Significance
This study reveals that impaired IKKα expression or activity in lung cancer enhances differentiation of protumorigenic Treg cells through a TNF/TNFR2/NF-κB signaling pathway in both human and mouse lung ADC. Depletion of one of the molecules that are required for Treg cell induction represses lung ADC development. Thus, the components that interfere with this particular Treg differentiation provide targets for the generation of TME-modifying therapies.
Keywords: immunosuppressive response, Treg cells, lung cancer, NK-κB signaling, inflammation
Abstract
The tumor microenvironment (TME) provides potential targets for cancer therapy. However, how signals originating in cancer cells affect tumor-directed immunity is largely unknown. Deletions in the CHUK locus, coding for IκB kinase α (IKKα), correlate with reduced lung adenocarcinoma (ADC) patient survival and promote KrasG12D-initiated ADC development in mice, but it is unknown how reduced IKKα expression affects the TME. Here, we report that low IKKα expression in human and mouse lung ADC cells correlates with increased monocyte-derived macrophage and regulatory T cell (Treg) scores and elevated transcription of genes coding for macrophage-recruiting and Treg-inducing cytokines (CSF1, CCL22, TNF, and IL-23A). By stimulating recruitment of monocyte-derived macrophages from the bone marrow and enforcing a TNF/TNFR2/c-Rel signaling cascade that stimulates Treg generation, these cytokines promote lung ADC progression. Depletion of TNFR2, c-Rel, or TNF in CD4+ T cells or monocyte-derived macrophages dampens Treg generation and lung tumorigenesis. Treg depletion also attenuates carcinogenesis. In conclusion, reduced cancer cell IKKα activity enhances formation of a protumorigenic TME through a pathway whose constituents may serve as therapeutic targets for KRAS-initiated lung ADC.
Cancer-related genomic aberrations generate neoantigens, which drive tumor-directed immune responses that affect tumor progression. Identifying cancer-cell-intrinsic alterations that shape tumor-directed immunity (TDI) is important for choosing and improving treatments for lung cancer, which remains the leading cause of cancer-related mortality (1). Immune checkpoint blockade has improved 5-y survival rates for patients with nonsmall cell lung carcinoma (NSCLC), which includes lung adenocarcinoma (ADC) and squamous cell carcinoma (SCC) (2, 3). Previously, we found that the CHUK gene, coding for IKKα, is a suppressor of lung ADC (4). Oncogenic KRAS mutations and CHUK nonsense mutations or homozygous and hemizygous deletions are present in ∼35% and 25% of human lung ADC patients, respectively (4–6). Patients with lung ADC whose tumors contain both CHUK hemizygous deletions and oncogenic KRAS mutations die earlier than patients with ADC with KRAS mutations alone (4, 6). IKKα inactivation in mouse lung epithelium results in spontaneous ADC formation and enhanced KrasG12D-initiated lung ADC development (4). Urethane-induced lung ADC incidence and ADC weights were also higher in mice lacking IKKα in type II lung epithelial cells compared to wild-type (WT) counterparts (7). Ikkα deletion in pancreatic epithelial cells enhances KrasG12D-induced pancreatic ductal ADC pathogenesis by interfering with the completion of autophagy, which results in chronic pancreatitis (8, 9).
Reduced IKKα activity increases expression of inflammatory cytokines and growth factors and enhances macrophage infiltration into sites of Ras-initiated pancreatic and skin carcinomas (10, 11). However, it remains to be determined how ADC cell-derived inflammatory responses influence cancer immunoediting to impact tumor elimination or escape (12). An adoptive CD4+ T cell TDI has the potential to eliminate Ras-initiated tumors at an early stage. Indeed, CD4+ T cells antagonize Kras-initiated lung ADC development, while tolerance-related Treg cells dampen TDI (13). We hypothesized that impaired IKKα expression or activity unleashes KrasG12D ADC-intrinsic signaling pathways that blunt TDI and allow ADC to escape immunosurveillance and undergo further progression. Because reduced IKKα expression or activity worsens survival of patients with lung ADC (4), we investigated how IKKα deficiency affects TDI in lung ADC with the hope of identifying new targets for therapeutic intervention.
Here we describe shared patterns of increased monocyte-derived macrophage and Treg-cell scores and regulators of macrophage and Treg differentiation in human and mouse lung ADC lacking IKKα. ADC-intrinsic Ikkα ablation up-regulated expression of regulators of macrophage recruitment and Treg differentiation, thereby converting the tumor microenvironment (TME) from cancer restrictive to cancer supportive. A major effector of ADC-intrinsic IKKα deficiency is a Treg-promoting TNF/TNFR2/NF-κB signaling cascade.
Results
IKKα Deficiency in Human Lung ADC Correlates with Increased FOXP3+ Treg and Monocyte-Derived Macrophage Scores.
Exploring the relationship between reduced IKKα expression and the TME in human lung ADC, we found that low IKKα correlated with increased intratumoral macrophage (CD68+, a monocyte lineage marker) and elevated Treg (FOXP3+) numbers in a tissue array of 90 human lung ADC (Fig. 1 A and B and SI Appendix, Fig. S1 A and B). Immunohistochemical (IHC) staining revealed an identical pattern of CD68+ and CD163+ macrophages in human lung ADC and adjacent tissues (Fig. 1C and SI Appendix, Fig. S1C), suggesting that either CD68 or CD163 can be used to detect intratumoral macrophages. We did not observe a significantly correlated distribution between CD8+ T cell numbers and IKKα expression in the lung ADC array, although some high IKKα ADC showed increased CD8+ T cell infiltration (SI Appendix, Fig. S1 D and E). IKKα expression, which was grouped by median and quartiles, was inversely correlated with Treg and macrophage (including monocyte) scores in human lung ADC cohorts (Fig. 1D, ref. 14, and SI Appendix, Fig. S1 F and G). Furthermore, IKBKB mRNA, which encodes IKKβ that interacts with IKKα and IKKγ to form the IKK complex (15), positively correlated with macrophage scores, though its expression did not show a significant correlation with Treg scores in the same ADC cohort (Fig. 1E) (14). Curiously, IKKα and IKKβ expressions were inversely correlated (SI Appendix, Fig. S1H).
Fig. 1.
Human lung ADCs lacking CHUK/IKKα are highly correlated with elevated TNF/TNFRSF1B-associated FOXP3 Treg-cell and macrophage scores. (A, Left) The array containing 90 human lung ADCs (BCS04017a; US Biomax, Inc.) was divided into two groups of low CD68 (L-CD68)- and high CD68 (H-CD68)-positive cells, analyzed by immunofluorescence (IF) staining. (Right) The correlation between macrophages (CD68) and Treg-cell (FOXP3) numbers was further analyzed. n, case numbers; ****P < 0.0001; Student’s t test. (B) Correlation of IKKα expression and macrophage numbers in the human lung ADC array (BCS04017a) was analyzed by IHC staining. The distribution of IKKα-stained intensities from strong (+++) to negative (−) staining was divided into two groups of L(low)-CD68 and H(high)-CD68. Data represent mean ± SEM. Data were analyzed by χ2 test. n, ADC numbers; ***P < 0.001. (C) Comparison of CD68 and CD163 expression patterns in human lung ADC and its adjacent tissue in human tissue arrays (HLung030PG02; US Biomax, Inc.), analyzed by IHC staining. Dark brown, positive cells; numbers on the Top, positions in the tissue array. (Scale bar, 50 μm.) (D) Correlation of CHUK expression levels with macrophage (Left) and Treg-cell (Right) scores in human lung ADCs determined by analyzing the data (14). Using quartile analysis, CHUK expression was divided into high (H-CHUK) and low (L-CHUK) expression levels. CHUK, IKKα; n, ADC numbers per group; P value at the Top of panels, Student’s t test. (E) Correlation of IKBKB expression levels with macrophage (Top) or Treg-cell (Bottom) scores in human lung ADCs determined by analyzing the data (14). Using quartiles and median analyses, IKBKB expression was divided into high IKBKB (H-IKBKB) and low IKBKB (L-IKBKB) expression levels. n, ADC numbers; P value at the Top of panels, Student’s t test. (F) Coexpression of the genes encoding molecules for macrophage and Treg-cell development in human lung ADCs determined by analyzing the data (6). P value at the Top of panels, two-tailed t test; n, ADC numbers; red lines, repression lines; r, Pearson correlation coefficient; −4 (−2 or −1), outliers that were removed from 230. (G) Correlation of H-CHUK or L-CHUK expression levels (using median and quartile analysis to divide CHUK expression groups) with the expression of CSF1, CSF1R, FOXP3, TNFRSF1B, TNF, or CCL22 genes in human lung ADCs (TCGA, PanCancer Atlas, cBioPortal). n, ADC numbers. P value at the Top of panels, Student’s t test.
