Abstract
Background:
Proteomic approaches allow measurement of thousands of proteins in a single specimen, which can accelerate biomarker discovery. However, applying these technologies to massive biobanks is not presently feasible due to the practical barriers and costs of implementing such assays at scale. To overcome these challenges, we employed a “virtual proteomic” approach, linking genetically-predicted protein levels to clinical diagnoses in >40,000 individuals.
Methods:
We used genome-wide association data from the Framingham Heart Study (n=759) to construct genetic predictors for 1,129 plasma protein levels. We validated the genetic predictors for 268 proteins and used them to compute predicted protein levels in 41,288 genotyped individuals in the eMERGE cohort. We tested associations for each predicted protein with 1,128 clinical phenotypes. Lead associations were validated using directly-measured protein levels and either LDL-C or subclinical atherosclerosis in the Malmö Diet and Cancer Study (MDCS) (n=651).
Results:
In the virtual proteomic analysis in eMERGE, 55 proteins were associated with 89 distinct diagnoses at false discovery rate (FDR) q<0.1. Among these, 13 associations involved lipid (n=7) or atherosclerosis phenotypes (n=6). We tested each association for validation in MDCS using directly-measured protein levels. At Bonferonni-adjusted significance thresholds, levels of apolipoprotein E isoforms were associated with hyperlipidemia, and circulating CLC1B and PDGFR-β predicted subclinical atherosclerosis. Odds ratios for carotid atherosclerosis were 1.31 (95% CI, 1.08–1.58; p=0.006) per SD increment in CLC1B, and 0.79 (0.66–0.94; p=0.008) per SD increment in PDGFR-β.
Conclusions:
In summary, we demonstrate a biomarker discovery paradigm to identify candidate biomarkers of cardiovascular and other diseases.
Keywords: biomarkers, proteomics, phenomics, electronic health record, atherosclerosis
INTRODUCTION
Biomarkers can facilitate early detection of disease and suggest potential therapeutic targets.1 A limitation of traditional biomarker studies is that the selection of candidates is constrained by existing knowledge of biological pathways that contribute to the disease in question. Hence, these markers often add little to the clinical data since they provide overlapping information.2,3 Further, such biomarkers may yield few novel insights regarding the biological mechanisms modulating or driving disease risk. Emerging technologies in metabolomics and proteomics allow the interrogation of large numbers of biomarkers from diverse pathways. The extended range of biomarkers that can be measured in biological specimens has enabled discovery-oriented biomarker methodologies. However, applying these approaches in large cohorts of individuals is typically not feasible due to the practical challenges and resource costs of implementing these assays at scale.
Circulating levels of many biomarkers are modulated by genetic variation that can be identified and quantified using approaches such as the genome-wide interrogation of variability attributable to common single nucleotide polymorphisms (SNPs). Genetic risk scores constructed from these analyses can be used to calculate a genetically-predicted biomarker level in a genotyped individual—a so-called “virtual biomarker.”4 Because the genetic contribution to many circulating biomarker levels is highly significant,5 a genetic predictor for each biomarker can be developed using a relatively small number of genotyped individuals and then used to compute genetically-predicted biomarker levels in readily-available, much larger genotyped populations. Further, this genetically-predicted biomarker can be used to test associations with any phenotype measured in the second population.4 In sum, genetic approaches can tremendously augment the effective sample sizes and the number of phenotypes that can be evaluated for a biomarker.
We hypothesized that we could apply these approaches to identify clinical phenotypes associated with plasma protein levels. We used a data set derived from participants in the Framingham Heart Study (FHS) Offspring cohort who had undergone profiling of 1,129 plasma proteins using an aptamer-based proteomic technology.6,7 We constructed genetic instruments that were then used to compute genetically-predicted protein levels in a large population of participants in the Electronic Medical Records and Genomics (eMERGE) network, a consortium of medical centers with EHR-linked DNA biobanks.8 We used these genetically-predicted values to interrogate a broad range of curated phenotypes in order to identify novel candidate biomarker-disease associations. We then validated two especially intriguing associations with atherosclerotic cardiovascular traits for which the directly-measured biomarkers were available in a traditional epidemiologic cohort. We anticipate that this novel approach will greatly accelerate the application of molecular profiling in clinical and research settings.
MATERIALS AND METHODS
The data, analytic methods, and study materials will be made available to other researchers for purposes of reproducing the results or replicating the procedure. Genetic data are available via data deposits made by the eMERGE Network to DBGAP9 and clinical phenotypes can be requested via Data Use Agreements through the eMERGE Network (https://emerge.mc.vanderbilt.edu/).8
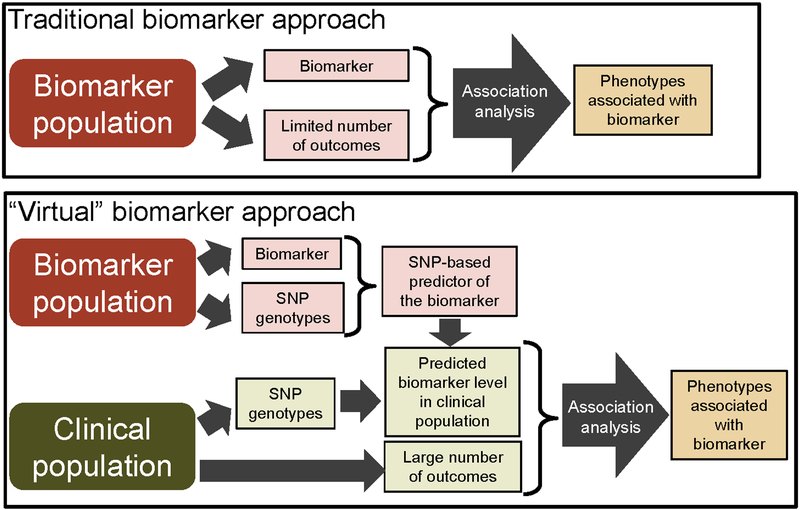
An overview of the study design is shown in Figure 1.
Figure 1. Comparison of a traditional and genetic biomarker study design.
In a traditional design (top panel), the biomarker and outcomes are measured in the same population and either prospective or cross-sectional associations are explored. In the genetics-based study design used in these analyses (bottom panel), genetic predictors of biomarker levels are constructed in one population. These predictors are then used to compute genetically-predicted biomarker levels for each individual in a second genotyped population. Associations between the predicted biomarker levels and a large number of outcomes are then tested within the clinical population.
Study populations
EHR populations:
The EHR populations (n=41,288) were from the eMERGE Network, a consortium of medical centers using EHRs as a tool for genomic research, and from Vanderbilt University Medical Center’s (VUMC) BioVU resource (Supplementary Table 1).10 The participating eMERGE sites were Geisinger Health System, VUMC, Marshfield Clinic, Northwestern University, Harvard University, Mayo Clinic, and Kaiser Permanente/University of Washington, Seattle. All participants were born prior to 1990 and clustered within 4 standard deviations of the first 2 principal components (PCs) based on a subset of individuals self-identified as “White, non-Hispanic” participants.
