Significance
Influenza hemagglutinin (HA) is a prototypical class I fusion protein and a major component of current flu vaccines. HA is a metastable glycoprotein and undergoes conformational changes to the so-called postfusion state. Stabilization of the prefusion conformations of fusion proteins has proven to be a key success factor for the induction of efficacious immune response and stabilization has become a grand challenge in structural vaccinology. The study shows that three stabilizing mutations in two important pH-sensitive switch regions involved in the early refolding process impede refolding of the prefusion HA. Based on the substitutions of these highly conserved and buried HA residues a generally applicable stabilization strategy for all subtypes of group 1 and 2 influenza A HA was developed.
Keywords: influenza, vaccine, protein design, fusion, protein stability
Abstract
For an efficacious vaccine immunogen, influenza hemagglutinin (HA) needs to maintain a stable quaternary structure, which is contrary to the inherently dynamic and metastable nature of class I fusion proteins. In this study, we stabilized HA with three substitutions within its pH-sensitive regions where the refolding starts. An X-ray structure reveals how these substitutions stabilize the intersubunit β-sheet in the base and form an interprotomeric aliphatic layer across the stem while the native prefusion HA fold is retained. The identification of the stabilizing substitutions increases our understanding of how the pH sensitivity is structurally accomplished in HA and possibly other pH-sensitive class I fusion proteins. Our stabilization approach in combination with the occasional back mutation of rare amino acids to consensus results in well-expressing stable trimeric HAs. This repair and stabilization approach, which proves broadly applicable to all tested influenza A HAs of group 1 and 2, will improve the developability of influenza vaccines based on different types of platforms and formats and can potentially improve efficacy.
The majority of influenza vaccines are based on whole, inactivated virus, but various alternatives have been developed such as adjuvanted hemagglutinin (HA) protein, membrane-extracted HAs forming multimeric rosette-like particles (1), or nanoparticles with spatially controlled HA display (2). The production of class I fusion proteins by recombinant protein expression is challenging because of their intrinsic instability, low expression levels, and failure to form correctly folded trimers. Uncleaved HA ectodomain (HA0) is monomeric and does not induce significant amounts of neutralizing antibodies. Although several antibodies that recognize the trimer interface have been described, suggesting some degree of reversible “breathing” of the HA trimer apex (3–6), the cleaved HA at the viral surface is predominantly in a closed trimeric conformation (7–9). For all current vaccines and emerging recombinant vaccine vectors or nucleic acid vaccines (10), the delivery of a conformationally correct prefusion HA trimer with improved expression, quality, and stability is crucial (11).
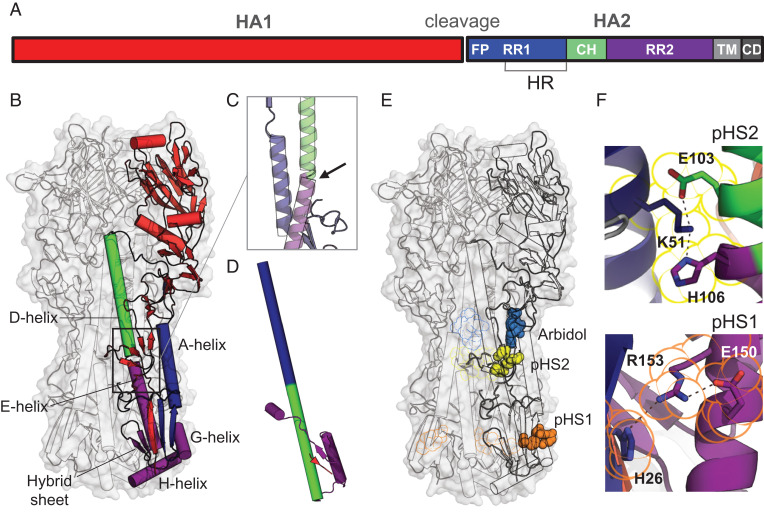
Like other class I fusion proteins, influenza A HA transforms from a high-energy, metastable prefusion state to a postfusion conformation, a transition, which in case of HA, is triggered by low pH. To become functional, HA0 needs to be proteolytically cleaved (12, 13) into the head domain (HA1) and the membrane-anchored fusion domain (HA2). HA2 consists of the N-terminal refolding region 1 (RR1, residues HA2 1 to 75) containing a heptad repeat motif, the central helix (residues HA2 76 to 105), and the C-terminal refolding region 2 (RR2, residues HA2 106 to 184) (Fig. 1A; residue numbering in SI Appendix, Fig. S1A). Early in the refolding process, the N-terminal part of HA2 containing the fusion peptide and two β-strands in the membrane-proximal region show disorder (14–17) (Fig. 1B). Another unstable region is located at the bend between helices D and E (Fig. 1C), just below the binding pocket of the stabilizing compound Arbidol (18). Early in the refolding process, after low pH triggered the release of the fusion peptide, this long helical structure is straightened, starting from H106 into the space freed by the fusion peptide (14, 15). H106 marks the beginning of RR2 and the location where the long helix subsequently breaks in the postfusion structure (Fig. 1D).
Fig. 1.
Influenza HA protein structure. (A) Schematic representation of HA with indicated fusion peptide (FP), refolding regions 1 and 2 (RR1 and RR2), heptad repeat (HR) motif, central helix (CH), transmembrane (TM) domain, and cytoplasmic domain (CD). (B) Structure of prefusion trimeric WT H3-HK68 [PDB identifier 4FNK (22)] in surface representation (gray). The internal protein structure is outlined in the cartoon with one monomer colored according to A. (C) Bend of the helical structure between helices D and E. (D) Conformation of the postfusion monomer [PDB identifier 1QU1 (50)]. (E) Location of histidine switches 1 and 2 (pHS1 and pHS2). The histidines and charged residues forming the switches are shown in space, filling representation in orange for pH switch 1 (pHS1) and yellow for pH switch 2 (pHS2). Fusion inhibitor Arbidol is plotted in blue based on PDB identifier 5T6N (18). pHS1, pHS2, and Arbidol binding to the other protomers were plotted as outline. (F) Details of the structure of the histidine switches pHS1 and pHS2 in the WT HA. H106-K51-E103 linked by hydrogen bonds in pHS2 (Top) and H26-R153-E150 triad in pHS1 (Bottom). Residues belonging to pHS1 and pHS2 are outlined in the same color as in E.