By analyzing a human lung ADC cohort (6), we found that expression levels of gene pairs encoding CSF1R/CSF1, TNFRSF1B/FOXP3, FOXP3/CSF1R, FOXP3/CSF1, TNF/FOXP3, and CSF1R/CCL22 were significantly correlated (Fig. 1F), indicating an association between TNF/TNFRSF1B-linked Treg and macrophage numbers in human lung ADC. Of note, IKKα amounts were inversely correlated with expression of CSF1, CSF1R, FOXP3, TNFRSF1B, TNF, CCL22, and IKKβ in a dose-dependent manner (Fig. 1G and SI Appendix, Fig. S1I). These results suggest that reduced IKKα expression is associated with increased macrophage and Treg numbers and their regulators in human lung ADC.
IKKα Loss Enhances KrasG12D-Induced Lung ADC Development and Macrophage and Treg Infiltration.
To investigate whether IKKα deficiency increases intratumoral macrophage and Treg numbers in mouse ADC, we expressed KrasG12D (16) and ablated Ikkα in lung epithelial cells (IkkαΔLU) by intratracheally injecting adenovirus–cyclization recombinase (Ad.Cre) (4) to generate KrasG12D and KrasG12D; IkkαΔLU mice on a C57BL/6 background. Ad.Cre is expressed only in the cytosolic compartment, disappearing after cell division. KrasG12D;IkkαΔLU mice developed heavier lungs with significantly higher ADC burden compared to KrasG12D mice (Fig. 2A and SI Appendix, Fig. S2A). Flow cytometry showed that ADC-associated F4/80+ cell numbers or F4/80+CD11b+ monocyte-derived macrophage numbers normalized to lung ADC weight were higher in IKKα-deficient tumors (Fig. 2 B and C). F4/80+ and F4/80+CD11b+ cell numbers versus CD45+ cells were also higher in IKKα-deficient tumors (SI Appendix, Fig. S2B). Treg cell numbers were elevated but CD8+ T cells were lower in KrasG12D;IkkαΔLU lung ADCs compared to KrasG12D lung ADCs (Fig. 2 B–E and SI Appendix, Fig. S2C).
Fig. 2.
IKKα loss promotes lung carcinogenesis associated with increased Foxp3 Treg-cell and macrophage numbers. (A) An approach (Top) of KrasG12D activation and Ikkα ablation in lungs of KrasG12D and KrasG12D;Ikkαf/f mice by adenovirus-Cre (Ad.Cre, or Cre) and the lung appearances (Bottom) of KrasG12D and KrasG12D;IkkαΔLU mice at 3 mo after Ad.Cre treatment. (B and C) Flow cytometric analyses of F4/80+CD11b+ (B) or F4/80+ (C) cells in KrasG12D (n = 5) and KrasG12D;IkkαΔLU (n = 6) lung ADCs. Representative images for flow cytometry on the Left and statistical analyses on the Right are shown. Data represent mean ± SEM (multiple experiments). This result is representative. **P < 0.01; ****P < 0.0001; Student’s t test. (D) Flow cytometric analyses of Treg-cell numbers in KrasG12D and KrasG12D;IkkαΔLU lung ADCs. Data represent mean ± SEM (three experiments). **P < 0.01; Student’s t test. (E) IHC staining for % CD8 T cells in KrasG12D and KrasG12D;IkkαΔLU lung ADCs, analyzed by n = 3 mice/group; three sections per mouse. Data represent mean ± SD (three repeats). ***P < 0.001; Student’s t test. (F) Lung appearances with ADC (Left) and ADC burden (Right) derived from Kras-CL and KrasIKKαL ADC cells in WT mice (n = 3/group). This is representative. Data represent mean ± SEM (three experiments). ***P < 0.001; Student’s t test. (G) IHC analysis of % F4/80 (Left), % Foxp3 (middle, n = 3), and % CD8 (Right, n = 4) cells in Kras-CL and KrasIKKαL ADCs. Data represent mean ± SD (three repeats). *P < 0.05; ***P < 0.001; Student’s t test. (H) Representative images of flow cytometric analyses for % macrophages (F4/80+CD11b+) in CD45+ cells isolated from KrasIKKαL ADCs in WT mice. (I) ROS (dichlorofluorescein [DCF]) levels in macrophages isolated from Kras-CL and KrasIKKαL lung ADCs in WT mice (n = 3/group). Data represent mean ± SEM (three repeats). *P < 0.05; Student’s t test. (J) A heat map analyzing the gene expression profiles in macrophages isolated from Kras-CL or KrasIKKαL lung ADC in WT mice. Statistical analysis for all gene comparison from two groups, P < 0.05; one-way ANOVA test. (K) A summary: increased IKKα-deficient KrasG12D ADC development is correlated with increased macrophage and Treg-cell numbers. Mϕ, macrophage.
To verify whether IKKα-deficient ADC cells cause similar TME alterations when growing in WT mice, we intratracheally injected mouse Kras-CL and KrasIKKαL cell lines, derived from KrasG12D-initiated ADC without or with IKKα deficiency, respectively (4) into lungs of WT mice. KrasIKKαL ADC cells express low amounts of IKKα and have increased tumorigenic activity compared to Kras-CL cells (4). KrasIKKαL tumor burden and KrasIKKαL ADC-associated F4/80+ and Treg numbers were higher, but CD8+ cells were lower than in Kras-CL ADC (Fig. 2 F and G and SI Appendix, Fig. S2D). Most intratumoral F4/80+ macrophages also expressed CD11b, a monocyte marker (Fig. 2H and SI Appendix, Fig. S2E).
Monocyte-derived macrophages isolated from KrasIKKαL ADC showed elevated reactive oxygen species (ROS) levels and expressed reduced antioxidant genes but more ROS-, inflammation-, and mitogenesis-related genes compared to macrophages isolated from Kras-CL ADC (Fig. 2 I and J and SI Appendix, Fig. S2 F and G). These results show that ADC-intrinsic IKKα deficiency alters the TME to become more immunosuppressive (Fig. 2K).
Bone Marrow–Derived Macrophages and Treg Cells Support Growth of IKKα-Deficient ADC.