Malmö Diet and Cancer Study (MDCS):
MDCS11 is a Swedish population-based, observational cohort was used to test for epidemiological evaluation of selected genetic associations identified in the primary analyses. The MDCS set comprised 651 participants from two nested case-control samples (163 cases of incident type 2 diabetes [T2D], 162 cases of incident CHD and 326 controls) (Supplementary Table 2). Participants with prevalent CHD or T2D were excluded.
Genetic data
SNP genotype data for the EHR populations were acquired on the Illumina Human660W-Quadv1_A, HumanOmni1-Quad, HumanOmni5-Quad, MEGA-EX, Human610, Human550, HumanOmniExpressExome-8v1.2A and MegArray platforms. Quality control (QC) steps for the EHR data sets were performed per previously published protocols.12 QC analyses used PLINK v 1.90β3.13 SNPs were pre-phased using SHAPEIT,14 imputed using IMPUTE215 in conjunction with the 10/2014 release of the 1000 Genomes cosmopolitan reference haplotypes. Imputed data were filtered for a sample missingness rate<2%, a SNP missingness rate<4% and a SNP deviation from Hardy-Weinberg<10−6. Principal components were generated using the SNPRelate package.16
Phenotype data
EHR diagnoses were based on phecodes (https://phewas.mc.vanderbilt.edu/), which are collections of related ICD-9-CM (International Classification of Disease, Ninth revision) diagnosis codes.17 Phecodes have been used extensively validated for use genetic studies.18–20 For each phecode, cases are participants with two or more instances of the code appearing in their medical record. Controls were participants without the clinical phenotype or any closely related phecodes and whose decade of birth fell within the range of birth decades observed among cases. Any eMERGE site that had fewer than 10 cases for a given phecode was excluded for that phenotype. Phenotypes that only affected a single sex were not included in these analyses. There were 1,128 clinical phenotypes with ≥300 cases in the EHR data set that were used in the PheWAS analyses.
Plasma protein levels in the MDCS were measured using SOMAscan technology (1.3k assay). Peripheral blood samples collected in EDTA-treated tubes and were assayed according to the manufacturer’s protocol, as previously described.7,21 Fasting LDL cholesterol (LDL-C) levels were measured, as previously described.22 The prevalent atherosclerosis phenotype was defined as a focal thickening of the carotid intima-media >1.2 mm with an area of =>10mm2, as previously described.
GWAS Summary statistics
Summary statistics from published GWAS of plasma proteins levels measured using the SOMAscan technology were used in these analyses, as described below.
Framingham Heart Study (FHS):
Proteomic profiling (n=1,129 proteins) was performed on peripheral blood samples among participants in the FHS Offspring cohort who attended the fifth examination (1991–1995) using the SOMAscan single-stranded DNA aptamer-based platform (1.1k assay in FHS and 1.3k assay in MDCS).7,21 A total of 759 participants without prevalent CVD and with genotyping on the Affymetrix 500K and 50K supplementary arrays, imputed to 1000 Genomes Phase I version 3 (August 2012), were used for GWAS of each protein.23 Summary statistics were obtained from Framingham investigators.23
KORA F4 study:
GWAS summary statistics for analyses of proteomic profiling of 1,124 proteins measured in the KORA F4 study population (n=997 participants ages 32–81 years) using the SOMAscan assay (V3.2) were downloaded from the website http://metabolomics.helmholtz-muenchen.de/pgwas/.5 KORA participants were genotyped on the Affymetrix Axiom Array (n=509,946 autosomal SNPs).
Statistical Analysis
Validating protein predictors:
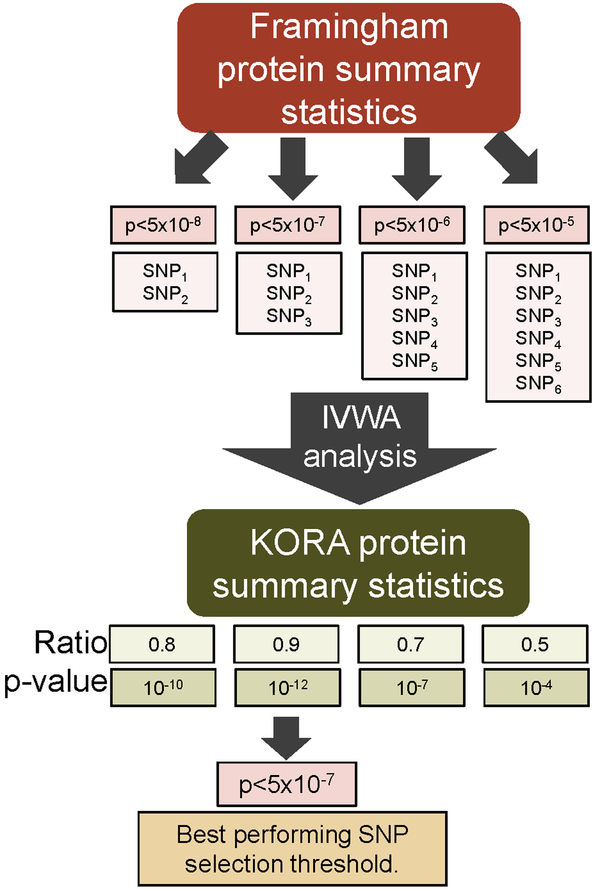
An overview of validation and construction of genetic predictors of protein levels is presented in Figure 2. There were 1,112 proteins with summary statistics available in both the FHS (used as the discovery set) and KORA (used as the validation set) data sets. To define a set of validated genetic predictors of protein levels, SNPs exceeding each of four p-value selection thresholds (5×10−8,5×10−7,5×10−6 and 5×10−5) were selected for each protein using the FHS set. The predictors based on lower thresholds incorporate additional SNPs with weaker associations, and potentially capture a broader polygenic signal. At each selection threshold, a LD-reduced (r-square=0.2) set of SNPs with a minor allele frequency (MAF) >5% that overlapped with the KORA set was selected using a clumping algorithm.24 To determine whether the SNPs at a given threshold predicted protein levels in KORA, we tested for consistency in the direction of the regression coefficients of the SNPs across data sets based on an inverse-variance weighted average (IVWA), using methods previously described.25 For each protein, we identified the SNP selection threshold that had the most significant association in the IVWA analysis and selected the ratio estimate and its p-value from the IVWA analysis. The association was considered significant if IVWA p-value<0.01. This significance threshold was determined empirically by examining the direction of the ratio estimates across p-values. For replicating proteins, the ratio estimate is expected to be positive. For proteins with p<0.01, 2 of 268 (0.7%) proteins had a negative ratio (Supplementary Figure 1). In contrast, 46% of proteins with p>0.01 had a negative ratio estimate, which is close to what would be expected by chance (50%).