Here, we describe the low stability and quality of wild-type (WT) influenza A HAs and their stabilization by a minimal substitution of three residues within the pH-sensitive switches involved in the refolding of the proteins. We show that the approach becomes applicable to a broader panel of HA subtypes when supplemented with a strain-specific mutation of rare amino acids back to subtype consensus, a process we call “repair,” previously applied to improving expression of HIV Env fusion proteins (19, 20).
Results
Stabilization of HA by Substitutions in Two pH Switch Regions.
To stabilize the prefusion conformation and to prevent refolding, substitutions were introduced in the two regions of instability indicated in Fig. 1E. Both regions where the refolding process begins (14, 15) contain HA2 histidine residues: H26 and H106. Histidines can form pH-sensitive switches, especially when in direct proximity to other histidines or positively charged residues, due to their protonation below pH 6. In both unstable regions, histidines form hydrogen bonds with arginine or lysine, additionally stabilized by a negative charge, and thus likely constitute such pH switches (Fig. 1F). In the first pH switch (pHS1), the half-buried H26 forms a hydrogen bond with R153 or K153, which in turn forms an ionic interaction with the neighboring E150 in group 1 HA (SI Appendix, Fig. S1B). The protonation of H26 results in an excess of positive charge and electrostatic repulsion with R153. The H26W substitution in pHS1 was introduced to stabilize the region and remove the dependence of stability on pH. H106 forms the second switch (pHS2) in group 2 HAs and hydrogen bonds to the buried K51, further stabilized by an ionic bond with E103 (Fig. 1F). Here, we mutated the conserved lysine and glutamic acid to aliphatic isoleucines to prevent the potential charge repulsion with the protonated H106 and stabilize the interaction of the central helix with the A helix in RR1. In group 1, histidine H106 is replaced by arginine (SI Appendix, Fig. S1B), but one turn down the central helix, another pH-sensitive aromatic cluster is formed, previously postulated to be a component of the triggering mechanism (21). Therefore, the conserved K51 and E103 can be destabilized indirectly or directly by the pHS2 of both group 1 and 2.
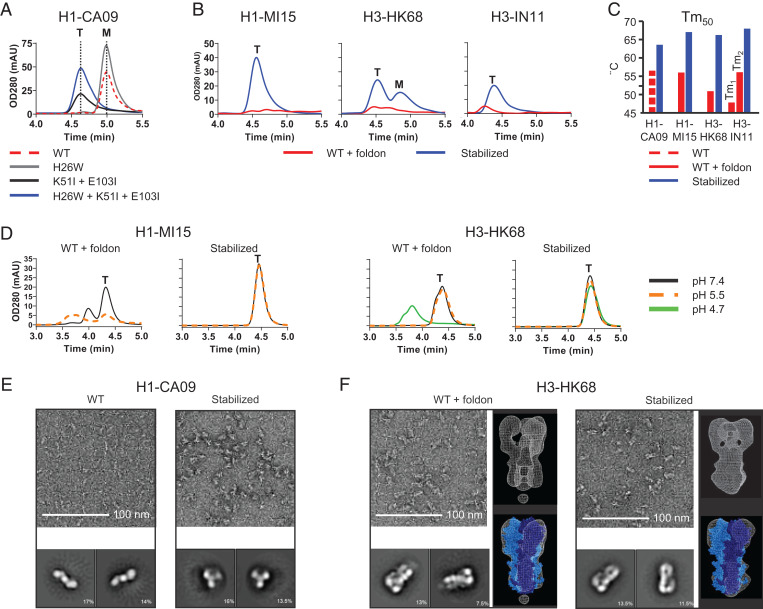
To study the impact of the substitutions H26W and K51I + E103I and their combination on the stability of soluble HA, the substitutions were introduced in the ectodomain of HA derived from group 1 strain H1N1 A/California/07/2009 (abbreviated as H1-CA09). Supernatants of cells transfected with plasmids encoding the HA variants were tested for trimer content (Fig. 2A), and molecular weights of purified HAs were confirmed by size-exclusion chromatography (SEC) multi-angle light scattering (MALS) (SI Appendix, Fig. S2A). The H26W substitution increased expression levels of monomeric HA, and the K51I + E103I substitutions caused a shift in the retention time that corresponds with a trimeric HA species, and the combination of substitutions resulted in an increased trimer expression. Since trimerization can also be accomplished by the fusion of a foldon domain to the HA C terminus, expression was tested by SEC of several HA variants of influenza A group 1 and 2 stabilized by either foldon or with the three substitutions (Fig. 2B). Variants with the three stabilizing substitutions showed a strong trimer peak around 4.5 min, and in contrast, the foldon-stabilized HAs were hardly detectable using SEC. Antibody binding with the purified proteins showed that the stabilizing substitutions did not affect antigenicity (SI Appendix, Table S1). Purified foldon HA trimers of H1N1 A/Michigan/45/2015 (H1-MI15) and H3N2 A/Hong Kong/1/1968 (H3-HK68) aggregated after incubation at 4 °C for 2 wk at low pH, but the stabilized HA trimers remained stable and showed an up to 15 °C increase in the melting temperature (Tm50) as determined by differential scanning fluorimetry (DSF) (Fig. 2 C and D and SI Appendix, Fig. S2C). Two-dimensional (2D) class averages obtained by an electron microscopy (EM) analysis of the negative-stained samples, using an acidic stain, of the purified HA revealed monomeric rods for the nonstabilized variant and regular homogenous trimers for the stabilized variant (Fig. 2E). The 2D class averages of the foldon-fused HA shows signs of splaying in the membrane-proximal region of the protein compared to the stabilized HA (Fig. 2F). Three-dimensional (3D) reconstructions using representative classes reveal a more closed conformation of the stabilized HA, similar to the X-ray structure of WT HA [Protein Data Bank (PDB) identifier 4FNK (22)] and reminiscent of the shape of virion-associated HA as shown by EM studies (7–9) (Fig. 2F).