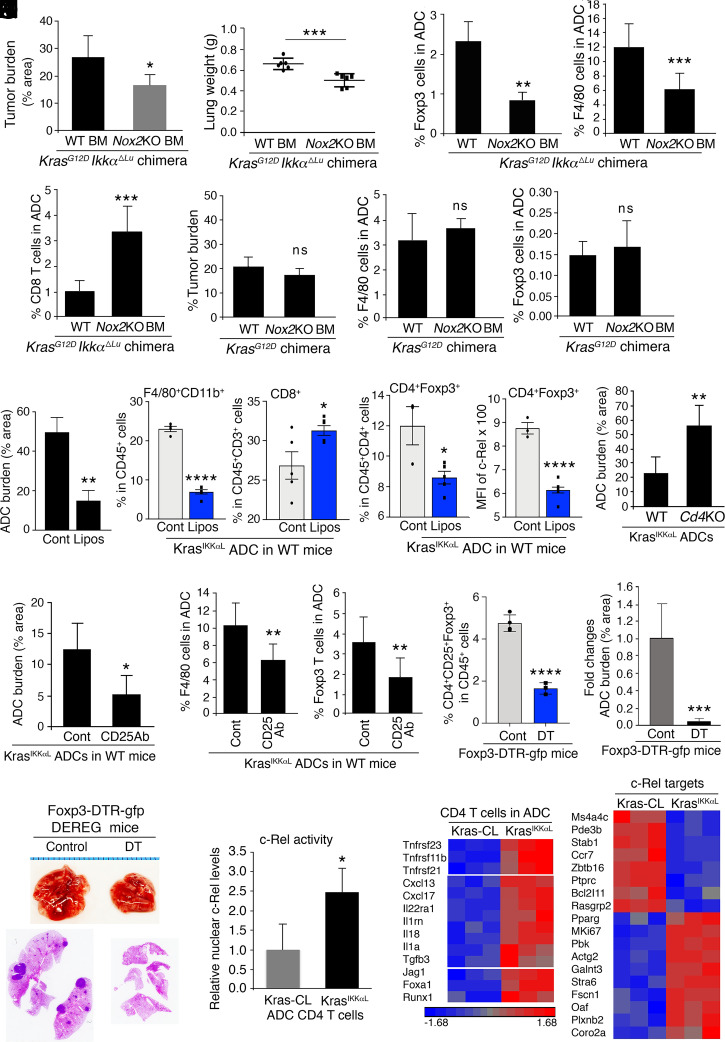
To determine the effect of macrophage-derived ROS on IKKα-deficient ADC development, we transferred WT or Nox2−/− (Nox2knockout [KO]) bone marrow (BM) into irradiated KrasG12D and KrasG12D;IkkαΔLU mice. Lung weights and ADC burden were decreased in chimeric KrasG12D;IkkαΔLU mice with Nox2KO BM compared to KrasG12D;IkkαΔLU mice with WT BM (Fig. 3 A and B). Interestingly, lung ADC with Nox2KO BM showed decreased macrophage and Treg numbers and elevated CD8+ T cell numbers compared to lung ADC with WT BM, in addition to the expected decrease in ROS (Fig. 3 C and D and SI Appendix, Fig. S3 A and B). By contrast, lung ADC burden and infiltrating BM-derived macrophages, Treg cells, and CD8+ T cell numbers in chimeric KrasG12D mice were not affected by the Nox2 status of the transplanted BM (Fig. 3 E and F and SI Appendix, Fig. S3C). Lung tumor burden and tumoral F4/80+ and Treg cell numbers were significantly reduced in Nox2−/− mice receiving KrasIKKαL cell injections compared to WT hosts transplanted with the same cells (SI Appendix, Fig. S3D). Thus, ROS produced by BM-derived macrophages may stimulate Treg differentiation to support the development and progression of IKKα-deficient lung ADC.
Fig. 3.
Macrophages paired with Foxp3 Treg cells specifically promote IKKα-deficient lung ADC development. (A and B) Lung ADC burden (% area, A) and lung weight (B) in KrasG12D;IkkαΔLU mice receiving WT or Nox2KO BM (n = 6 mice/group). g, grams. Data represent mean ± SD. *P < 0.05; ***P < 0.001; Student’s t test. (C and D) IHC analyses of % Foxp3 Treg cells (C, Left, n = 3), % F4/80 macrophages (C, Right, n = 4), and % CD8 cells (D, n = 4) in lung ADCs of KrasG12D;IkkαΔLU mice receiving WT BM or Nox2KO BM. Data represent mean ± SEM (three repeats). **P < 0.01; ***P < 0.001; Student’s t test. (E) Lung ADC burden (% area) in KrasG12D mice receiving WT BM (n = 5 mice) or Nox2KO BM (n = 6 mice). ns, not significant; Student’s t test. (F) IHC analyses of % F4/80 macrophages (Left) and % Foxp3 Treg cells (Right) in lung ADCs of chimeric KrasG12D mice receiving WT BM or Nox2KO BM (n = 3 mice/group; three sections per mouse). Data represent mean ± SD. ns, not significant; Student’s t test. (G) KrasIKKαL lung ADC burden (% area, Left) in WT mice treated with clodronate-loaded liposomes (Lipos) or a vehicle control (n = 4 for each group). Flow cytometric analyses of % F4/80+CD11b+ cells (Middle) and % CD8+ cells (Right) in CD45+ cells associated with KrasIKKαL lung ADCs derived from WT mice treated with clodronate-loaded Lipos (n = 5/group). Data represent mean ± SEM *P < 0.05; **P < 0.01; ****P < 0.0001; Student’s t test. (H) Flow cytometric analyses of % Treg cells (Left) in CD4+ T cells and c-Rel levels (MFI, median fluorescence intensity, Right) in CD4+Foxp3+ Treg cells associated with KrasIKKαL lung ADCs of WT mice treated with clodronate-loaded Lipos or a vehicle control (n = 5/group). Data represent mean ± SD. *P < 0.05; ****P < 0.0001; Student’s t test. (I) KrasIKKαL lung ADC burden in WT and Cd4−/− (Cd4KO) mice (n = 5/group). *P < 0.05; Student’s t test. (J) KrasIKKαL lung ADC burden in WT mice treated with an anti-Treg antibody (anti-CD25Ab) or a vehicle control (n = 5/group). Ab, antibody. Data represent mean ± SD. *P < 0.05; Student’s t test. (K) IHC analyses of % F4/80 macrophages (Left) and % Foxp3 Treg cells (Right) in lung ADCs derived from WT mice treated with an anti-CD25 Ab or a vehicle control (Cont) (n = 3/group; three sections per mouse). Data represent mean ± SEM (three repeats). ***P < 0.001; **P < 0.01; Student’s t test. (L) Flow cytometric analyses of % Treg (CD4+CD25+Foxp3+) cells (Left) in CD45+ cells isolated from lung ADCs of Foxp3-DTR-gfp mice (DEREG) treated with or without DT. Lung ADC burden of Foxp3-DTR-gfp mice treated with or without DT (Right). Data represent mean ± SEM (three repeats). ***P < 0.001; ****P < 0.0001; Student’s t test. (M) Lungs (Top) and hematoxylin and eosin slides (Bottom) representing lung ADCs with dark spots in Foxp3-DTR-gfp mice with DT or a vehicle control. (N) Western blotting analyses of nuclear c-Rel protein levels of CD4 T cells isolated from Kras-CL ADCs or KrasIKKαL lung ADCs (SI Appendix, Fig. S3L). The relative activity was measured by comparing the nuclear c-Rel protein intensity with LaminB1 intensity, a nuclear protein loading control. Each sample was isolated from the ADCs obtained from multiple mice. Data represent mean ± SEM (three repeats). *P < 0.05; Student’s t test. (O) Heat maps analyzing gene expression profiles related to c-Rel targets (Right) and to inflammation and Foxp3 regulators (Left) in CD4 T cells isolated from Kras-CL or KrasIKKαL ADCs in WT mice. Statistical analysis for all listed genes from two groups, P < 0.05; one-way ANOVA test.