Figure 2. Approach to developing and validating a genetic predictor for a protein in the KORA population.
For each protein, up to 4 predictors are created by incorporating SNPs meeting increasingly higher p-value thresholds. Each predictor is then tested for validation in the KORA data set using an inverse-variance weighted average (IVWA) association approach. The predictor with the lowest IVWA association p-value is then selected for further analysis.
PheWAS:
A PheWAS analysis was performed for each of the 268 proteins. PheWAS tests associations between a variable and a large collection of clinical diagnoses extracted from an EHR data set.17,19 Genetic predictors were constructed for each protein by selecting an LD-reduced (r-square=0.2) set of SNPs exceeding the best p-value selection threshold identified in the validation step (above). A new set of SNPs was selected because the SNP overlap between the FHS and the KORA and EHR data sets differed. SNPs and their weightings used to construct each protein predictor are shown in Supplementary Table 3. For each predictor, the genetically-predicted protein level among participants in the EHR population represented a weighted genetic risk score and was computed using the equation:
Logistic regression, adjusting for 3 PCs, birth decade, sex, eMERGE site, and genotyping platform was used to test associations with each PheWAS phenotype and predicted protein levels. The genetically-predicted phenotype values were standardized to have a standard deviation of 1, so odds-ratios (ORs) reflect the risk per standard deviation (s.d.) increase in the genetically-predicted biomarker value. A Benjamini-Hochberg (B-H) false discovery rate (FDR) adjustment was applied across all analyses to adjust for multiple testing.26 PheWAS analyses used SAS v9.3 (SAS Institute, Cary, NC).
To ascertain the contribution of SNPs located near the gene locus for the protein product, genetic predictors were also constructed using only SNPs located within 1 Mb of the transcription start and stop sites for the gene.
Validation studies
For each protein, the proportion of phenotypic variation explained by the genetically predicted level was determined in the MDCS cohort by computing the partial Pearson’s correlation coefficient, adjusting for age and sex, between the genetically predicted and measured levels of the protein.
The genetic predictors for all proteins associated with a clinical diagnosis related to elevated LDL cholesterol levels (n=7 proteins) and atherosclerotic cardiovascular diseases (peripheral vascular disease, stroke, carotid stenosis, and coronary heart disease [CHD]) (n=6 proteins) in the discovery EHR cohort were validated in the MDCS cohort by testing associations with either baseline LDL levels (n=615 subjects) or the carotid plaque (n=168) phenotype, respectively. LDL differences were estimated using linear regression and odds ratios (ORs) for the presence of carotid atherosclerosis were estimated using logistic regression models. All models analyzed log-transformed protein levels as the independent variable and adjusted for age, sex, and plate. LDL changes and ORs are per standard deviation increase in the log-transformed protein level. Associations with a Bonferroni adjusted p-value<(0.05/7=0.0071) for LDL and p<(0.05/6=0.008) for atherosclerosis were considered significant. The CLC1B and PDGFR-β associations were also examined using a logistic model that further adjusted for body mass index, fasting glucose levels, hypertension status, smoking status, LDL levels and C-reactive protein levels.
Study approval
The eMERGE study was approved by the Institutional Review Board (IRB) at each site.8 The study protocols were approved by the IRB of Boston University Medical Center, Beth Israel Deaconess Medical Center, and Lund University, Sweden, and all participants provided written informed consent.
RESULTS
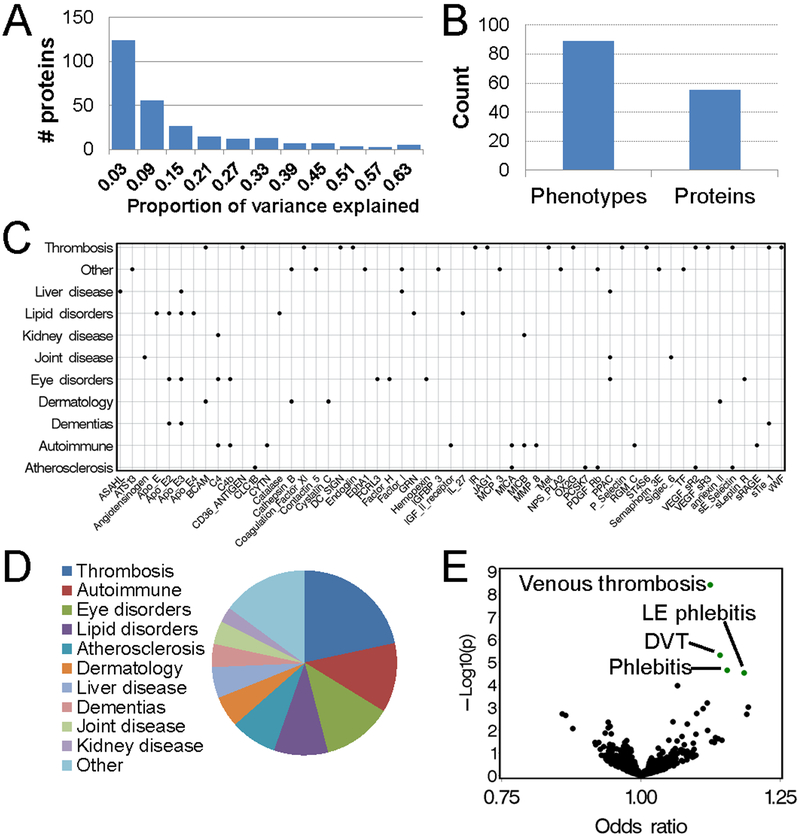
We employed a two-step approach to identify novel biomarker-disease associations (Figure 1). We constructed genetic predictors for each of 268 plasma proteins that were measured in the FHS Offspring cohort and successfully validated in the KORA data set (Figure 2). In cross-validation analyses using the MDCS cohort, the median proportion of the variability in plasma protein levels explained by the predictors was 0.08 (interquartile range [IQR] 0.04 – 0.22) (Figure 3A and Supplementary Table 4). Genetically-predicted protein levels were then imputed into the EHR population (n=41,288), and associations were tested with each of 1,128 clinical phenotypes. There were 223 associations among 55 proteins and 89 clinical diagnoses that were significant at FDR q<0.1 (Figure 3B and Supplementary Table 5).
Figure 3. Summary of associations between genetic predictors and clinical phenotypes.