Fig. 2.
Characterization of HA-containing stabilizing mutations. (A) Analytical SEC profiles of cell culture supernatant of Expi293F cells expressing H1-CA09 HA with combinations of stabilizing mutations; WT, H26W substitution, K51I + E103I substitution, and fully stabilized H26W, K51I + E103I. Data are representing mean of three independent transfections in one experiment. Peaks representing monomeric and trimeric species are indicated by an “M” and “T,” respectively. (B) Analytical SEC culture supernatant profiles of Expi293F cells expressing WT HA including foldon trimerization domain and fully stabilized HA (H26W, K15I, and E103I) for H1-MI15, H3-HK68, and H3-IN11. (C) Temperature stability of purified HA determined by DSF. Shown are Tm50 values for WT H1-CA09 and stabilized H1-CA09 HA and for WT foldon-fused and stabilized HA of H1-MI15, H3-HK68, and H3-IN11. (D) Two-week pH stability at 4 °C of purified HA analyzed by analytical SEC. Shown are profiles of WT and stabilized HA of H1-MI15 (pH 5.5 and 7.4) and H3-HK68 (pH 4.7, 5.5, and 7.4). (E) Negative-stained EM of WT H1-CA09 (Left) and stabilized HA (Right) with representative 2D-averaged classes. (F) Negative-stained EM of WT H3-HK68 HA foldon-fused (Left) and stabilized HA (Right) with representative 2D-averaged classes (each box is 27.4 nm) and 3D density map (white mesh) superimposed on the X-ray structure of H3-HK68 HA trimer (PDB identifier 4FNK) represented in blue spheres with one monomer plotted in dark blue.
Crystal Structure of Stabilized H3-HK68.
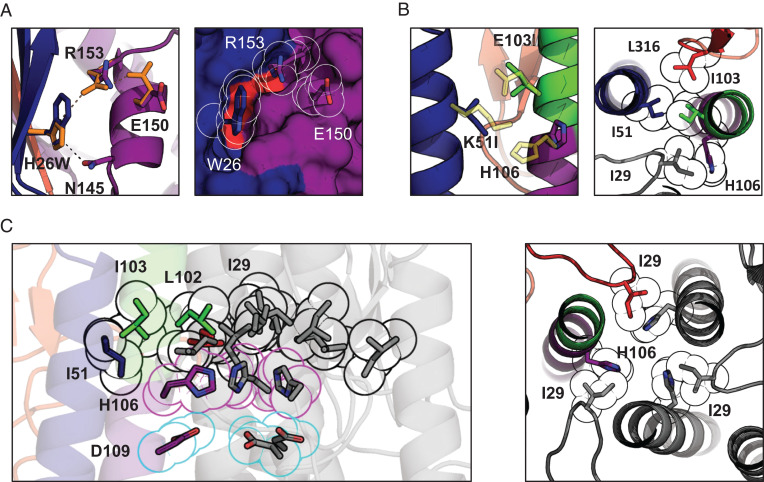
To understand how the substitutions stabilize the prefusion HAs and to determine their impact on its conformation, the X-ray crystal structure of the stabilized apo ectodomain of group 2 H3-HK68 was determined at 2.2-Å resolution (crystallographic statistics are provided in SI Appendix, Tables S2 and S3). The stabilized HA is within the RMSD of 0.54 Å to the foldon-trimerized H3-HK68 [PDB identifier 4FNK (22)] (SI Appendix, Fig. S3). H26 is located in a five-strand intersubunit β-sheet (here called hybrid sheet) composed of three sequence-distant regions: N terminus of HA1, RR1, and RR2. The H26W mutation preserves the WT hydrogen bond between the imidazole group and N145 in helix G and slightly displaces the guanidine group of R153 to form a π-cation interaction. The bulkier tryptophan fills the small neighboring hydrophobic cavity and binds with HA1 L13 of the hybrid sheet (red patch in Fig. 3A), effectively strengthening the interactions of three sequence-distant regions of HA: N terminus of HA1, G helix, and RR1. The substitutions in pHS2 (K51I + E103I) become part of a larger aliphatic cluster composed of neighboring HA1 L316 and I29 (Fig. 3B). This cluster merges with its equivalents from the neighboring protomers and forms an intersubunit aliphatic layer spanning the entire horizontal cross-section of the HA stem (Fig. 3C). No longer stabilized by their respective pHS2 H-bond networks, the three histidines flip toward the trimer’s threefold axis below the aliphatic layer and hydrogen bond with D109. Additionally, I29 from the 30-Loop (HA1 residues 22 to 37) (14) locks the histidine in the inwards-facing conformation by shifting toward the threefold axis by almost 2 Å.
Fig. 3.