Because the BM contains multiple haemopoietic cell types, we verified that the effects of NOX2-dependent ROS on lung ADC development were exerted by macrophages. We intratracheally injected KrasIKKαL cells into WT mice and then depleted host macrophages with clodronate-loaded liposomes, which kill monocyte-derived macrophages (17). In this setting, we could not use Kras-CL cells because they generated very small ADCs with a low number of filtrating macrophages and Treg cells and were poorly responsive to macrophage-generated ROS. Clodronate-loaded liposome treatment significantly reduced lung ADC burden, ADC-associated F4/80+CD11b+ macrophages, and Treg cell numbers, as well as c-Rel amounts in CD4+Foxp3+ Treg cells, but increased CD8+ T cell numbers compared to vehicle control (Fig. 3 G and H and SI Appendix, Fig. S3E). To determine the role of CD4+ T cells, we used Rag1−/− and Cd4−/− mice as recipients, which developed heavier lungs with increased ADC burden compared to WT mice after intratracheal KrasIKKαL cell injection (Fig. 3I and SI Appendix, Fig. S3 F and G). These results suggest that CD4+ T cells have antitumor activity. Intratumoral F4/80+ cell numbers were elevated in Rag1−/− and Cd4−/− hosts compared to WT hosts (SI Appendix, Fig. S3 H and I), suggesting that CD4+ T cells may inhibit macrophage infiltration.
To examine the effect of Treg cells on tumorigenesis, we intratracheally injected KrasIKKαL cells into WT mice and treated the mice with an anti-CD25 antibody to deplete Treg cells (18). Treg reduction attenuated ADC development and reduced intratumoral monocyte-derived macrophages (Fig. 3 J and K and SI Appendix, Fig. S3J). We also depleted Foxp3+ cells using diphtheria toxin (DT) in Foxp3-DTR-GFP (DEREG) mice (19) that were intratracheally injected with KrasIKKαL cells, resulting in dampened lung ADC development (Fig. 3 L and M). These data indicate that Treg cells support lung tumorigenesis in this setting.
TGFβ, a strong Treg-cell inducer, was not highly expressed in KrasG12D;IkkαΔLU ADC. Because c-Rel promotes Treg cell generation (20) and macrophage depletion reduced its expression in CD4+Foxp3+ cells (Fig. 3H), we analyzed nuclear c-Rel and expression of its target genes in CD4+ T cells isolated from KrasIKKαL and Kras-CL lung ADC. KrasIKKαL ADC–derived CD4+ T cells showed elevated nuclear c-Rel (Fig. 3N and SI Appendix, Fig. S3 K and L), expressed higher amounts of c-Rel target mRNAs (20, 21), Foxp3 up-regulating molecules, TNF-family–related pathways, IL-1 signaling, and fewer apoptosis-related genes compared to CD4+ cells from Kras-CL ADC (Fig. 3O). These results suggest that intratumoral CD4+ T cells of IKKα-deficient ADC have higher c-Rel–driven NF-κB activity.
CD4+ T Cell c-Rel Signaling and Macrophages Promote Generation of Intratumoral Treg Cells.
To establish a link between intratumoral monocyte-derived macrophages and Treg generation, we cocultured ADC-associated macrophages with CD4+CD25− T cells (22) and found that macrophages from KrasIKKαL ADCs induced higher CD4+Foxp3+ Treg cell numbers than macrophages from Kras-CL ADCs (Fig. 4A). The macrophages enhanced Treg cell induction in a dose-dependent manner and the induced Foxp3+ cells expressed less CD4 (Fig. 4B), which was probably due to elevated CD25 expression (data not shown). Since Nox2KO macrophages decreased intratumoral Treg cell numbers, we examined the role of macrophage-produced ROS in Treg induction. We cocultured CD4+CD25− T cells with Nox2KO macrophages, which produced fewer ROS than WT macrophages and were less potent in Treg induction (Fig. 4 C, Left and Middle). To confirm the role of ROS in Treg cell induction, we added N-acetylcysteine (NAC) or apocynin, a ROS inhibitor, to the coculture experiments and found them to attenuate Treg-cell induction by WT macrophages (Fig. 4 C, Right). NAC treatment also reduced the burden of KrasIKKαL-generated ADC and decreased the number of ADC-associated Treg cells, macrophages, macrophage ROS amounts, and c-Rel expression in intratumoral CD4+ T cells (Fig. 4D and SI Appendix, Fig. S4A). Reduced ADC IKKα expression is associated with increased TNF expression and Treg-cell scores (Fig. 1G), and TNF activates NF-κB (23). We therefore tested whether TNF enhances Treg induction in the coculture system by binding to TNFR2 and activating NF-κB in CD4+ T cells. Addition of either TNF or H2O2 to the macrophage-CD4+CD25− T cell coculture system enhanced Treg-cell induction and increased c-Rel expression in both parental CD4+CD25+ T cells and CD4+CD25+Foxp3+ Treg cells (Fig. 4E).
Fig. 4.
A TNF/TNFRSF1B/c-Rel signaling cascade promotes Foxp3 Treg-cell induction. (A) Flow cytometric analyses of % Treg cells in CD4 T cells from the coculture of WT CD4+CD25− T cells with macrophages (macro) that were isolated from Kras-CL or KrasIKKαL lung ADCs (n = 3/group). Data represent mean ± SEM (three repeats). *P < 0.05; Student’s t test. (B) Flow cytometric analyses of % Treg cells in CD4 T cells from the coculture of WT CD4+CD25− T cells and peritoneal macrophages (PM) in a dose-dependent manner (ratio, 2:1 and 1:1) (Right). n = 4. Data represent mean ± SEM (three repeats). ***P < 0.001; ****P < 0.0001; Student’s t test (Left). (C) ROS (dichlorofluorescein [DCF]) levels in WT and Nox2KO PM (Left). Flow cytometric analyses of % Treg cells in CD4 T from the coculture of WT or Nox2KO PM with WT CD4 T cells (n = 3/group, Middle) and from the coculture of WT PM and WT CD4 T cells treated with NAC (n = 4), apocynin (n = 4), and vehicle control (n = 3) (Right). Data represent mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test. (D) Lung ADC burden (% area, Left) in WT mice receiving KrasIKKαL cells, treated with NAC (n = 5) and vehicle control (n = 4). ROS (DCF) levels in ADC-associated macrophage (Middle), and cytosolic and nuclear c-Rel levels in ADC-associated CD4 T cells detected by Western blotting, from NAC- and vehicle (Cont)-treated WT mice receiving KrasIKKαL cells (Right). LaminB1, a nuclear protein loading control; α-tubulin, a cytosolic protein loading control. Data represent mean ± SEM (three repeats); **P < 0.01; ***P < 0.001; Student’s t test. (E) Flow cytometric analyses of % Treg cells in CD4 T cells from the coculture of WT PM and WT CD4+CD25− T cells supplemented with TNF, H2O2, or a vehicle control (n = 3/group, Left), and c-Rel intensity (median fluorescence intensity, MFI) in these CD4 cells and CD25+Foxp3+ cells (Right). ns, not significant; **P < 0.01; ***P < 0.001. Data represent mean ± SEM (three repeats); Student’s t test. (F) Flow cytometric analyses of % Treg-cell induction in CD4 T cells from the coculture of WT PM and WT CD4 T cells treated with PTXF (50 μg/mL) and control. *P < 0.05. Data represent mean ± SEM (three repeats); Student’s t test. (G) Lung ADC burden (% area, Left) in WT mice receiving KrasIKKαL cells, treated with PTXF (50 mg/kg) or a vehicle control (n = 