 RNA polymerase (purified in house) from two complementary DNA primers containing a T7 promoter. Magnesium concentration was optimized for in vitro transcription reactions with both commercially available unlabeled nucleoside triphosphates (NTPs) (Applichem) and 13C,15N-labeled NTPs (produced in house). The RNAs were purified by anion exchange chromatography in denaturing conditions (46). The purified RNA was precipitated by butanol extraction to eliminate urea and salts (47). Lyophilized RNA was resuspended in NMR buffer. RNA was refolded by denaturing 5 min at 95 °C and incubation on ice. The 5′-Cy5–labeled RNA and 5′-biotinylated RNAs for EMSA and SPR experiments, respectively, were ordered from Integrated DNA Technologies. U1 snRNA was essentially prepared as described previously (21). The U1 snRNA used in this study contains an optimal 5′-ss fused to the U1 snRNA sequence. This duplex is stabilized by an apical GNRA-type SL (5′-GGGUAAGUAUCGCAAGAU ACUUACCUGGCAGGGGAGAUACCAUGAUCACGAAGGUGGUUUUCCCAGGGCGAGGCUUAUCCAUUGCACUCCGGAUGUGCUGACCCCUGCGAUUUCCCCAAAUGUGGGAAACUCGACUGCAUAAUUUGUGGUAGUGGGGGACUGCGUUCGCGCUUUCCCCUGucga-3′).
NMR Spectroscopy.
NMR measurements for the free protein, free RNA, and RNA–protein complexes were performed in NMR buffer [10 mM sodium phosphate (pH 6), 50 mM NaCl] at 303 K, unless otherwise noted, with Bruker AVIII-600 MHz, AVIII-700 MHz and Avance-900 MHz spectrometers all equipped with cryoprobes. Data were processed using Topspin (Bruker) and analyzed with CARA (48). For more details on NMR titrations, protein and RNA resonance assignment, and {1H}-15N hetNOE experiments, see SI Appendix.
Solution Structure Calculation.
Chemical shifts and NOESY spectra were used as input for automatic peak picking, NOE assignment, and structure calculation with the ATNOS/CANDID/CYANA suite (49) followed by automated assignments within the NOE-ASSIGN module of CYANA 3.0 (50). In addition to NOE-derived distance constraints, dihedral angle constraints were generated by TALOS+ (51) using backbone chemical shifts as input to predict secondary structures. The structures were refined in the Cartesian space using the SANDER approach of AMBER20 (52). Analysis of refined structures was performed using AMBER20 and PROCHECK-NMR (53).
Crystallization and Structure Determination.
For crystallization, RNA–protein complexes were assembled at a 1:1 molar ratio and passed over a HiLoad 16/60 Superdex 75 pg gel-filtration column (GE Healthcare) with 10 mM Hepes (pH 7.5), 50 mM NaCl as gel-filtration buffer. Fractions corresponding to the 1:1 complex, as confirmed by native polyacrylamide gel electrophoresis (PAGE), were pooled and concentrated to 0.65 mM (about 11 mg/mL) with centrifugal filters.
Crystals of the SF3A1-UBL/U1-SL4 complex were obtained by mixing 200 nL of complex (0.65 mM) with 200 nL of reservoir of Wizard Classic 3&4 crystallization screen (Rigaku) using the sitting-drop vapor diffusion method. Crystals appeared within one day in several different conditions at 18 °C. The best diffracting crystal was obtained in 100 mM Tris⋅HCl (pH 8.5), 200 mM Li2SO4, and 40% polyethylene glycol 400 (PEG400). Crystals were cryoprotected using reservoir solution supplemented with 15% (vol/vol) glycerol and flash frozen in liquid nitrogen.
Native diffraction data were recorded at a wavelength of 0.999987 Å on an EIGER 16Mdetector and DA+ software at the PXI beamline of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) at a temperature of 100 K. The data were processed, and the structure was solved and refined as described in SI Appendix. The structural model of SF3A1-UBL bound to U1 snRNP was generated in Pymol by superimposing the coordinates of U1-SL4 of the crystal structure with those of U1 snRNP of the precatalytic spliceosome (pre-B complex, PDB ID: 6QX9).
CLIR-MS/MS.
For CLIR-MS/MS experiments, samples of SF3A1-UBL bound to U1-SL4 and U1 snRNP were reconstituted as described in SI Appendix. Preparation of the CLIR-MS/MS samples, including cross-linking by UV irradiation, protease and RNase digestion, TiO2 metal oxide affinity chromatography, C18 solid phase extraction, and subsequent liquid chromatography coupled to tandem mass spectrometry and data analysis are described in detail in SI Appendix.
Surface Plasmon Resonance.