Crystal structure and suggested mechanism of action of the HA-stabilizing mutations. (A) Changes in the sidechain orientation induced by the H26W mutation in pHS1 (Left). The structure of the WT pHS1 is plotted in orange. The structure of the mutant is colored according to the color scheme introduced in Fig. 1A. The mutated pHS1 is also plotted in space-filling representation (Right). W26 sidechain tightly packs between the β-sheet (blue) and R153. (B) Changes in the sidechain orientation induced by the K51I mutation in pHS2 (Left). The structure of the WT pHS2 is plotted in yellow. The structure of the mutant is colored according to the color scheme introduced in Fig. 1A. pHS2 together with two neighboring residues I103 and I29 is also shown in a top cross-section view (Right). All four aliphatic residues form a tight hydrophobic cluster with H106 turning away. (C) The hydrophobic cluster from B is shown for the whole trimer (black outline), with the other two protomers colored in gray. The cluster encompasses the three H106 residues facing each other (magenta outline) and stabilized by negatively charged D109 residues (cyan outline). The histidines form most tight interactions with I29 (Right), shown in cross-section perpendicular to the threefold axis from the top.
Stabilization of Other Group 1 and 2 HAs.
The stabilizing strategy was applied to representative HAs from group 1 and 2 influenza A strains covering a wide range in year of isolation and subtype, including potentially pandemic strains. SEC analysis of cell culture supernatants of transiently transfected cells indicated that WT HA proteins express mostly monomeric and at lower level. In contrast, predominantly trimeric protein was observed for stabilized HAs of all tested subtypes (Fig. 4A). We examined the sequences of poorly expressing HAs for the presence of rare amino acids and mutated them back to subtype consensus, a process we call “repair” (19, 20). We repaired all rare amino acids (SI Appendix, Table S4) with frequencies below 1% within the H3 subtype, which could be either sequencing errors or naturally occurring mutations. We also considered mutations with frequencies between 1 and 3%, but these were only tested if buried and considered problematic after visual inspection. We applied the repair strategy to HAs that showed no or very low expression levels. The introduction of the three stabilizing mutations results in expression of HA trimers for H3-WI05 and H3-NE93, and a 2- to 2.5-fold titer increase was observed after additional application of the repair mutations specific to the two strains V186G + D122N and T138A + K135T, respectively. The other two HAs, H3-SI16 and H3-BR07, did not express, and even introduction of the stabilizing substitutions did not rescue expression. Only introduction of the repair substitution P194L in HA1, common for both strains, completely restored expression. Interestingly, L194P has been described as an egg-adaptive substitution that substantially increases the mobility of a region in the head domain (23). For all low-expressing HA variants shown in Fig. 4B, a “repair and stabilize” approach is needed to obtain well-expressing, high-quality trimers.
Fig. 4.
Stabilizing mutations applied to group 1 and 2 representative HAs. (A) Phylogeny of influenza A and B adapted from ref. 51; maximum likelihood tree representing amino acid sequences of HA. Analytical SEC profiles of culture supernatant of Expi293F cells expressing selected group 1 and 2 WT and fully stabilized HA (H26W, K15I, and E103I) subtype HAs. Peaks representing monomeric and trimeric species are indicated by an “M” and “T,” respectively. Due to differences in experimental setup, retention times are different from B, C, and D. (B) Analytical SEC profiles of culture supernatant of Expi293F cells expressing WT, stabilized and repaired of selected H3 HAs; H3N2 A/Netherlands/179/1993 (H3-NE93), H3N2 A/Singapore/INFIMH/16/0019/2016 (H3-SI16), H3N2 A/Wisconsin/67/2005 (H3-WI05), and H3N2 A/Brisbane/10/2007 (H3-BR07). (C) Analytical SEC profiles of Expi293F cell culture supernatants of H1N1 A/South Carolina/1/1918 I53_dn5B (H1-SC18), H1-CA09-I53_dn5B, H3-HK68-I53_dn5B, and H3-WI05-I53_dn5B. For the H1 HAs, the molecular weight determined by MALS is shown. (D) Analytical SEC profiles of purified WT and stabilized H1-CA09 HA-I53_dn5B kept at pH 5.5 and 7.4 for 2 wk (Top Left). Temperature stability as determined by DSF for WT and stabilized (Top Right). Negative-stained EM micrograph of WT and stabilized (Bottom) HA-I53_dn5B fusions. Representative 2D-averaged classes (box is 35 nm) could be generated for the stabilized variant.
An interesting development for influenza vaccines is the application of antigen fusion to self-assembling proteins that form nanoparticles. HA fusion to I53_dn5B, the trimeric component of the two-component nanoparticle combined with the pentameric component (I53_dn5A), can self-assemble in icosahedral particles displaying 20 trimeric HA spikes in an ordered array. These nanoparticle immunogens increase immunogenicity and improve breadth of antibody responses (2). We investigated whether the three stabilizing substitutions would improve the quality of the trimeric HA-I53_dn5B fusion protein component. For most of the HA fusions, stabilization increased protein expression (Fig. 4C). The SEC profile of cell culture supernatants showed a peak at lower retention times for nonstabilized H1 fusions. Since the molecular weight as determine by MALS was similar for both HAs, this is indicative for a more open trimer structure of the nonstabilized fusions compared to the stabilized variants (SI Appendix, Fig. S4A). To study the nature of the shifted retention time in SEC, the stabilized and nonstabilized H1-CA09-I53_dn5B were purified (SI Appendix, Fig. S4B), and the hydrodynamic radius was measured using in-line dynamic light scattering (DLS). The nonstabilized H1-I53_dn5B showed a radius of 7.7 nm, and the stabilized H1-I53_dn5B showed a radius of 6.8 nm, which agrees with the difference in retention time in SEC (SI Appendix, Table S5). The purified nonstabilized trimers were unstable after incubation at 4 °C for 2 wk at low pH as measured by SEC analysis (Fig. 4 D, Top Left), but the stabilized protein remained stable and showed an up to 14 °C increase in the melting temperature (Tm50) as determined by DSF (Fig. 4 D, Top Right and SI Appendix, Fig. S4C). 2D class averages obtained by negative stain transmission electron microscopy revealed regular homogenous trimers for the stabilized HA-I53_dn5B, but the nonstabilized variants were mostly aggregated (Fig. 4 D, Bottom).