Panel (A): Frequency histogram of the variances in the plasma levels of a protein explained by the genetic predicted biomarker in the MDCS validation set. Values are adjusted for age and sex. Panel (B): Frequency histogram showing the number of distinct clinical phenotypes and proteins with associations. Panel (C): Significant associations between genetically predicted protein levels and a phenotype category. Panel (D): Volcano plot summarizing associations between a genetic predictor for the protein F11 and the clinical phenotypes. Each dot represents a clinical phenotype, and dots highlighted in green had a significant association (FDR q<0.1). Panel (E): Pie-chart showing the proportions proteins with an association with a clinical phenotype belonging to the one of 11 categories.
A number of proteins were associated with multiple similar phenotypes. For instance, the protein Coagulation Factor XI (F11) was associated with 4 diagnoses related to venous thrombosis (Figures 3C and 3D). Among the 55 proteins with associations, 16 (22.6%) (Basal Cell Adhesion Molecule [BCAM], Platelet glycoprotein 4 [CD36 Antigen], F11, CD209 Molecule [DC SIGN], Endoglin, Insulin Receptor [IR], Protein Jagged-1 [JAG1], Hepatocyte Growth Factor Receptor [Met], OX-2 Membrane Glycoprotein [OX2G], P-selectin, sE-Selectin, Carbohydrate Sulfotransferase 15 [CHST15], Tyrosine-protein kinase receptor Tie-1 [Tie-1], Vascular Endothelial Growth Factor Receptor 2 [VEGF-sR2], Vascular Endothelial Growth Factor Receptor 3 [VEGF-sR3], von Willebrand Factor [vWF]) were associated with a thrombosis phenotype (Figure 3E and Supplementary Table 6). For all but 4 of these proteins (F11, DC SIGN, ST4S6, sTie-1), the association signal was driven by SNPs comprising the genetic predictor that were located near the ABO gene locus. These findings are consistent with prior observations that levels for a large number of proteins are regulated by the ABO gene.5,27
Other phenotype categories with multiple protein associations included autoimmune disorders (soluble MAPK/MAK/MRK overlapping kinase [MOK,RAGE], Cystatin-SN, Complement C4-A [C4A], Hemojuvelin, Cation-independent Mannose-6-phosphate Receptor [IGF2R], MHC Class I Polypeptide-related Sequence A [MICA], MHC Class I Polypeptide-related Sequence B [MICB], Neutrophil collagenase [MMP8]), atherosclerotic phenotypes (C-type Lectin Domain Family 1 Member B [CLC1B], MICA, Proprotein Convertase Subtilisin/Kexin Type 7 [PCSK7], Platelet Derived Growth Factor Receptor Beta [PDGFR-β], soluble E-Selectin, VEGF-sR2), lipid disorders (Apolipoprotein E [APO-E] isoforms, Catalase, Interleukin 27 [IL-27], Granulin) and dementia (Tie-1 and APO-E isoforms) (Figure 3E).
In order to validate lead predicted biomarker associations, we tested the association between directly-measured plasma protein levels and validation phenotypes in the MDCS cohort. We tested all proteins associated with either a diagnosis of elevated cholesterol levels (n=7 proteins) or an atherosclerosis phenotype (n=6 proteins). The genetic predictors for the 7 cholesterol-associated proteins accounted for 3% (median, IQR=1%−7%) of the variance in the measured protein levels in the MDCS cohort (Supplementary Table 7). Four the 7 proteins were apolipoprotein E (ApoE) isoforms. Directly measured levels of each isoform were significantly associated with LDL-C levels in the MDCS cohort, and had a consistent direction of association as the genetic association (Table 1). There was not an epidemiological association with LDL-C for the other 3 proteins (Catalase, IL-27, Granulin).
Table 1.
Associations between genetically predicted and measured levels of a protein and LDL cholesterol levels.
| Protein | CIS* | Odds-ratio† | 95% CI | p-value | Beta‡ | 95% CI | p-value | |
|---|---|---|---|---|---|---|---|---|
| ApoE | Y | 1.07 | (1.04–1.11) | 6.2×10−6 | 0.43 | (0.36 – 0.50) | 4.9 ×10−30 | |
| ApoE2 | Y | 1.10 | (1.07–1.14) | 4.1×10−10 | 0.21 | (0.14 – 0.29) | 1.2 ×10−7 | |
| ApoE3 | Y | 1.12 | (1.08–1.15) | 4.5×10−12 | 0.37 | (0.30 – 0.45) | 7.0 ×10−22 | |
| ApoE4 | Y | 1.07 | (1.04–1.11) | 5.7×10−6 | 0.29 | (0.21 – 0.36) | 6.6 ×10−13 | |
| Catalase | N | 1.07 | (1.04–1.10) | 1.1×10−5 | 0.05 | (−0.03 – 0.13) | 0.26 | |
| Granulin | N | 1.09 | (1.06–1.13) | 1.0×10−8 | −0.02 | (−0.10 – 0.06) | 0.66 | |
| IL-27 | N | 1.07 | (1.03–1.10) | 3.6×10−5 | 0.00 | (−0.07 – 0.08) | 0.90 |
Footnotes:
Indicates whether the association was significant (p<0.01) when analyses were limited to SNPs located within 1 Mb of the gene locus.
EHR genetic association based on logistic regression analyses for an association with a diagnosis of “Hyperlipidemia” (n=8,511 cases and 13,436 controls) adjusting for birth year, gender, eMERGE site, genotyping platform and 3 principal components.
MDCS epidemiological validation showing the change in LDL-C levels based on a linear regression model adjusting for age, sex and plate.
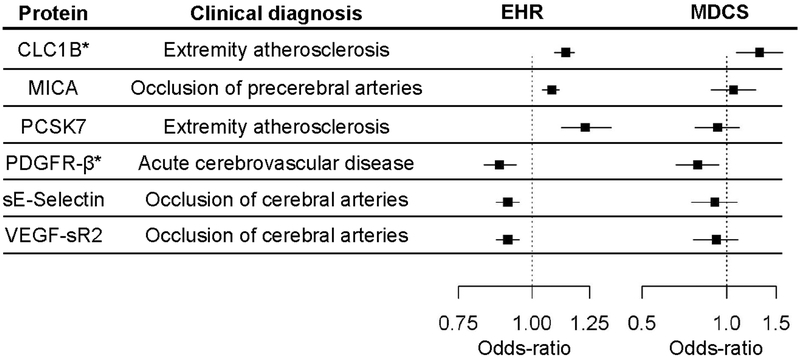
The 6 proteins with atherosclerosis associations involved peripheral vascular disease (CLC1B, PCSK7) and cerebrovascular disease (MICA, PDGFR-β, soluble E-Selectin, VEGF-sR2) (Table 2). The genetic predictors for these proteins accounted for 27% (median, IQR=7%−48%) of the variance in the measured levels in the MDCS cohort (Supplementary Table 7). We tested these proteins for an association with the presence of carotid plaque in the MDCS cohort (Table 3). For five of the 6 proteins, the direction of association was consistent between the discovery and validation cohorts (Figure 4). Two of the 6 lead proteins met a conservative, Bonferroni-adjusted level of significance (p < 0.008). Measured CLC1B levels were associated with an increased risk of carotid plaque in MDCS (OR=1.31 per SD increment in biomarker level [95% CI:1.08–1.58], p=0.006), consistent with the direction of effect found in the discovery cohort for the predicted levels (OR=1.14 [1.09–1.18], p=3.0×10−9). Similarly, we found an inverse association between measured plasma PDGFR-β levels and carotid plaque in MDCS (OR= 0.79 [0.66–0.94], p=0.008), consistent with the association of predicted PDGFR-β and acute stroke in the EHR cohort (OR=0.88 [0.83–0.94], p=4.6×10−5). The carotid plaque associations for CLC1B and PDGFR-β persisted despite further adjustment for established atherosclerosis risk factors and high-sensitivity C-reactive protein levels. The multivariable-adjusted ORs per SD increment in CLC1B and PDGFR-β were 1.27 (95% CI: [1.05–1.54], p=0.015) and 0.81 (95% CI [0.67–0.97], p=0.02), respectively. The model c-statistic increased from 0.68 to 0.69 with the addition of each protein level as a covariate.