4/g/roup). IHC analyses of % Treg cells (Middle) in ADCs and ROS (DCF) levels in ADC-associated macrophages (Right) from WT mice receiving KrasIKKαL cells, treated with PTXF or a vehicle control (n = 4/group). Data represent mean ± SEM (three repeats); *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test. (H) Western blotting analyses of cytosolic and nuclear c-Rel levels in CD4 T cells isolated from KrasIKKαL lung ADCs in WT mice treated with PTXF or a vehicle control (Cont). Each example was obtained from multiple mice. LaminB1, a nuclear protein loading control; α-tubulin, a cytosolic protein loading control. (I) KrasIKKαL ADC burden (% area, Left) in WT and Tnf−/− (KO) mice (n = 6/group). Data represent mean ± SD. ****P < 0.0001; Student’s t test. Images of WT and Tnf KO lungs are representative (Right). Dark pink stained spots in hematoxylin and eosin slides represent tumors in the lungs. (J) IHC analyses of % Foxp3 Treg-cell (Left) and % F4/80 macrophage (Right) numbers in KrasIKKαL ADCs from WT and Tnf KO mice (n = 4/group; three sections per mouse). Data represent mean ± SD. ****P < 0.0001; Student’s t test. (K) TNFR2/TNFRSF1B expression in CD4 T cells isolated from lung ADCs of WT and Tnf KO mice, analyzed by RT-PCR. Data represent mean ± SEM (three repeats). ***P < 0.001; Student’s t test. (L–M) Flow cytometric analyses of % Treg cells in CD4 T cells (L) and c-Rel levels (MFI, M) in Treg cells from the coculture of WT PM with WT or Tnfrsf1b−/− (KO) CD4+CD25− T cells, treated with or without TNF. Data represent mean ± SEM (three repeats). *P < 0.05; **P < 0.01; ***P < 0.001; Student’s test. (N) ROS (DCF) levels in WT and Tnf KO peritoneal macrophages, analyzed by RT-PCR. Data represent mean ± SEM (three repeats). ****P < 0.0001; Student’s t test. (O) TNF expression in WT peritoneal macrophages treated with H2O2 or vehicle control. Data represent mean ± SEM (three repeats). *P < 0.05; Student’s t test. (P) Flow cytometric analyses of % Treg cells in CD4 T cells from the coculture of WT PM with WT or Tnfrsf1b−/− CD4 T cells as well as from the coculture of Tnf KO PM with WT or Tnfrsf1b−/− CD4 T cells, analyzed by flow cytometry. Data represent mean ± SEM (three repeats). ns, not significant; *P < 0.05; ***P < 0.001; Student’s test. (Q) Lung weight (Left) and KrasIKKαL ADC burden (Right) in irradiated CD45.1 mice receiving CD45.2 WT or Tnf−/− (KO) BM (n = 4). Data represent mean ± SD. *P < 0.05; Student’s test. (R) Flow cytometric analyses of ADC-associated CD4+Foxp3+ cells in CD45+ cells (Left) and c-Rel levels (MFI) in Foxp3 cells (Right) in CD45.1 mice receiving CD45.2 WT or Tnf KO BM cells (n = 3). Data represent mean ± SEM (three repeats). *P < 0.05; **P < 0.01; Student’s test. (S) Lung appearances of Tnfrsf1b−/−;IkkαΔLU;KrasG12D and IkkαΔLU;KrasG12D mice. (T) Lung ADC burden (% area, Left) of Tnfrsf1b−/−;IkkαΔLU;KrasG12D and IkkαΔLU;KrasG12D mice (n = 4/group). Flow cytometric analyses of Foxp3+ Treg-cell numbers (Middle, n = 3) and CD4 T cell c-Rel levels (Right, n = 3) in lung ADCs of Tnfrsf1b−/−;IkkαΔLU;KrasG12D and IkkαΔLU;KrasG12D mice. Data represent mean ± SEM (three experiments). *P < 0.05; **P < 0.01; Student’s test. (U) Survival rates for lung ADC patients expressing CHUK hemizygous deletions and TNFRSF1B (TNFR2) gain versus patients with ADC expressing CHUK and TNFRSF1B double diploid. The RNA-sequence data were obtained from cBioPortal, TCGA, ref. 6. Logrank test for P value analyses.
We examined c-Rel’s involvement in Treg generation and tumorigenesis by using pentoxifylline (PTXF), which inhibits c-Rel activity, although it is not a c-Rel–specific inhibitor (20). PTXF treatment significantly reduced ex vivo Treg induction (Fig. 4F) and reduced KrasIKKαL-generated lung ADC burden, as well as intratumoral Treg and macrophage numbers, macrophage ROS amounts, and CD4+ T cell c-Rel activity, while increasing CD8+ T cell numbers (Fig. 4 G and H and SI Appendix, Fig. S4 B–D). These results suggest that NF-κB/c-Rel enhances Treg-cell induction and contributes to tumorigenesis. To rule out an effect of PTXF on macrophages or ADC cells, we treated Rag1−/− mice inoculated with KrasIKKαL cells with PTXF. The inhibitor did not alter lung weights and ADC burden in Rag1−/− mice (SI Appendix, Fig. S4 E and F), suggesting that PTXF acts on T cells.
To test TNF’s involvement in Treg differentiation and tumorigenesis, we intratracheally injected KrasIKKαL cells into lungs of WT and Tnf−/− mice. Both lung ADC burden and intratumoral Treg-cell and macrophage numbers were reduced in Tnf−/− mice along with increased CD8+ T cell numbers compared to WT (Fig. 4 I and J and SI Appendix, Fig. S4G). Notably, CD4+ T cells isolated from Tnf−/− ADC expressed reduced amounts of Tnfrsf1b, which encodes TNFR2 (Fig. 4K), suggesting that TNFR2 is the main CD4+ T cell TNF receptor mediating Treg-cell induction. Notably, TNFRSF1B mRNA is highly correlated with CD4 mRNA expression in human lung ADC (SI Appendix, Fig. S4H) (24, 25). To determine the role of TNFR2 in Treg-cell induction, we cocultured WT macrophages with WT or Tnfrsf1b−/− CD4+CD25− T cells. Tnfrsf1b−/− CD4+ T cells gave rise to fewer Treg cells and lower c-Rel expression in Foxp3+ cells compared to WT CD4+ T cells (Fig. 4 L and M). In addition, Tnfrsf1b−/− CD4+ T cells responded poorly to TNF, exhibiting reduced Treg-cell induction and lower c-Rel expression than WT CD4+ T cells (Fig. 4 L and M). Nonetheless, residual TNF-stimulated Treg induction in cocultures of WT macrophage and Tnfrsf1b−/− CD4+ T cells suggested that TNF may also work via TNFR1 in the absence of TNFR2. Because expression of CD4 and TNFRSF1A, which encodes TNFR1, was not highly correlated in human lung ADCs (6), TNFR2 may be the more important TNFR in this setting.
Because TNF and H2O2 showed similar ability to enhance Treg induction in the coculture system, we hypothesized that TNF and ROS up-regulate each other’s production. Indeed, ROS levels were lower in Tnf−/− than WT macrophages (Fig. 4N), and H2O2 enhanced TNF expression (Fig. 4O). Accordingly, Tnf−/− macrophages cocultured with WT CD4+ T cells induced fewer Treg cells compared to WT macrophages, and the coculture of Tnf−/− macrophages with Tnfrsf1b−/− CD4+ T cells led to a further reduction in Treg induction (Fig. 4P). To determine the role of macrophage-produced TNF in lung ADC development, we injected CD45.2 WT or Tnf−/− BM cells into irradiated CD45.1 WT mice that were inoculated with KrasIKKαL lung ADC cells. The results showed decreased ADC burden, lung weights, and ADC-associated Treg cells in mice reconstituted with Tnf−/− BM (Fig. 4 Q and R and SI Appendix, Fig. S4I). These experiments also suggested that ADC-associated monocyte-derived F4/80+CD11bHigh and F4/80+CD11bLow macrophages originate from the BM (SI Appendix, Fig. S4J), further supporting the role of macrophage ROS in TNF production at a level needed for Treg induction.