Stabilization of the pH Switch Region Renders HA Nonfusogenic.
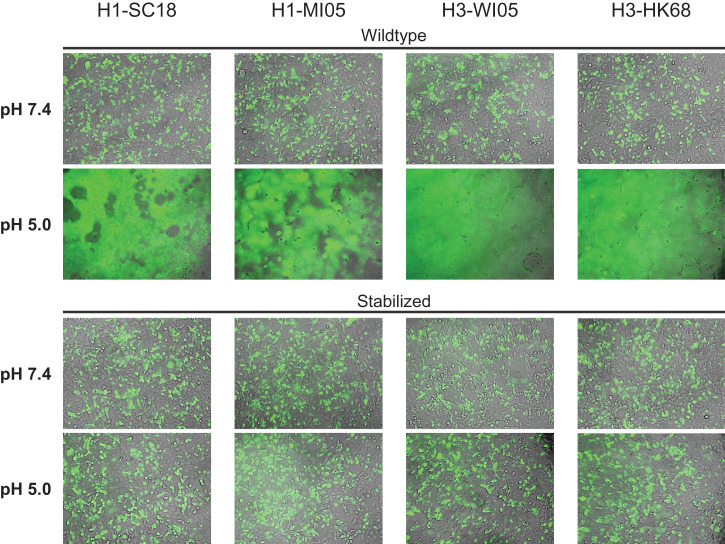
To determine whether the stabilizing mutations prevent HA from triggering membrane fusion, we performed cell–cell fusion experiments using full-length WT and stabilized HAs. Plasmids encoding human TMPRSS2 and green fluorescent protein (GFP) were cotransfected to allow proteolytic processing of HA and visualization of syncytia, respectively. At pH 7.4, none of the WT nor stabilized HAs showed syncytia formation (Fig. 5). Next, to simulate the fusion-triggering, low-pH environment that the infectious virus encounters during endosomal uptake, we exposed the cells to low-pH medium. The low pH pulse triggered syncytia formation in all four tested WT HAs, whereas for stabilized HAs, no syncytium formation was observed.
Fig. 5.
Stabilization of pH switch regions prevents low-pH–triggered cell–cell fusion. HA fusogenicity as measured in a cell–cell fusion assay in HEK293 cells by cotransfection of plasmids encoding WT or stabilized HA (H1-SC18, H1-MI05, H3-WI05, and H3-HK68), human TMPRSS2, and GFP. Overlays of GFP and brightfield channels 24 h after transfection are shown before (pH 7.4) and 1 h after (pH 5.0) exposure to low-pH medium to trigger fusion.
Discussion
Recent advances in vaccinology which are accelerated by the SARS-CoV-2 pandemic like the success of RNA-based vaccines or the emergence of particle-based platforms may change future vaccine approaches (24, 25). For most of the established vaccine platforms and novel approaches, expression of high-quality antigens in their most-relevant conformation is a key success factor. The production of influenza A–based vaccines can be challenging because of HA’s unstable quaternary structure and low expression levels (Figs. 2 and 4) (11, 26). Stress conditions like heat or long-term storage can reduce the potency of protein-based vaccines, and stability improvement can prolong vaccine shelf life and alleviate cold-chain issues often encountered in remote or poorer areas of the world.
HA stability and the pH values that trigger conformational transformation vary among strains, and few mutations have been identified with a broad cross-subtype–stabilizing effect (27–30). pH-sensitive regions that initiate the HA’s large conformational changes have been previously described (14, 15, 21, 30–33). In this study, we targeted the regions involved in early structural rearrangements to stabilize the HA trimers. We show that the substitution of residues in two pH-sensitive regions, H26W in pHS1 and K51I + E103I in pHS2, stabilize trimeric HA for all tested group 1 and 2 subtypes of influenza A. We demonstrate a broad applicability of the stabilization approach when combined with the occasional repair of rare mutations to subtype consensus and hence resolved a major hurdle for influenza vaccine development.
pHS1 is located in a conserved structural element at the base of the HA stem, which comprises an intersubunit β-sheet, the hybrid sheet, composed of β-strands of the head domain and the highly dynamic RR1 and RR2 in the fusion domain. Previously, we described that the hybrid sheet is a common element in many class I fusion proteins, and its stabilization proved a successful design strategy for HIV and SARS-CoV-2 antigens (19, 34). Substitutions in the conserved salt bridge in the pHS2 switch (Fig. 1F) were previously shown to stabilize a ferritin particle displaying group 1 HA stem (35). Furthermore, pHS2 is in direct vicinity of the binding site of Arbidol, a group 2 hemagglutinin fusion inhibitor that binds and stabilizes the prefusion conformation. The close proximity of pHS2 to the Arbidol-binding site suggests that the area exhibits an intrinsic instability and therefore is interesting for increasing the protein’s refolding energy barrier (18). Also, for RSV F protein, the binding site of a fusion inhibitor and the position of stabilizing substitutions are overlapping, indicating that inhibitor screening and vaccine design can be mutually guiding (36, 37). It appears that the strategy to stabilize the refolding regions via substitutions or ligand binding is successful in these genetically distant yet structurally and functionally related proteins. Histidine clusters or histidines interacting with salt bridges seem to be good indicators of such unstable regions as in the example of pHS1, which is highly conserved in both group 1 and group 2 HAs. pHS2 is conserved in group 2 and replaced in group 1 with an aromatic cluster in the direct vicinity of the K51-E103 salt bridge, one turn down the central helix. The likely pH-sensitive aromatic cluster is formed with H111, H18, H38, and W21 and was postulated to be an alternative component of the triggering mechanism (21). Even though the pHS2 is not completely conserved between group 1 and group 2, the pH sensitivity of the region is conserved, and mutation of the salt bridge to isoleucines has a stabilizing effect for HA from both groups.