Table 2.
Associations between genetically predicted levels of a protein and clinical diagnoses related to atherosclerotic disease.
| Protein | Clinical diagnosis | Cases | Controls | CIS* | Odds-ratio† | 95% CI | p-value |
|---|---|---|---|---|---|---|---|
| CLC1B | Atherosclerosis of the extremities | 2,683 | 24,460 | Yes | 1.14 | (1.09–1.18) | 3.0 ×10−9 |
| MICA | Occlusion/stenosis of precerebral arteries | 3,784 | 27,696 | Yes | 1.08 | (1.04–1.11) | 7.9 ×10−5 |
| PCSK7 | Atherosclerosis of the extremities | 487 | 26108 | Yes | 1.23 | (1.12–1.36) | 1.8 ×10−5 |
| PDGFR-β | Acute cerebrovascular disease | 1,184 | 28,518 | Yes | 0.88 | (0.83–0.94) | 4.6 ×10−5 |
| sE-Selectin | Occlusion of cerebral arteries | 1,772 | 29,416 | No | 0.91 | (0.87–0.95) | 8.0 ×10−5 |
| VEGF-sR2 | Occlusion of cerebral arteries | 1,772 | 29,416 | No | 0.91 | (0.87–0.95) | 7.5 ×10−5 |
Footnotes:
Indicates whether the association was significant (p<0.01) when analyses were limited to SNPs located within 1 Mb of the gene locus.
Based on logistic regression analyses adjusting for birth year, gender, eMERGE site, genotyping platform and 3 principal components.
Table 3.
Associations of candidate proteins with carotid plaque in the MDCS cohort (n=651).
| Protein | Odds ratio, per SD increment in biomarker* | 95% CI | p-value |
|---|---|---|---|
| CLC1B | 1.31 | (1.08–1.58) | 0.006 |
| MICA | 1.06 | (0.88–1.27) | 0.53 |
| PCSK7 | 0.93 | (0.77–1.12) | 0.42 |
| PDGFR-β | 0.79 | (0.66–0.94) | 0.008 |
| sE-Selectin | 0.91 | (0.75–1.09) | 0.29 |
| VEGF-sR2 | 0.92 | (0.76–1.10) | 0.35 |
Footnotes:
From multivariable logistic regression models adjusting for age, sex and plate.
Figure 4. Summary of atherosclerosis associations in the EHR discovery and MDCS validation sets.
Odds-ratios and 95% CI for associations between the genetic predictors and the indicated PheWAS clinical diagnoses in the EHR data set (on left) and between measured levels of the protein and carotid atherosclerosis (on right). An asterisk denotes that the association in MDCS was significant.
DISCUSSION
We employed a “virtual proteomic” approach to identify novel biomarker-disease associations in a large (>40,000 person) clinical dataset. Because direct measurement of >1,000 proteins in this many individuals would be impractical, we leveraged genomic data to construct predicted biomarker levels for each individual and identified clinical associations with 55 proteins. We selected candidate associations related to hyperlipidemia and atherosclerotic cardiovascular disease for direct validation in an epidemiologic cohort and demonstrated significant associations between directly-measured protein levels and LDL-C (ApoE) and carotid atherosclerosis (CLC1B and PDGFR-β).
There are multiple challenges related to identifying and validating novel disease biomarkers using standard epidemiological approaches. These include logistical, throughput, cost and resource challenges inherent in measuring a candidate biomarker in large study populations. Proteomic discovery in large EHR-based cohorts is further complicated by sample processing challenges, particularly time to appropriate storage, that do not affect genetic analyses. These limitations may compel investigators to rely on candidate biomarker approaches that have less potential for discovery. Traditional biomarker study designs are also limited by the fact that a biomarker can only be tested against other phenotypes measured in the same cohort, and cannot be rapidly repurposed for use in other populations. We evaluated an approach designed to overcome these limitations by using genetic predictors as surrogates for circulating biomarkers. This approach is similar to the PrediXcan and TWAS methods that have been used to associate RNA expression levels with diseases.28,29 The utility of the PrediXcan and TWAS approaches lends general support to the strategy outlined in the current investigation. One distinction, and a potential advantage of the virtual proteomics method, is the ability to validate the genetic findings with directly-measured levels. This approach enabled us to test for biomarker associations in a significantly larger population than the biomarker was actually measured in, and to screen the biomarker against a large number of clinical diagnoses. Hence, we identified associations with phenotypes not ascertained in the original cohort.
For these analyses, our candidate biomarkers were plasma protein levels. One advantage of protein biomarkers is that their levels are modulated by common SNP variants thereby making them amenable to a genetic-based study design.5 A second advantage is that an association potentially highlights a mechanism of disease, as the protein may be a mediator of the disease process. We note that a virtual proteomics approach cannot detect all of the associations that would be captured with direct proteomic measurements on the same study population. The intent of the approach is to enable protein discovery in study populations not currently accessible to a traditional approach. Some associations have immediate face-value interpretability. For instance, we observed that higher genetically-predicted levels of the blood coagulation Factor XI (F11) were associated with an increased risk of deep vein thrombosis, consistent with previously reported associations between F11 levels and venous thrombosis.30 Apolipoprotein E and its isoforms were associated with hyperlipidemia and dementia, consistent with the known biology of this protein.31 A third advantage is that proteomics technologies are rapidly advancing and increasingly large numbers of proteins can be measured by these technologies. Hence, there is great potential for ongoing novel biomarker discovery in this domain. Lastly, predicted biomarker associations can be validated using biospecimens obtained from conventional clinical or epidemiologic studies.