We compared ADC development in Tnfrsf1b−/−; IkkαΔLU;KrasG12D and IkkαΔLU;KrasG12D mice and found that lung ADC burden was significantly lower in Tnfrsf1b−/−; IkkαΔLU;KrasG12D mice than IkkαΔLU;KrasG12D mice (Fig. 4 S and T, Left). Treg numbers and c-Rel levels in Foxp3+ Treg cells were lower in lung ADCs of Tnfrsf1b−/−;IkkαΔLU;KrasG12D mice compared to IkkαΔLU;KrasG12D mice, which were consistent with the result obtained from the coculture system (Fig. 4 T, Middle and Right and SI Appendix, Fig. S4K). Patients with lung ADC with CHUK hemizygous deletions and TNFRSF1B gain die earlier compared to patients doubly diploid for CHUK and TNFRSF1B (Fig. 4U). Patients with lung ADC with CHUK hemizygous deletions and FOXP3 gain showed a lower survival trend compared to patients with ADC who are double diploid for CHUK and FOXP3 (SI Appendix, Fig. S4L). These results suggest that TNFR2 activates c-Rel and stimulates Foxp3+ Treg cell differentiation and that immunosuppressive Foxp3+ Treg induction accelerates lung carcinogenesis. Accordingly, we postulated that low IKKα lung ADC generates a TNF-rich TME. Indeed, TNF expression was higher in KrasIKKαL ADC than Kras-CL ADCs (SI Appendix, Fig. S4M).
IKKα Suppresses TNF, CSF1, CCL22, and IL-23A Expression in Lung ADC.
To investigate how IKKα loss up-regulates expression of cytokines and chemokines that shape the immunosuppressive TME (iTME), we used bead-based flow cytometry and detected higher TNF, CCL22, IL-23A, and CSF amounts in sera of ADC-bearing IkkαΔLU;KrasG12D mice than in KrasG12D mice with ADC (SI Appendix, Fig. S5A). We confirmed that KrasIKKαL ADC cells expressed significantly higher levels of Csf1, Tnf, Ccl22, Il23a, and Adam8 mRNAs compared to Kras-CL cells (Fig. 5A). Likewise, low CHUK mRNA was correlated with elevated IL23A and ADAM8 mRNAs in human lung ADC (Fig. 5B). IL-23A inhibits CD8+ T cell infiltration, enhances inflammation, and increases tumor incidence (26, 27), and ADAM8 promotes cell proliferation, invasion, and metastasis in human cancers (28, 29). Reduced IKKα also correlated with increased expression of CCL7, CCL8, and ADAM9 in mouse lung ADC cells (SI Appendix, Fig. S5B). Together, these results suggest that IKKα deficiency correlates with elevated expression of cytokines that are important for generation of Treg cells and iTME.
Fig. 5.
IKKα reduction up-regulates transcription of cytokine genes in human and mouse lung ADC cells. (A) Expression of CCL2, CSF1, TNF, ADAM8, and IL-23A in Kras-CL and KrasIKKαL ADC cells, analyzed by RT-PCR. Data represent mean ± SEM (three repeats). ***P < 0.001; **P < 0.01; Student’s t test. (B) Correlation of CHUK (IKKα) expression levels (using quartiles’ analysis to divide IKKα expression groups) with the expression levels of IL-23A or ADAM8 genes in human lung ADCs by analyzing the TCGA data (PanCancer Atlas, cBioPortal). H-CHUK, high CHUK; L-CHUK, low CHUK; P value at the Top of panels, Student’s t test; n, ADC numbers. (C–D) Expression of CHUK (IKKα), IL-23A (IL-23a), CCL22, CSF1, and TNF in human A549 (C) and mouse Kras-CL (D) lung ADC cells treated with Si-control (Si-Cont) or Si-IKKα RNA, analyzed by RT-PCR. Data represent mean ± SEM (three repeats). *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test. (E–F) ChIP analyses for binding of IKKα to the promoter regions of TNF (Tnf), CSF1 (Csf1), CCL22 (Ccl22), and IL-23A (Il-23a) genes by using an anti-IKKα antibody for immunoprecipitation, followed by PCR with primers for these genes in A549 cells (E) and in Kras-CL cells (F) treated with Si-control (Si-Cont) or Si-IKKα RNA. Data represent mean ± SEM (three repeats). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; Student’s t test. (G) ChIP-seq analyses for IKKα enrichment on the TNF gene in A549 cells. The enrichment peaks are marked by red lines. bp, nucleotide base pair; red arrow, 6,066 bp; Ab, antibody for immunoprecipitation. TNF gene: thickest lines, exons; second-thickest lines, untranslated sequences; thin lines, introns; red line, consensus sequences containing eight nucleotides. (H) A working model showing that ADC-IKKα reduction up-regulates the expression of TNF, CSF1, CCL-22, and IL-23A and macrophage recruitment, which generate a TNF/TNFRSF1B/c-Rel pathway for CD4 T cells to stimulate Treg-cell induction, accelerating KrasG12D ADC development. Arrows, promotion; lines, inhibition; Mϕ, monocyte-derived macrophage.
To determine how IKKα modulates cytokine expression, we silenced IKKα in either human or mouse lung ADC cells and found it to increase expression of IL23A, CCL22, CSF1, and TNF mRNAs (Fig. 5 C and D). In addition to being a cytoplasmic protein kinase, IKKα was reported to act in the nucleus (30–32). We therefore hypothesized that IKKα may directly or indirectly suppress expression of these cytokine genes. We conducted chromatin immunoprecipitation (ChIP) assays with an IKKα antibody and found enrichment for IKKα on the IL23A, CCL22, CSF1, and TNF promoter regions in both human A549 and mouse Kras-CL cells, and this was attenuated by IKKα silencing (Fig. 5 E and F and SI Appendix, Fig. S5C). Of note, the correlation between IKKα enrichment at these cytokine genes and their low expression in IKKα-deficient cells, suggests that IKKα act as a transcriptional suppressor of these particular genes. Because an interaction between IKKα and SMAD3/4 regulates expression of certain genes in keratinocytes (33–35), we tested whether IKKα suppresses the above cytokine gene through SMAD transcription factors. We performed a ChIP assay with SMAD3 or SMAD4 antibodies and could not detect SMAD3/4 enrichment on the IKKα-bound genes (SI Appendix, Fig. S5D). We further performed unbiased ChIP-sequencing (ChIP-seq) experiments with an IKKα antibody (Fig. 5G). Sequence analysis of the regions at which IKKα was enriched revealed an 8-bp-long consensus sequence on the TNF, CSF1, CCL22, and IL-23A genes (Fig. 5G and SI Appendix, Fig. S5E). Future studies will probe the regulatory importance of this consensus sequence and whether it is recognized by IKKα or another transcription factor or chromatin protein with which IKKα interacts.
Based on the above findings, we propose a working model that explains how IKKα deficiency in lung ADC generates iTME that dismantles immune surveillance and accelerates tumor progression. Key to this model is the up-regulation of TNF, CSF1, CCL22, and IL-23A in IKKα-deficient lung ADC, which enhances macrophage recruitment and Treg differentiation (Fig. 5H).