In this study, we describe a stabilization approach that is generally applicable to all group 1 and 2 HA trimers to obtain high levels of stable, high-quality trimers by three stabilizing substitutions and, if needed, additional strain-specific repair of rare amino acid. The identification and arresting of the two pH-sensitive switches located in regions involved in early steps of the HA-refolding process increases our understanding of how the pH triggering is structurally accomplished in pH-sensitive class I fusion proteins. This design strategy provides a means to obtaining high-quality reagents for diagnostics, isolation of B-cells and mAbs, and robust reagents for influenza structural research. Most importantly, stabilizing HAs may improve developability and efficacy of influenza vaccines based on a variety of platforms.
Materials and Methods
Expression of HA Proteins.
DNA fragments encoding histidine-tagged HA proteins were synthesized (Genscript) and cloned in the pcDNA2004 expression vector, a modified pcDNA3 plasmid with an enhanced cytomegalovirus promotor. Culture supernatants for analytical SEC analysis were generated by a transient transfection of Expi293F cells at a 200-µL scale in 96-half deep-well plates at a cell density of 2.5E + 06 vc/mL using the ExpiFectamine 293 Transfection Kit (Gibco, Thermo Fisher Scientific). At day 3 posttransfection, culture supernatants were harvested and clarified by centrifugation (10 min at 400 × g) followed by filtration (96-well filter plates, 0.22-µm polyvinylidene difluoride membrane, Corning). Protein batches generated for purification were produced in ExpiCHO or Expi293F suspension cells (350 mL scale). ExpiCHO cells cultured in ExpiCHO expression medium were transiently transfected using industrial-grade DNA using ExpiFectamine Chinese hamster ovary (CHO) transfection reagent (Gibco, Thermo Fisher Scientific). At day 1 posttransfection, ExpiFectamine CHO Enhancer and ExpiCHO Feed (Gibco, Thermo Fisher Scientific) were added to the cell cultures, and transfected cells were incubated at 32 °C, 5% CO2. Supernatants were harvested when cell viability dropped below 50% (typically between days 7 to 11). Expi293F cells were cultured in Expi293F Expression medium [+] GlutaMAX (Gibco, Thermo Fisher Scientific) and transiently transfected using ExpiFectamine 293 (Gibco, Thermo Fisher Scientific) according to the manufacturer’s instructions, and 18 h posttransfection, enhancers 1 and 2 were added (Gibco, Thermo Fisher Scientific). Depending on the cell viability, the culture supernatants were harvested between days 5 and 7. Culture supernatants were clarified by centrifugation followed by filtration over a 0.2-µm bottle-top filter (Corning).
Protein material generated for structural studies by protein crystallography was generated by transient transfection in Expi293 GnTI- cells. At day 1 posttransfections the feed, comprising enhancers 1 and 2 (Gibco, Thermo Fisher Scientific), was added, and cultures were harvested and clarified at day 7.
Purification of HA Proteins.
From the harvested culture supernatants, HA proteins were purified by a two-step protocol using an ÄKTA Avant 25 system (GE Healthcare Life Sciences). For his-tagged proteins, the material was applied to either a prepacked cOmplete His-tag purification column (Roche) or self-packed HiScale 26/60 column with Ni Sepharose High Performance (GE Healthcare Life Sciences). Following a wash with 1 mM imidazole, the bound proteins were eluted with a step gradient to 300 mM imidazole. For C-tagged protein, the clarified supernatant was loaded on a HiScale 16/20 column (GE Healthcare) packed with an affinity resin that consisted of a C-tag–specific single domain antibody immobilized on Agarose-based beads (Thermo Fisher Scientific). The elution of the C-tagged proteins was performed using a Tris buffer containing 2 M MgCl2. The His-tag– and C-tag–containing elution fractions were pooled and filtered through a Millex-GV 0.22-µM filter membrane (Millipore Sigma). To further polish the purified protein, SEC was performed by running a HiLoad Superdex 200-pg 26/60 column (GE Healthcare Life Sciences). Peak fractions were analyzed on a sodium dodecyl-sulfate polyacrylamide gel electrophoresis, pooled, and, when the concentration was <1 mg/mL, concentrated by centrifugation using Amicon Ultra-15 centrifugal filters with a 10-kDa cutoff.
SEC, MALS, and DLS Analysis.
The presence of expressed HA in the Expi293F cell culture harvests and the purity of produced proteins were analyzed by analytical SEC. A high-performance liquid chromatography (HPLC) Infinity 1260 series setup (Agilent) in combination with a TSK gel GS3000SWxl column (Sigma-Aldrich) and an in-line miniDAWN Treos MALS detector and Optilab T-rEx differential refractometer (Wyatt Technology) or an Ultra HPLC Vanquish system (ThermoFisher Scientific) in combination with a Unix-C SEC-300 column (Sepax Technologies Inc.) and an in-line µDAWN instrument (Wyatt Technology), µ T-rEx differential refractometer (Wyatt Technology), and Nanostar DLS reader (Wyatt Technology) were run in 150 mM sodium phosphate, 50 mM sodium chloride, pH 7.0. The ultraviolet (UV) signal of supernatants of nontransfected cells was subtracted from the UV signal of HA transfected cells. SEC profiles were analyzed by the Astra software package (Wyatt Technologies), and molecular weight calculations were derived from the light scattering and refractive index signal using a dn/dc value of 0.185.
Enzyme-Linked Immunosorbent Assay.