Although the discovery phase of our approach employs a genetic association study, it is not a Mendelian Randomization (MR), a study design that seeks to infer causality between a biomarker and outcome.32 MR requires explicit knowledge and modeling of known mediators and risk factors associated with a biomarker and a disease. The primary goal of this study was to identify new protein biomarkers, not to establish causal relationships. In order for a biomarker to be an effective predictor of an outcome of interest, its level must vary with respect to risk of that outcome. Hence, we constructed genetic predictors designed to capture genetic signals modulating biomarker levels. However, a significant association with a genetic predictor can occur in situations in which the biomarker does not vary with respect to the risk of the outcome. For instance, if a SNP in the predictor tags a pleiotropic genetic locus that is associated with an outcome through a mechanism unrelated to that regulating biomarker levels, it could give rise to a false positive association with the outcome.33 The definitive approach to discriminate these possibilities is to directly test the candidate association between the measured biomarker level and outcome in an independent cohort, as done in the present study.
We selected hyperlipidemia and atherosclerosis traits for the independent validation. Among the hyperlipidemia associations, there was strong replication among the ApoE isoforms, consistent with previous epidemiological studies examining ApoE levels measured using the SOMAscan platform.7 The genetic associations for the ApoE isoforms were driven by SNPs located near the ApoE gene locus. In contrast, for the proteins that did not have an epidemiological replication, the genetic association was driven by trans-acting SNPs. Among these, granulin (GRN) is perhaps the most interesting. The genetic association we observed was driven by SNPs in chromosome 1 located near the CELSR2-PSRC1-MYBPHL-SORT1 gene loci, and have been previously reported.34,35 Prior protein-wide association studies (PWAS) have linked GRN with coronary artery disease through SNPs located at this gene locus5,27 and GWAS have also identified SNPs with strong associations to LDL-C levels in this same region.36 Mutations in GRN are associated with the neurodegenerative disease frontotemporal dementia (FTD). However, GRN does not have a known role in LDL homeostasis. The genetic association between GRN and hyperlipidemia is likely explained by SORT1, which is the main neuronal receptor for GRN. SORT1 also reduces LDL levels by increasing uptake and catabolism of LDL.37 Thus, while genetic variation may influence SORT1 activity in these 2 different contexts, this does not translate into an epidemiological association that supports the observation that GRN may be a LDL-C biomarker.
For proteins with an association with an atherosclerotic disease, we tested their association with the presence of carotid plaque, a well-established marker of atherosclerotic disease burden. Among 6 candidate associations, 2, PDGFR-β and CLC1B, met the conservative Bonferroni significance threshold. The genetic associations for both of these proteins were driven by cis-acting SNPs, which may also account for the higher proportion of variability explained for the protein levels.
Lower serum levels of PDGFR-β (Platelet Derived Growth Factor Receptor Beta) were associated with an increased risk of acute stroke and the presence of carotid plaques in our analyses. PDGFR-β is a tyrosine kinase cell-surface receptor involved in development and signaling in a range of contexts. The contribution of PDGFR-β to the formation and promotion of atherosclerotic plaque formation has been described in both in vitro and in vivo studies in model systems. During development, this receptor is involved in recruitment and migration of pericytes and smooth muscle cells to endothelial cells.38 Inhibition of this receptor in ApoE-deficient mice markedly decreased atherosclerotic lesion size in the aorta39, while constitutive activation of signaling promotes leukocyte accumulation in the adventitia and media of arteries.40 In human cadaver studies, PDGFR-β was mostly expressed in smooth muscle cells.41 Macrophages, which are important mediators of atherosclerotic plaque formation secrete ligands, also secrete ligands which activate this receptor.42 Based on the known biology of this receptor, the inverse association between PDGFR-β levels in plasma and atherosclerosis risk that we observed suggests that the regulation of receptor levels in plasma may differ from regulation at the site of an atherosclerotic lesion. Consistent with this, PDGFRB mRNA levels are low in whole blood samples, but high in the aorta and coronary arteries.43 It may also be possible that our observed genetic association with acute stroke in the EHR data set may be related to non-atherosclerotic mechanisms. PDGFRB expression levels are decreased by micro-RNAs (miR-223, miR-339 and miR-21), and this decrease is associated with atherothrombosis.44 Thus, low plasma levels of PDGFR-β may be promoting arterial occlusion through a direct thrombotic mechanism.
Increased predicted plasma concentration of CLC1B (C-type lectin domain family 1 member B) was associated with peripheral vascular disease and epidemiologically-associated with carotid plaque formation. CLC1B is a cell surface receptor initially identified on platelets, where it was found to promote aggregation in response to the snake venom rhodocytin.45 As a platelet receptor, it was also shown to mediate blood and lymphatic vessel separation during development, modulate lymphatic endothelial cell signaling, and promote lymph node development and maintenance.46 Circulating neutrophils also express CLC1B, which can be activated to promote phagocytosis and production of proinflammatory cytokines.47 The receptor is also expressed on other myeloid cells and can modulate inflammatory responses.48 While the receptor can be cleaved to generate a soluble product, regulators of this process have not been identified. A specific role related to atherosclerosis has not been defined.
Another protein, MICA, was genetically associated with a diagnosis of carotid stenosis, but its protein levels were not associated with carotid plaque in the validation set. The MICA genetic predictor was also associated with a diagnosis of psoriasis, an inflammatory condition associated with increased rates of vascular disease.49 Hence, it is possible that the EHR data set may capture atherosclerotic disease mechanisms not represented in the validation cohort and could explain the failure to replicate that association.
There are several limitations to this study. Although the FHS Offspring cohort represents one of the larger studies of SOMAscan measured plasma proteins to date, larger sample sizes will facilitate the development of stronger genetic predictors, conferring more power to identify disease associations. The SOMAscan platform also contains a discrete sampling of proteins, so many candidate protein biomarkers are not interrogated by this study. Plasma proteins with weak genetic effects or whose levels fluctuate abruptly in response to acute events (such as troponin levels in myocardial infarction) are not well-suited to the discovery approaches used here. Furthermore, there is low power to detect associations with many phenotypes for predictors that only account for small amounts of the phenotypic variance. Inaccurate predictors will contribute to the rate of false positives. The PheWAS phenotypes are based EHR billing codes, rather than a systematic ascertainment of a diagnosis, which can lead to misclassification causing both false positive and false negative associations.50 These considerations highlight the importance of prospective epidemiological and molecular studies using well-curated phenotypes to validate candidate associations.