Discussion
Human lung ADC and SCC, which originate from different cell types, show distinct histological features and genomic alterations (cBioPortal, The Cancer Genome Atlas [TCGA], refs. 6 and 36, PanCancer Atlas). KRAS mutations are common in human lung ADC but rare in lung SCC. Previous studies conducted by other investigators had focused on the role of canonical, IKKβ-dependent, NF-κB signaling in mouse Kras-initiated lung ADC (37, 38). We, on the other hand, had focused on the role of IKKα, which unlike IKKβ, activates noncanonical NF-κB signaling (39) and has several other, NF-κB unrelated, functions (31, 32, 40, 41). Previously, we reported that Ikkα ablation in mice promotes KrasG12D-initiated lung ADC development and results in spontaneous SCC formation associated with increased macrophage infiltration (4, 17, 42). In the present study we followed on the significant correlation between monocyte-derived macrophages and Treg scores and reduced IKKα expression in human lung ADC cohorts. Low IKKα expression also correlated with elevated expression of cytokines and chemokines (CSF1R, CSF1, TNFRSF1B, TNF, CCL2, FOXP3, and CCL22) that regulate macrophage recruitment and Treg-cell development (24, 25, 43–45). Similar correlations were observed in the mouse lung ADC model we have investigated.
Although macrophage-produced ROS support Treg-cell differentiation in vitro (22), the mechanism by which macrophage ROS production stimulates tumorigenesis is unknown. Our results suggest that ROS are required for maintaining high macrophage TNF expression, which is needed for stimulation of Treg-cell differentiation via a TNF/TNFR2/c-Rel signaling cascade. Ablation of TNF or the ROS-producing enzyme Nox2 inhibited lung ADC pathogenesis and reduced tumoral monocyte-derived macrophage and Treg infiltration. Moreover, in human lung ADC cohorts (TCGA, ref. 6 and PanCancer Atlas, cBioPortal), TNFR2 expression is significantly correlated with CD4 expression. Also, Tnfrsf1b−/− CD4+ T cells gave rise to fewer Treg cells than WT CD4+ T cells when cocultured with macrophages. Correspondingly, lung ADC burden and intratumoral Treg-cell numbers were reduced in KrasG12D;IkkαΔLU;Tnfrsf1b−/− mice compared to KrasG12D; IkkαΔLU mice. However, TNF still led to residual Treg induction in cocultures of Tnfrsf1b−/− CD4+ T cells and macrophages, suggesting that in the absence of TNFR2, TNF may stimulate Treg differentiation via TNFR1, even though TNFR1 expression is not significantly correlated with CD4 expression in human lung ADC. Even in the complete absence of TNF, we still observed a basal level of Treg induction in the coculture system, suggesting that other macrophage-generated stimuli can drive Treg differentiation. Regardless of the underlying mechanism, our results demonstrate that Treg cells are key components of the iTME that accompanies IKKα-deficient lung ADC.
Together with IKKβ, IKKα is one of the two catalytic subunits of the IKK complex. Curiously, however, in human lung ADC, the expression patterns of IKKα and ΙΚΚβ associated with expression of macrophage- and Treg cell-regulating cytokines and chemokines are reciprocal. Ours and other animal studies are consistent with the findings in human lung ADC (4, 7, 38, 46, 47). However, a recent report showed that Ikkα, but not Ikkβ, deletion in lung epithelial cells attenuated Kras-initiated lung ADC development (48). The cause of this discrepancy is not clear, but it may be due to unknown microenvironmental conditions. Consistent with the results described herein, transgenic Tg-K5.IKKα and Tg-Lori.IKKα mice develop normally and are resistant to carcinogen-induced tumorigenesis and metastasis (15, 49, 50), whereas transgenic Tg-K5.IKKβ and Tg-EDL-2.IKKβ mice develop epidermal and esophageal hyperplasia and oral carcinomas (51–53). Our results are also consistent with previous studies showing that while IKKβ is the critical IKK catalytic subunit responsible for NF-κB activation, IKKα has numerous other functions. IKKα ablation in human and mouse lung ADC cells results in up-regulation of the CSF1, CCL22, TNF, and IL-23A genes, to whose promoter regions IKKα is recruited, suggesting that in lung ADC, IKKα works in the nucleus as a transcriptional repressor. However, ChIP assays cannot determine whether IKKα directly recognizes specific DNA sequences or interacts with transcription factors that bind to these sequences, a question that needs to be addressed in future studies. In past studies IKKα was reported to interact with SMAD transcription factors to determine expression of Myc antagonists (33–35) and was shown to modulate methylation of histone proteins (17, 31, 54). Whether these mechanisms apply to lung ADC cells remains to be determined.
Overall, Ikkα ablation promotes KrasG12D-initiated lung ADC development in mice. In previous studies we showed that increased tumor-intrinsic ROS are associated with KrasG12DIkkαΔLU ADC progression (4). We now extend these results to show that Ikkα-deficient lung ADC generates an iTME by up-regulating Treg-cell induction and that ROS produced by intratumoral macrophage facilitate Treg differentiation through the TNF/TNFR2/c-Rel pathway. Given the present findings, tumor cell–intrinsic ROS may also contribute to iTME generation through potentiation of TNF signaling.
Materials and Methods
Mice, Human Tissue Array, and Cell Lines.
All mice used in this study were cared for in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the NIH. All animal experiments (Protocols 17-051 and 17-052) were approved by the IACUC. The following mice were of a C57BL/6 background: Ikkαf/f, Nox2−/− (stock No. 002365; The Jackson Laboratory), Rag1−/−, KrasLSL-G12D (KrasG12D, stock No. 008179; The Jackson Laboratory), Cd4−/− (stock No. 002663; The Jackson Laboratory), Tnf−/− (stock No. 003008; The Jackson Laboratory), Tnfr2−/− (stock No. 002620; The Jackson Laboratory), CD45.1 B6 (stock No. 002014; The Jackson Laboratory), and B6.Cg-Foxp3 sf/J (Foxp3-DTR-gfp/DEREG, stock No. 32050; The Jackson Laboratory). Human lung ADC tissue arrays (BCS04017a and HLug03PG02) were purchased from US Biomax, Inc. We used an A549 human lung ADC cell line carrying a KRASG12S mutation, a Kras-CL mouse lung ADC cell line carrying a KrasG12D mutation, and a KrasIKKαL mouse lung ADC cell line carrying a KrasG12D mutation, and multiple Ikkα mutations were used (4).
Antibodies.
For Western blotting and immunostaining, we used antibodies to: Lamin B (sc-6216), c-Rel (sc-6955), NOX2 (sc-5827), IKKα (sc-7183 and sc-7606), SMAD4 (sc-7154), TNFα (sc-1348), and normal mouse IgG (sc-2025) from Santa Cruz Biotechnology, Inc.; CD68 (M087629-2) from Dako Products; β-actin (A5441) from Sigma-Aldrich; α-tubulin (ab4074) and Foxp3 (ab10901) from Abcam; c-Rel (rabbit, No. 12707) and IKKβ (No. 2678) from Cell Signaling Technology; IKKα (No. NB100-56704) from Novus Biologicals; IKKγ (No. 559675) from BD Biosciences; and SMAD3 (ab28379) from Abcam.
Supplementary Material
Acknowledgments
This work was supported by funding from the NCI (ZIA BC011212 and ZIA BC011212) to Y.H., from NCI (U01AA027681 and R37AI043477) to M.K., and from the National Research Foundation of Korea (NRF-2016R1A5A2008630) to N.-Y.S.
Footnotes
Reviewers: Y.B.-N., Universidad Hebraica; S.G., Columbia University; and G.N., Istituto Europeo di Oncologia.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120956119/-/DCSupplemental.
Data Availability
Tumor-associated macrophage data have been deposited in National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) from mouse lung adenocarcinoma-associated macrophages (GSE114501). Anonymized tumor-associated CD4 T cells and ChIP-seq data have been deposited in GEO from mouse lung adenocarcinoma-associated CD4 cells (GSE122419 and GSE132460). All other study data are included in the article and/or supporting information.