The antigenicity of the HA variants was assessed by enzyme-linked immunosorbent assay. Purified proteins were coated at a concentration of 10 nM and incubated with a dilution series of monoclonal antibodies: CR6261 [group 1 specific (38)], CR8020 [group 2 specific (39)], CR9114 (40), and CT149 (41) (both group 1 and 2 specific) using 70 nM as a starting concentration. Antibody binding was determined by incubation with a secondary antibody anti-human Fc horseradish peroxidase (mouse anti-human IgG, Jackson ImmunoResearch) and visualized by the addition of peroxidases substrate. An EnSight multimode plate reader (PerkinElmer) was used for plate read out. The halfmaximal effective concentration values of two independent experiments were calculated using Spotfire suite (Tibco Software, Inc.).
DSF.
Melting temperatures for HA proteins were determined by DSF by monitoring the fluorescent emission of Sypro Orange Dye (Thermo Fisher Scientific) added to 6 µg HA protein in solution. The measurement was performed with a starting temperature of 25 °C and a final temperature of 95 °C (54 °C increase per hour). Melting curves were measured using a ViiA7 real-time PCR machine (Applied Biosystems), and Tm50 values were derived from the negative first derivative as described previously (42).
Negative-Stain EM.
Negative-stain EM was performed on WT H1-CA09, stabilized H1-CA09, WT H3-HK68-folon, stabilized H3-HK68, WT H1-CA09-I53_dn5B, and stabilized H1-CA09-I53_dn5B (NeCen, Leiden University). HA samples were diluted to a concentration of 5 to 20 µg/mL in 20 mM Tris and 150 mM sodium chloride, pH 7.8, and a 4-µL sample was adhered onto a carbon-coated 200-mesh copper grid (Electron Microscopy Sciences) that had been glow discharged (Pelco easiGlow, 25 mA for 45 s) prior to use. The sample drop was applied for 1 min and subsequently blotted with a filter paper (Whatman no. 1 or 4). Following 1-min drying, grids were stained with 3 µL 2% uranyl acetate in filtered Milli-Q (filtered with Millipore filter 0.22 µm) for 60 s and blotted with filter paper (Whatman no. 1 or 4). Data were collected using a Talos L120C electron microscope operating at 120 keV with a magnification of 73,000×. Images were acquired with a Gatan Bm ultrascan. For 2D class-averaged images, 80 to 200 images were collected, and ∼60,000 to 240,000 particles were picked, classified, and averaged using RELION 3.1 (43). For the WT H3-HK68-foldon and the stabilized H3-HK68 HA with foldon, representative particles were selected and used to generate an ab initio model that was classified in 3D. C3 symmetry was applied to generate the initial model and then relaxed for 3D refinement.
pH Stability Evaluation.
Purified HA proteins were diluted to a concentration of 0.1 mg/mL in pH 4.7 and pH 5.5 sodium acetate buffers (20 mM NaAc, 75 mM NaCl) and in phosphate-buffered saline pH 7.4 (Gibco; 1.06 mM KH2PO4, 2.97 mM Na2HPO4-7 H2O, 155 mM NaCl). Prior to an analysis by DSF and analytical SEC, the samples were incubated at 4 °C.
HA Protein Crystallization.
Structural analysis using crystallography was performed on stabilized H3 A/Hong Kong/1/1968 HA (Proteros biostructures GmbH). Crystals of trimeric HA protein were grown using the sitting drop vapor diffusion method. Equal volumes of protein (20 mg/mL in 20 mM Tris HCL, 150 mM NaCl, pH 7.8) and reservoir solution (20% [wt/vol] PEG3350, 0.2 M LiNO3) were mixed and incubated at 293 K. The quality of the obtained crystals was improved by dehydration using the free mounting system. Crystals were flash cooled in a 100-K N2 stream.
X-ray Data Collection, Structure Solution, and Model Building.
Crystals were frozen in liquid nitrogen and measured in a cryogenic stream at 100 K. Diffraction data have been collected at the SWISS LIGHT SOURCE using cryogenic conditions (SI Appendix, Table S2). Data were processed using the autoPROC (44), XDS (45), and AIMLESS (46). Phase information was obtained by molecular replacement using Phaser (47) and the structure of WT H3-HK68 [5KAQ.pdb (48)] as a search model. Model building and refinement was performed using COOT (49) and the CCP4 software suite ( SI Appendix, Table S3).
Cell–Cell Fusion Assay.
To test the effect of stabilization of the pH switch regions on HA fusogenicity, we performed cell–cell fusion experiments. To this end, plasmids encoding full-length WT HA or stabilized variants thereof, human TMPRSS2 and GFP, were coexpressed from pcDNA2004 plasmids in human embryonic kidney (HEK) 293 cells using Trans-IT transfection reagent according to the manufacturer’s instructions. Transfections were performed on 80% confluent cell monolayers in 24-well plates. After 24 h incubation at 37 °C and 10% CO2, the transfected cells were imaged on an EVOS cell imaging system (Thermo Fisher). The wells were then exposed to pH 5.0 Dulbecco’s modified Eagle’s medium for 10 min at 37 °C, after which the medium was aspirated and replaced with normal culture medium. The cells were incubated for an additional 1 h at 37 °C to allow the cytoskeleton rearrangements involved in syncytia formation before being imaged again to visualize the GFP redistribution associated with syncytia formation. Overlays between brightfield and GFP channels were made in ImageJ.
Supplementary Material
Acknowledgments
We thank Ruud van Meersbergen, Daan van Overveld, and Patrick Wanningen for technical support; Indigo King and Yifan Song from Cyrus Biotechnology for computational protein modeling; and Stephan Krapp and Klaus Maskos from Proteros for protein crystallization and model building. We thank Ludo Renault from the Netherlands Centre for Electron Nanoscopy at Leiden University, an Instruct–European Research Infrastructure Consortium center, for negative-stain EM and image processing. This project has been funded in whole or in part with federal funds from the Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, under Other Transaction Agreement HHSO100201700018C.