Epitope effects (i.e. missense SNPs that alter the epitope for the DNA aptamer, causing reduced detection by the Somalogic assay) may create associations between genotypes and proteins that do not represent variation in the level of the protein. If the genetic variant creates an assay-related alteration rather than a functional change, it should not contribute to a clinical phenotype and should not lead to false positive results with regard to clinical endpoints. On the other hand, if these missense variants are also associated with a clinical phenotype, then the implied biomarker association may be a false positive. Validating candidate associations using an independent data set and testing for direct associations between the measured protein level and the phenotype should mitigate the risk of spurious associations caused by epitope effects. In specific support of the PDGFR-β and CLC1B findings, prior studies analyzing binding with homologous proteins for these SOMAscan aptamers did not observe cross-reactivity27 and the SNP variants underlying the association with the genetic predictors are located at the gene locus. However, de novo measurements of these proteins using antibody-based assays would provide further independent validation. To ensure high quality findings, we only reported associations for predictors that cross-validated in the KORA cohort. Hence, we did not report clinical associations for some proteins with strong SNP associations in the discovery set that did not cross-validate. Extension of these approaches and our findings to other ancestral groups is also needed to define their broader utility. Finally, our validation cohort was moderate in size, and the validation phenotype was not identical to the clinical phenotypes, which can lead to false negative associations. While these limitations may have decreased the number of associations that could be identified and validated, the confirmed findings provide support to this approach. Studies in large well-characterized cohorts are necessary to further refine and validate the use of this approach for discovery.
In summary, we identified and validated 2 biomarkers of atherosclerotic disease, using a virtual proteomic approach applied to large EHR datasets. The biomarkers include one (PDGFR-β) with strong supporting biological evidence and one (CLC1B) which may represent a novel disease mechanism. This discovery-oriented study design is efficient and rapid, and overcomes important limitations inherent to traditional approaches to biomarker discovery. This approach may accelerate and broaden the process of biomarker discovery.
Supplementary Material
Clinical Perspective
What Is New?
High-throughput proteomic approaches can measure large numbers of proteins, some of which may serve as biomarkers of disease, in human biospecimens.
Given the practical barriers to conducting large-scale proteomic studies, we developed a “virtual biomarker” strategy that leverages genetic data and clinical phenotypes extracted from electronic health records.
We identified CLC1B and PDGFR-β as potential circulating biomarkers of atherosclerosis, and validated them in an epidemiologic cohort.
What Are the Clinical Implications?
A virtual biomarker study can efficiently identify potential biomarker-disease associations.
These associations can subsequently be validated in targeted studies.
Genetically-driven biomarker studies have the potential to accelerate biomarker discovery for a broad range of diseases.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the expert technical support of the VANTAGE and VANGARD core facilities.
FUNDING SOURCES
This work was supported by a career development award from the Vanderbilt Faculty Research Scholars Fund (JDM), American Heart Association (16FTF30130005) (JDM), Pharmacogenomics Research Network/NIH (P50-GM115305) and R01LM010685 (JCD). Proteomics analyses were supported by NIH R01HL133870-01A1 and NIH R01HL132320-01 (REG, TJW and RSV) and 5T32HL007208 and John S. LaDue Memorial Fellowship in Cardiology at Harvard Medical School (MDB). BioVU is supported by institutional funding, the 1S10RR025141-01 instrumentation award, and by the Clinical and Translational Science Award grant UL1TR000445 from the National Center for Advancing Translational Sciences/NIH and analytic support is provided through P30CA068485 and P30EY08126. VANTAGE and VANGARD core facilities are supported, in part, by the Vanderbilt-Ingram Cancer Center and Vanderbilt Vision Center. The eMERGE Network was initiated and funded by National Human Genome Research Institute/NIH through the following grants: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG008657 (Kaiser Permanente/University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center) and U01HG8685 (Brigham and Women’s Hospital).
Footnotes
DISCLOSURES
None.
REFERENCES
- 1.Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation. 2006;113:2335–2362. [DOI] [PubMed] [Google Scholar]
- 2.Wang TJ. Assessing the role of circulating, genetic, and imaging biomarkers in cardiovascular risk prediction. Circulation. 2011;123:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature. 2008;451:949–952. [DOI] [PubMed] [Google Scholar]
- 4.Maher BS. Polygenic Scores in Epidemiology: Risk Prediction, Etiology, and Clinical Utility. Curr Epidemiol Rep. 2015;2:239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, Gold L, Pezer M, Lauc G, El-Din Selim MA, Mook-Kanamori DO, Al-Dous EK, Mohamoud YA, Malek J, Strauch K, Grallert H, Peters A, Kastenmüller G, Gieger C, Graumann J. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. [DOI] [PubMed] [Google Scholar]
- 7.Ngo D, Sinha S, Shen D, Kuhn EW, Keyes MJ, Shi X, Benson MD, O’Sullivan JF, Keshishian H, Farrell LA, Fifer MA, Vasan RS, Sabatine MS, Larson MG, Carr SA, Wang TJ, Gerszten RE. Aptamer-Based Proteomic Profiling Reveals Novel Candidate Biomarkers and Pathways in Cardiovascular Disease. Circulation. 2016;134:270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, Sanderson SC, Kannry J, Zinberg R, Basford MA, Brilliant M, Carey DJ, Chisholm RL, Chute CG, Connolly JJ, Crosslin D, Denny JC, Gallego CJ, Haines JL, Hakonarson H, Harley J, Jarvik GP, Kohane I, Kullo IJ, Larson EB, McCarty C, Ritchie MD, Roden DM, Smith ME, Böttinger EP, Williams MS. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berglund G, Elmstähl S, Janzon L, Larsson SA. The Malmo Diet and Cancer Study. Design and feasibility. J Intern Med. 1993;233:45–51. [DOI] [PubMed] [Google Scholar]
- 12.Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes MG, Jarvik GP, Jiang L, Kullo IJ, Li R, Ling H, Manolio TA, Matsumoto ME, McCarty CA, McDavid AN, Mirel DB, Olson LM, Paschall JE, Pugh EW, Rasmussen LV, Rasmussen-Torvik LJ, Turner SD, Wilke RA, Ritchie MD. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. [DOI] [PubMed] [Google Scholar]