References
- 1.Reck M., Rabe K. F., Precision diagnosis and treatment for advanced non-small-cell lung cancer. N. Engl. J. Med. 377, 849–861 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Brahmer J., et al. , Nivolumab versus Docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia L., Liu Y., Wang Y., PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: Current status and future directions. Oncologist 24 (suppl. 1), S31–S41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song N. Y., et al. , IKKα inactivation promotes Kras-initiated lung adenocarcinoma development through disrupting major redox regulatory pathways. Proc. Natl. Acad. Sci. U.S.A. 115, E812–E821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray Z., Shi G., Wang X., Hu Y., Macrophage inducible nitric oxide synthase promotes the initiation of lung squamous cell carcinoma by maintaining circulated inflammation. Cell Death Dis. 9, 642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network, Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavdoula E., et al. , CHUK/IKK-α loss in lung epithelial cells enhances NSCLC growth associated with HIF up-regulation. Life Sci. Alliance 2, e201900460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todoric J., et al. , Stress-Activated NRF2-MDM2 Cascade Controls Neoplastic Progression in Pancreas. Cancer Cell 32, 824–839.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su H., et al. , Cancer cells escape autophagy inhibition via NRF2-induced macropinocytosis. Cancer Cell 39, 678–693.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N., et al. , Loss of acinar cell IKKα triggers spontaneous pancreatitis in mice. J. Clin. Invest. 123, 2231–2243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park E., et al. , Reduction in IkappaB kinase alpha expression promotes the development of skin papillomas and carcinomas. Cancer Res. 67, 9158–9168 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Ward J. P., Gubin M. M., Schreiber R. D., The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv. Immunol. 130, 25–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi N. S., et al. , Regulatory T cells in tumor-associated tertiary lymphoid structures suppress anti-tumor T cell responses. Immunity 43, 579–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorsson V., et al. ; Cancer Genome Atlas Research Network, The immune landscape of cancer. Immunity 48, 812–830.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Hu Y., Attribution of NF-κB activity to CHUK/IKKα-involved carcinogenesis. Cancers (Basel) 13, 1411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DuPage M., Dooley A. L., Jacks T., Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Z., et al. , The pivotal role of IKKα in the development of spontaneous lung squamous cell carcinomas. Cancer Cell 23, 527–540 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lahl K., et al. , Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 204, 57–63 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galdino N. A. L., et al. , Depletion of regulatory T cells in ongoing paracoccidioidomycosis rescues protective Th1/Th17 immunity and prevents fatal disease outcome. Sci. Rep. 8, 16544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grinberg-Bleyer Y., et al. , NF-κB c-Rel is crucial for the regulatory T cell immune checkpoint in cancer. Cell 170, 1096–1108.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh H., et al. , An NF-κB transcription-factor-dependent lineage-specific transcriptional program promotes regulatory T cell identity and function. Immunity 47, 450–465.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraaij M. D., et al. , Induction of regulatory T cells by macrophages is dependent on production of reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 107, 17686–17691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Neriah Y., Karin M., Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12, 715–723 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Chen X., Oppenheim J. J., Contrasting effects of TNF and anti-TNF on the activation of effector T cells and regulatory T cells in autoimmunity. FEBS Lett. 585, 3611–3618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S., Wang J., Brand D. D., Zheng S. G., Role of TNF-TNF receptor 2 signal in regulatory T cells and its therapeutic implications. Front. Immunol. 9, 784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langowski J. L., et al. , IL-23 promotes tumour incidence and growth. Nature 442, 461–465 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Baird A. M., et al. , IL-23 is pro-proliferative, epigenetically regulated and modulated by chemotherapy in non-small cell lung cancer. Lung Cancer 79, 83–90 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Yim V., et al. , Synthesis and biological evaluation of analogues of the potent ADAM8 inhibitor cyclo(RLsKDK) for the treatment of inflammatory diseases and cancer metastasis. Bioorg. Med. Chem. 24, 4032–4037 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Conrad C., et al. , ADAM8 in invasive cancers: Links to tumor progression, metastasis, and chemoresistance. Clin. Sci. (Lond.) 133, 83–99 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Anest V., et al. , A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature 423, 659–663 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Zhu F., et al. , IKKalpha shields 14-3-3sigma, a G(2)/M cell cycle checkpoint gene, from hypermethylation, preventing its silencing. Mol. Cell 27, 214–227 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Liu B., et al. , IKKalpha is required to maintain skin homeostasis and prevent skin cancer. Cancer Cell 14, 212–225 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Descargues P., et al. , IKKalpha is a critical coregulator of a Smad4-independent TGFbeta-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 105, 2487–2492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinari B., et al. , The tumor suppressor activity of IKKalpha in stratified epithelia is exerted in part via the TGF-beta antiproliferative pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 17091–17096 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu F., et al. , NLRP3 inhibition ameliorates severe cutaneous autoimmune manifestations in a mouse model of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy-like disease. J. Invest. Dermatol. 141, 1404–1415 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cancer Genome Atlas Research Network, Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meylan E., et al. , Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature 462, 104–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Y., et al. , Reduced cell proliferation by IKK2 depletion in a mouse lung-cancer model. Nat. Cell Biol. 14, 257–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senftleben U., et al. , Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 293, 1495–1499 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Hu Y., et al. , IKKalpha controls formation of the epidermis independently of NF-kappaB. Nature 410, 710–714 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Hu Y., et al. , Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science 284, 316–320 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Wang X., et al. , Macrophage inducible nitric oxide synthase circulates inflammation and promotes lung carcinogenesis. Cell Death Discov. 4, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cannarile M. A., et al. , Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer 5, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anz D., et al. , Suppression of intratumoral CCL22 by type i interferon inhibits migration of regulatory T cells and blocks cancer progression. Cancer Res. 75, 4483–4493 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Scheu S., Ali S., Ruland C., Arolt V., Alferink J., The C-C chemokines CCL17 and CCL22 and their receptor CCR4 in CNS autoimmunity. Int. J. Mol. Sci. 18, 2306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelzer C., Thome M., IKKα takes control of canonical NF-κB activation. Nat. Immunol. 12, 815–816 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Lawrence T., Bebien M., Liu G. Y., Nizet V., Karin M., IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature 434, 1138–1143 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Vreka M., et al. , IκB kinase α is required for development and progression of KRAS-mutant lung adenocarcinoma. Cancer Res. 78, 2939–2951 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu B., et al. , A critical role for I kappaB kinase alpha in the development of human and mouse squamous cell carcinomas. Proc. Natl. Acad. Sci. U.S.A. 103, 17202–17207 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia X., et al. , Reduction of IKKalpha expression promotes chronic ultraviolet B exposure-induced skin inflammation and carcinogenesis. Am. J. Pathol. 176, 2500–2508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Page A., et al. , IKKbeta leads to an inflammatory skin disease resembling interface dermatitis. J. Invest. Dermatol. 130, 1598–1610 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Page A., et al. , IKKβ overexpression leads to pathologic lesions in stratified epithelia and exocrine glands and to tumoral transformation of oral epithelia. Mol. Cancer Res. 9, 1329–1338 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Tetreault M. P., et al. , Esophageal expression of active IkappaB kinase-beta in mice up-regulates tumor necrosis factor and granulocyte-macrophage colony-stimulating factor, promoting inflammation and angiogenesis. Gastroenterology 150, 1609–1619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu F., et al. , Autoreactive T cells and chronic fungal infection drive esophageal carcinogenesis. Cell Host Microbe 21, 478–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tumor-associated macrophage data have been deposited in National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) from mouse lung adenocarcinoma-associated macrophages (GSE114501). Anonymized tumor-associated CD4 T cells and ChIP-seq data have been deposited in GEO from mouse lung adenocarcinoma-associated CD4 cells (GSE122419 and GSE132460). All other study data are included in the article and/or supporting information.