Footnotes
Competing interest statement: F.J.M., M.J., T.R., B.B., and J.P.M.L. are coinventors on related vaccine patents. F.J.M., M.J., T.R., P.B., I.J.M.B., M.d.M., D.V., L.L., B.K., M.J.G.B., J.J., B.B., and J.P.M.L. are employees of Janssen Vaccines & Prevention. M.J., J.J., B.B., P.B., and J.P.M.L. hold stock of Johnson & Johnson.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115379119/-/DCSupplemental.
Data Availability
The coordinates and structure factors for the crystal structure of stabilized H3 A/Hong Kong/1/1968 HA were deposited in PDB (accession no. 7QA4).
References
- 1.Feshchenko E., et al. , Pandemic influenza vaccine: Characterization of A/California/07/2009 (H1N1) recombinant hemagglutinin protein and insights into H1N1 antigen stability. BMC Biotechnol. 12, 77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyoglu-Barnum S., et al. , Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592, 623–628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe A., et al. , Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177, 1124–1135.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zost S. J., et al. , Canonical features of human antibodies recognizing the influenza hemagglutinin trimer interface. J. Clin. Invest. 131, 146791 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bangaru S., et al. , A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177, 1136–1152.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy K. R., et al. , A prevalent focused human antibody response to the influenza virus hemagglutinin head interface. MBio 12, e0114421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCraw D. M., et al. , Structural analysis of influenza vaccine virus-like particles reveals a multicomponent organization. Sci. Rep. 8, 10342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böttcher C., Ludwig K., Herrmann A., van Heel M., Stark H., Structure of influenza haemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 463, 255–259 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Harris A., et al. , Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 103, 19123–19127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei C. J., et al. , Next-generation influenza vaccines: Opportunities and challenges. Nat. Rev. Drug Discov. 19, 239–252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weldon W. C., et al. , Enhanced immunogenicity of stabilized trimeric soluble influenza hemagglutinin. PLoS One 5, e12466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limburg H., et al. , TMPRSS2 is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. J. Virol. 93, e00649-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell C. J., Hu M., Okda F. A., Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol. 26, 841–853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benton D. J., Gamblin S. J., Rosenthal P. B., Skehel J. J., Structural transitions in influenza haemagglutinin at membrane fusion pH. Nature 583, 150–153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J., Gui M., Xiang Y., Structural intermediates in the low pH-induced transition of influenza hemagglutinin. PLoS Pathog. 16, e1009062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia N. K., Guttman M., Ebner J. L., Lee K. K., Dynamic changes during acid-induced activation of influenza hemagglutinin. Structure 23, 665–676 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das D. K., et al. , Direct visualization of the conformational dynamics of single influenza hemagglutinin trimers. Cell 174, 926–937.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadam R. U., Wilson I. A., Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U.S.A. 114, 206–214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutten L., et al. , A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep. 23, 584–595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawi R., et al. , Automated design by structure-based stabilization and consensus repair to achieve prefusion-closed envelope trimers in a wide variety of HIV strains. Cell Rep. 33, 108432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha Y., Stevens D. J., Skehel J. J., Wiley D. C., H5 avian and H9 swine influenza virus haemagglutinin structures: Possible origin of influenza subtypes. EMBO J. 21, 865–875 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekiert D. C., et al. , Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu N. C., et al. , A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog. 13, e1006682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho W., et al. , Next-generation vaccines: Nanoparticle-mediated DNA and mRNA delivery. Adv. Healthc. Mater. 10, e2001812 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brisse M., Vrba S. M., Kirk N., Liang Y., Ly H., Emerging concepts and technologies in vaccine development. Front. Immunol. 11, 583077 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei C. J., et al. , Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J. Virol. 82, 6200–6208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell C. J., Hemagglutinin stability and its impact on influenza A virus infectivity, pathogenicity, and transmissibility in avians, mice, swine, seals, ferrets, and humans. Viruses 13, 746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byrd-Leotis L., Galloway S. E., Agbogu E., Steinhauer D. A., Influenza hemagglutinin (HA) stem region mutations that stabilize or destabilize the structure of multiple HA subtypes. J. Virol. 89, 4504–4516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee P. S., Zhu X., Yu W., Wilson I. A., Design and structure of an engineered disulfide-stabilized influenza virus hemagglutinin trimer. J. Virol. 89, 7417–7420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R., Wilson I. A., Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 85, 5172–5182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antanasijevic A., Durst M. A., Lavie A., Caffrey M., Identification of a pH sensor in influenza hemagglutinin using X-ray crystallography. J. Struct. Biol. 209, 107412 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mair C. M., et al. , A histidine residue of the influenza virus hemagglutinin controls the pH dependence of the conformational change mediating membrane fusion. J. Virol. 88, 13189–13200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trost J. F., et al. , A conserved histidine in group-1 influenza subtype hemagglutinin proteins is essential for membrane fusion activity. Virology 536, 78–90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juraszek J., et al. , Stabilizing the closed SARS-CoV-2 spike trimer. Nat. Commun. 12, 244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yassine H. M., et al. , Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 21, 1065–1070 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Battles M. B., et al. , Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat. Chem. Biol. 12, 87–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krarup A., et al. , A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 6, 8143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekiert D. C., et al. , Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekiert D. C., et al. , A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dreyfus C., et al. , Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., et al. , A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat. Commun. 6, 7708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutten L., et al. , Structure-based design of prefusion-stabilized filovirus glycoprotein trimers. Cell Rep. 30, 4540–4550.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joyce M. G., et al. , NISC Comparative Sequencing Program, Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166, 609–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J., Skehel J. J., Wiley D. C., N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. U.S.A. 96, 8967–8972 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carnell G. W., Ferrara F., Grehan K., Thompson C. P., Temperton N. J., Pseudotype-based neutralization assays for influenza: A systematic analysis. Front. Immunol. 6, 161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for the crystal structure of stabilized H3 A/Hong Kong/1/1968 HA were deposited in PDB (accession no. 7QA4).