SPR experiments were essentially performed as described previously (16) with small modifications. In two replicates (Experiments 2 and 3), protein stocks were diluted to a 7.5 μM (wild-type), 10 μM (G780I), 15 μM (K717A), 20 μM (F763A), 25 μM (K765A, KK2AA, R788A, R788K), 40 μM (K754A), or 50 μM (K786A, RGG2AAA, G789I) concentration in SPR running buffer and nine twofold serial dilutions and were injected over all flow cells. Another replicate (Experiment 1) was performed using protein stocks diluted to 1 μM (wild-type) and 20 μM (mutants) concentration and four to five twofold serial dilutions. Dissociation constants (Kd) shown in Table 2 are averages from three experiments (SI Appendix, Table S2), and fold change was calculated for mutant proteins relative to WT. For more details, see SI Appendix.
Electrophoretic Mobility Shift Assays.
EMSA experiments were performed as described previously (16), and more details can be found in SI Appendix.
In Vivo Splicing Assays.
The three-exon/two-intron reporter pDUP51p and the U1 snRNA expression plasmid pNS6U1 have been described previously (4). A construct expressing U1-5a snRNA carrying U1-SL3 mutation M1d (U1-5a/SL3-M1d) was used in all splicing assays. In HeLa cells, endogenous SF3A1 was silenced by treatment with SF3A1-targeting siRNA (siSF3A1) and exon 2 inclusion of the reporter was rescued by cotransfection with RNAi-resistant SF3A1 constructs (SF3A1-RNAiR). Total RNA from transfections were used in primer extension reactions, and exon 2 inclusion of Dup51p was monitored by separation of reaction products on urea-PAGE gels. Expression of RNAi-resistant SF3A1 protein in HeLa cells under knockdown conditions was confirmed by Western blot. For details on the U1-5a/SL3-M1d variant, cell culture, transfections, siRNA-mediated knockdown, rescue experiments, primer extension, Western blot, and immunoprecipitation, see SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Alvar Gossert, Dr. Simon Ruedisser, and Dr. Fred Damberger for the maintenance of the Biomolecular NMR spectroscopy platform of ETH Zürich. We thank the beamline scientists at the Swiss Light Source PXI and PXIII beamlines, in particular Vincent Olieric (PSI, Switzerland) for support during data collection and Nenad Ban for access to crystallization equipment. This work was supported by ETH Zürich (Research Grant ETH-24 16-2 to A.L., F.H.-T.A.), the Swiss National Science Foundation through National Center of Competence in Research, RNA & Disease (NCCR RNA & Disease, grants to F.H.-T.A., A.L., and S.J.), project grant no. 310030B_189379 (to F.H.-T.A.), and 31003A_179498 (to S.J.). K.S. was supported by a PhD fellowship of the German Academic Scholarship Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114092119/-/DCSupplemental.
Data Availability
Coordinates of the SF3A1-UBL solution structure and the SF3A1-UBL/U1-SL4 crystal structure have been deposited at the Protein Data Bank under accession numbers 7P08 and 7P0V, respectively. The chemical shifts of SF3A1-UBL have been deposited in the Biological Magnetic Resonance Bank (BMRB ID: 34643). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (54) with the dataset identifier PXD027189. All other study data are included in the article and/or SI Appendix.
References
- 1.Nesic D., Krämer A., Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP, and prespliceosome formation. Mol. Cell. Biol. 21, 6406–6417 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z., et al. , Molecular architecture of the human 17S U2 snRNP. Nature 583, 310–313 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A., The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell 33, 509–518 (1983). [DOI] [PubMed] [Google Scholar]
- 4.Sharma S., Wongpalee S. P., Vashisht A., Wohlschlegel J. A., Black D. L., Stem-loop 4 of U1 snRNA is essential for splicing and interacts with the U2 snRNP-specific SF3A1 protein during spliceosome assembly. Genes Dev. 28, 2518–2531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan X., Yan C., Zhang X., Lei J., Shi Y., Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res. 28, 1129–1140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charenton C., Wilkinson M. E., Nagai K., Mechanism of 5′ splice site transfer for human spliceosome activation. Science 364, 362–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaschka C., Lin P. C., Charenton C., Nagai K., Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 559, 419–422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Feltz C., Hoskins A. A., Structural and functional modularity of the U2 snRNP in pre-mRNA splicing. Crit. Rev. Biochem. Mol. Biol. 54, 443–465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pomeranz Krummel D. A., Oubridge C., Leung A. K. W., Li J., Nagai K., Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458, 475–480 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., et al. , CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing. Nat. Commun. 8, 1035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plaschka C., Lin P. C., Nagai K., Structure of a pre-catalytic spliceosome. Nature 546, 617–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jutzi D., et al. , Aberrant interaction of FUS with the U1 snRNA provides a molecular mechanism of FUS induced amyotrophic lateral sclerosis. Nat. Commun. 11, 6341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S., Maris C., Allain F. H. T., Black D. L., U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol. Cell 41, 579–588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martelly W., et al. , Synergistic roles for human U1 snRNA stem-loops in pre-mRNA splicing. RNA Biol. 18, 2576–2593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C. J., Ferfoglia F., Raleff F., Krämer A., Interaction domains and nuclear targeting signals in subunits of the U2 small nuclear ribonucleoprotein particle-associated splicing factor SF3a. J. Biol. Chem. 286, 13106–13114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martelly W., Fellows B., Senior K., Marlowe T., Sharma S., Identification of a noncanonical RNA binding domain in the U2 snRNP protein SF3A1. RNA 25, 1509–1521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lescrinier E. M. H. P., et al. , Structure of the pyrimidine-rich internal loop in the poliovirus 3′-UTR: The importance of maintaining pseudo-2-fold symmetry in RNA helices containing two adjacent non-canonical base-pairs. J. Mol. Biol. 331, 759–769 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Tolbert B. S., et al. , Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement. J. Biomol. NMR 47, 205–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohlenschläger O., et al. , The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12, 237–248 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Dorn G., et al. , Structural modeling of protein-RNA complexes using crosslinking of segmentally isotope-labeled RNA and MS/MS. Nat. Methods 14, 487–490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campagne S., et al. , An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands. Nucleic Acids Res. 49, e63 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ke Chang C., et al. , The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem. Biophys. Res. Commun. 