- 15.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei W-Q, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER, Cox NJ, Roden DM, Denny JC. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS ONE. 2017;12:e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosley JD, Witte JS, Larkin EK, Bastarache L, Shaffer CM, Karnes JH, Stein CM, Phillips E, Hebbring SJ, Brilliant MH, Mayer J, Ye Z, Roden DM, Denny JC. Identifying genetically driven clinical phenotypes using linear mixed models. Nat Commun. 2016;7:11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5:e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borgquist S, Butt T, Almgren P, Shiffman D, Stocks T, Orho-Melander M, Manjer J, Melander O. Apolipoproteins, lipids and risk of cancer. Int J Cancer. 2016;138:2648–2656. [DOI] [PubMed] [Google Scholar]
- 23.Benson MD, Yang Q, Ngo D, Zhu Y, Shen D, Farrell LA, Sinha S, Keyes MJ, Vasan RS, Larson MG, Smith JG, Wang TJ, Gerszten RE. Genetic Architecture of the Cardiovascular Risk Proteome. Circulation. 2018;137:1158–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International Schizophrenia Consortium, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majumdar A, Haldar T, Witte JS. Determining Which Phenotypes Underlie a Pleiotropic Signal. Genet Epidemiol. 2016;40:366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, GTEx Consortium, Nicolae DL, Cox NJ, Im HK. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, Jansen R, Geus EJC de, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimäki T, Raitoharju E, Kähönen M, Seppälä I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B. Integrative approaches for large-scale transcriptome-wide association studies. Nature Genetics. 2016;48:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meijers JC, Tekelenburg WL, Bouma BN, Bertina RM, Rosendaal FR. High levels of coagulation factor XI as a risk factor for venous thrombosis. N Engl J Med. 2000;342:696–701. [DOI] [PubMed] [Google Scholar]
- 31.Zlokovic BV. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol. 2013;70:440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 33.Gianola D, de los Campos G, Toro MA, Naya H, Schön C-C, Sorensen D. Do Molecular Markers Inform About Pleiotropy? Genetics. 2015;201:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, Hunter TA, DeJesus-Hernandez M, Bisceglio GD, Mackenzie IR, Singleton A, Cookson MR, Crook JE, Dillman A, Hernandez D, Petersen RC, Graff-Radford NR, Younkin SG, Rademakers R. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet. 2010;87:890–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tönjes A, Scholz M, Krüger J, Krause K, Schleinitz D, Kirsten H, Gebhardt C, Marzi C, Grallert H, Ladenvall C, Heyne H, Laurila E, Kriebel J, Meisinger C, Rathmann W, Gieger C, Groop L, Prokopenko I, Isomaa B, Beutner F, Kratzsch J, Fischer-Rosinsky A, Pfeiffer A, Krohn K, Spranger J, Thiery J, Blüher M, Stumvoll M, Kovacs P. Genome-wide meta-analysis identifies novel determinants of circulating serum progranulin. Hum Mol Genet. 2018;27:546–558. [DOI] [PubMed] [Google Scholar]
- 36.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang H-Y, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson Å, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen L-P, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen A-K, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Döring A, Elliott P, Epstein SE, Ingi Eyjolfsson G, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen A-L, Hayward C, Hernandez D, Hicks AA, Holm H, Hung Y-J, Illig T, Jones MR, Kaleebu P, Kastelein JJP, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linsel-Nitschke P, Heeren J, Aherrahrou Z, Bruse P, Gieger C, Illig T, Prokisch H, Heim K, Doering A, Peters A, Meitinger T, Wichmann H-E, Hinney A, Reinehr T, Roth C, Ortlepp JR, Soufi M, Sattler AM, Schaefer J, Stark K, Hengstenberg C, Schaefer A, Schreiber S, Kronenberg F, Samani NJ, Schunkert H, Erdmann J. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 2010;208:183–189. [DOI] [PubMed] [Google Scholar]
- 38.Raines EW. PDGF and cardiovascular disease. Cytokine Growth Factor Rev. 2004;15:237–254. [DOI] [PubMed] [Google Scholar]
- 39.Sano H, Sudo T, Yokode M, Murayama T, Kataoka H, Takakura N, Nishikawa S, Nishikawa SI, Kita T. Functional blockade of platelet-derived growth factor receptor-beta but not of receptor-alpha prevents vascular smooth muscle cell accumulation in fibrous cap lesions in apolipoprotein E-deficient mice. Circulation. 2001;103:2955–2960. [DOI] [PubMed] [Google Scholar]
- 40.He C, Medley SC, Hu T, Hinsdale ME, Lupu F, Virmani R, Olson LE. PDGFRβ signalling regulates local inflammation and synergizes with hypercholesterolaemia to promote atherosclerosis. Nat Commun. 2015;6:7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karvinen H, Rutanen J, Leppänen O, Lach R, Levonen A-L, Eriksson U, Ylä-Herttuala S. PDGF-C and -D and their receptors PDGFR-alpha and PDGFR-beta in atherosclerotic human arteries. Eur J Clin Invest. 2009;39:320–327. [DOI] [PubMed] [Google Scholar]
- 42.Demoulin J-B, Montano-Almendras CP. Platelet-derived growth factors and their receptors in normal and malignant hematopoiesis. Am J Blood Res. 2012;2:44–56. [PMC free article] [PubMed] [Google Scholar]
- 43.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan M, Yan H-B, Li J-N, Li W-K, Fu Y-Y, Chen W, Zhou Z. Thrombin Stimulated Platelet-Derived Exosomes Inhibit Platelet-Derived Growth Factor Receptor-Beta Expression in Vascular Smooth Muscle Cells. Cell Physiol Biochem. 2016;38:2348–2365. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki-Inoue K, Inoue O, Ozaki Y. The novel platelet activation receptor CLEC-2. Platelets. 2011;22:380–384. [DOI] [PubMed] [Google Scholar]
- 46.Osada M, Inoue O, Ding G, Shirai T, Ichise H, Hirayama K, Takano K, Yatomi Y, Hirashima M, Fujii H, Suzuki-Inoue K, Ozaki Y. Platelet activation receptor CLEC-2 regulates blood/lymphatic vessel separation by inhibiting proliferation, migration, and tube formation of lymphatic endothelial cells. J Biol Chem. 2012;287:22241–22252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerrigan AM, Dennehy KM, Mourão-Sá D, Faro-Trindade I, Willment JA, Taylor PR, Eble JA, Reis e Sousa C, Brown GD. CLEC-2 is a phagocytic activation receptor expressed on murine peripheral blood neutrophils. J Immunol. 2009;182:4150–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mourão-Sá D, Robinson MJ, Zelenay S, Sancho D, Chakravarty P, Larsen R, Plantinga M, Van Rooijen N, Soares MP, Lambrecht B, Reis e Sousa C. CLEC-2 signaling via Syk in myeloid cells can regulate inflammatory responses. Eur J Immunol. 2011;41:3040–3053. [DOI] [PubMed] [Google Scholar]
- 49.Katsiki N, Anagnostis P, Athyros VG, Karagiannis A, Mikhailidis DP. Psoriasis and Vascular Risk : An Update. Curr Pharm Des. 2014;20:6114–6125. [DOI] [PubMed] [Google Scholar]
- 50.Ahmad FS, Chan C, Rosenman MB, Post WS, Fort DG, Greenland P, Liu KJ, Kho AN, Allen NB. Validity of Cardiovascular Data From Electronic Sources: The Multi-Ethnic Study of Atherosclerosis and HealthLNK. Circulation. 2017;136:1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.