425, 219–224 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Qin H., Lim L. Z., Wei Y., Song J., TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA. Proc. Natl. Acad. Sci. U.S.A. 111, 18619–18624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J., Bean R. L., Vogt V. M., Summers M., Solution structure of the Rous sarcoma virus nucleocapsid protein: muPsi RNA packaging signal complex. J. Mol. Biol. 365, 453–467 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thapar R., Denmon A. P., Nikonowicz E. P., Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip. Rev. RNA 5, 49–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maris C., et al. , A transient α-helix in the N-terminal RNA recognition motif of polypyrimidine tract binding protein senses RNA secondary structure. Nucleic Acids Res. 48, 4521–4537 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lunde B. M., Moore C., Varani G., RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong P. A., Vernon R. M., Forman-Kay J. D., RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Castello A., et al. , Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D., Arginine-mediated RNA recognition: The arginine fork. Science 252, 1167–1171 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Chavali S. S., Cavender C. E., Mathews D. H., Wedekind J. E., Arginine forks are a widespread motif to recognize phosphate backbones and guanine nucleobases in the RNA major groove. J. Am. Chem. Soc. 142, 19835–19839 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan A. T., et al. , Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 18, 796–804 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasilyev N., et al. , Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U.S.A. 112, E5391–E5400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghisolfi L., Joseph G., Amalric F., Erard M., The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 267, 2955–2959 (1992). [PubMed] [Google Scholar]
- 35.Gao F., et al. , β-turn formation by a six-residue linear peptide in solution. J. Pept. Res. 60, 75–80 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Corbin-Lickfett K. A., Chen I. H. B., Cocco M. J., Sandri-Goldin R. M., The HSV-1 ICP27 RGG box specifically binds flexible, GC-rich sequences but not G-quartet structures. Nucleic Acids Res. 37, 7290–7301 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozdilek B. A., et al. , Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 45, 7984–7996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loughlin F. E., et al. , The solution structure of FUS bound to RNA reveals a bipartite mode of RNA recognition with both sequence and shape specificity. Mol. Cell 73, 490–504.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Rogalska M. E., et al. , Therapeutic activity of modified U1 core spliceosomal particles. Nat. Commun. 7, 11168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hegele A., et al. , Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell 45, 567–580 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Wu J. Y., Maniatis T., Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Shao W., Kim H.-S., Cao Y., Xu Y.-Z., Query C. C., A U1-U2 snRNP interaction network during intron definition. Mol. Cell. Biol. 32, 470–478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y. Z., et al. , Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23, 376–385 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loerch S., Maucuer A., Manceau V., Green M. R., Kielkopf C. L., Cancer-relevant splicing factor CAPERα engages the essential splicing factor SF3b155 in a specific ternary complex. J. Biol. Chem. 289, 17325–17337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thandapani P., O’Connor T. R., Bailey T. L., Richard S., Defining the RGG/RG motif. Mol. Cell 50, 613–623 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Duss O., Maris C., von Schroetter C., Allain F. H. T., A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res. 38, e188 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cathala G., Brunel C., Use of n-butanol for efficient recovery of minute amounts of small RNA fragments and branched nucleotides from dilute solutions. Nucleic Acids Res. 18, 201 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keller R., The Computer Aided Resonance Assignment Tutorial (Cantina Verlag, 2004). [Google Scholar]
- 49.Herrmann T., Güntert P., Wüthrich K., Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24, 171–189 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Güntert P., “Automated NMR structure calculation With CYANA” in Protein NMR Techniques, Downing A. K., Ed. (Humana Press, 2004), pp. 353–378. [DOI] [PubMed] [Google Scholar]
- 51.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Case D. A., et al. , The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M., AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Vizcaíno J. A., et al. , The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss M., Global indicators of X-ray data quality. J. Appl. Cryst. 34, 130–135 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates of the SF3A1-UBL solution structure and the SF3A1-UBL/U1-SL4 crystal structure have been deposited at the Protein Data Bank under accession numbers 7P08 and 7P0V, respectively. The chemical shifts of SF3A1-UBL have been deposited in the Biological Magnetic Resonance Bank (BMRB ID: 34643). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (54) with the dataset identifier PXD027189. All other study data are included in the article and/or SI Appendix.