Significance
Hypothalamic oscillators that generate pulsatile patterns of hormone secretion represent a fundamental physiological feature regulating homeostatic systems. How individual cells within these neural ensembles generate and coordinate episodic activity and resultant pulse secretion is unknown. Recently, arcuate KNDy (kisspeptin/neurokinin B/dynorphin) cells were identified as a critical component of the gonadotrophin-releasing hormone (GnRH) pulse generator required for reproduction. Using in vivo calcium imaging of KNDy neurons in freely moving mice, we reveal that, prior to each GnRH pulse, individual KNDy cells demonstrate synchronized activity with striking temporal order, with subsets of cells behaving as “leaders” or “followers.” Future work to distinguish these novel subpopulations and define mechanisms underlying the temporal ordering of cellular synchronization may provide avenues to regulate pulse secretion.
Keywords: fertility, luteinizing hormone, KNDy, kisspeptin, pulses
Abstract
A hypothalamic pulse generator located in the arcuate nucleus controls episodic release of gonadotropin-releasing hormone (GnRH) and luteinizing hormone (LH) and is essential for reproduction. Recent evidence suggests this generator is composed of arcuate “KNDy” cells, the abbreviation based on coexpression of kisspeptin, neurokinin B, and dynorphin. However, direct visual evidence of KNDy neuron activity at a single-cell level during a pulse is lacking. Here, we use in vivo calcium imaging in freely moving female mice to show that individual KNDy neurons are synchronously activated in an episodic manner, and these synchronized episodes always precede LH pulses. Furthermore, synchronization among KNDy cells occurs in a temporal order, with some subsets of KNDy cells serving as “leaders” and others as “followers” during each synchronized episode. These results reveal an unsuspected temporal organization of activation and synchronization within the GnRH pulse generator, suggesting that different subsets of KNDy neurons are activated at pulse onset than afterward during maintenance and eventual termination of each pulse. Further studies to distinguish KNDy “leader” from “follower” cells is likely to have important clinical significance, since regulation of pulsatile GnRH secretion is essential for normal reproduction and disrupted in pathological conditions such as polycystic ovary syndrome and hypothalamic amenorrhea.
Reproduction in mammals depends on a hypothalamic pulse generator that regulates the episodic release of gonadotropin-releasing hormone (GnRH) from the hypothalamus (1, 2). Regulation of the frequency and amplitude of GnRH pulses, and, in turn, that of the gonadotropins luteinizing hormone (LH) and follicle-stimulating hormone from the anterior pituitary gland, is essential for steroid hormone production and gamete development at the gonads. Although the first observations of the pulsatile nature of GnRH and LH release were made in the 1970s, until recently the precise location and cellular identity of the neural pulse generator responsible for episodic GnRH release remained a major unanswered question.
The 2003 discovery that mutations in the gene encoding the kisspeptin receptor (G protein-coupled protein 54 [GPR54]) result in hypogonadotropic hypogonadism delivered compelling evidence that kisspeptin-positive cells in the brain are required for maintaining GnRH release (3, 4). Subsequent studies testing the role of kisspeptin in animal models confirmed activation of GnRH neurons via GPR54 to potently stimulate GnRH and LH release (5, 6). Multilabeling experiments in sheep later revealed that kisspeptin cells in the arcuate nucleus of the hypothalamus (ARC) also coexpressed two other important mediators of GnRH release, the tachykinin neurokinin B (NKB) and the endogenous opioid peptide dynorphin (7); as an abbreviation, these cells were termed KNDy (kisspeptin/neurokinin B/dynorphin) neurons. Colocalization of KNDy peptides was subsequently demonstrated in the mouse, rat, cow, goat, and nonhuman primate (8–12). Anatomical characterization of KNDy neurons revealed reciprocal connections and the expression of postsynaptic receptors for NKB and dynorphin, indicating KNDy cells form an interconnected population potentially capable of synchronization (9, 13–16). These characteristics provided the basis for the “KNDy hypothesis” of GnRH pulse generation, in which NKB acts as the signal responsible for pulse onset by triggering activation of reciprocally connected KNDy neurons and driving the kisspeptin-mediated secretion of GnRH (13, 17–20). In support of this, bilateral infusions of NKB or dynorphin antagonists into the mediobasal hypothalamus enhances and suppresses LH pulsatile release, respectively, in sheep and goats (12, 21). Further, the conditional inhibition and brief activation of kisspeptin-expressing cells in the ARC using optogenetic tools in mice suppresses and elicits LH pulses, respectively (22, 23). Finally, in vivo measurement of KNDy neuron population activity using GCaMP6 fiber photometry in awake and freely moving mice revealed transient increases in intracellular calcium by KNDy neurons before an LH pulse, indicative of episodic activity within the KNDy neuron population that drives pulsatile LH release (22). However, as these studies either manipulate or record the activity of large proportions of the KNDy population, they do not identify individual cells which are responsible for GnRH/LH pulse generation, or whether those cells are activated homogeneously during an individual pulse. To address these questions, we conducted in vivo calcium imaging using miniature microscopy in order to visualize KNDy cell activity at the single-cell level in freely behaving mice, and, combined this with serial blood sampling, to examine the pattern of activation of KNDy cells during an LH pulse. Using this imaging approach, we provide in vivo evidence that directly demonstrates the synchronization among individual KNDy cells that was first theorized by the KNDy hypothesis and demonstrate that synchronized activation always precedes an individual LH pulse. Unexpectedly, we also found that not all KNDy cells are activated simultaneously during a pulse; rather, KNDy cells are recruited in a recurring temporal order composed of distinct subpopulations of “leader” cells, which activate and reach peak activity at the onset of synchronized episodes, and “follower” cells, which reach peak activity during either the maintenance or termination phase of the episode. These results provide a major advance to our understanding of oscillatory neuroendocrine systems by demonstrating that LH pulse secretion is preceded by highly synchronized activity among individual KNDy cells in an in vivo setting. In addition, the existence of temporally defined subpopulations of “leader” and “follower” cells in each synchronized event is strikingly similar to results described recently in pancreatic beta cells (24), suggesting that this temporal organization may be a common feature of endocrine pulse generators in the brain as well as in the periphery.
Results
Confirmation of GCaMP6 Expression in ARC Kisspeptin Cells.
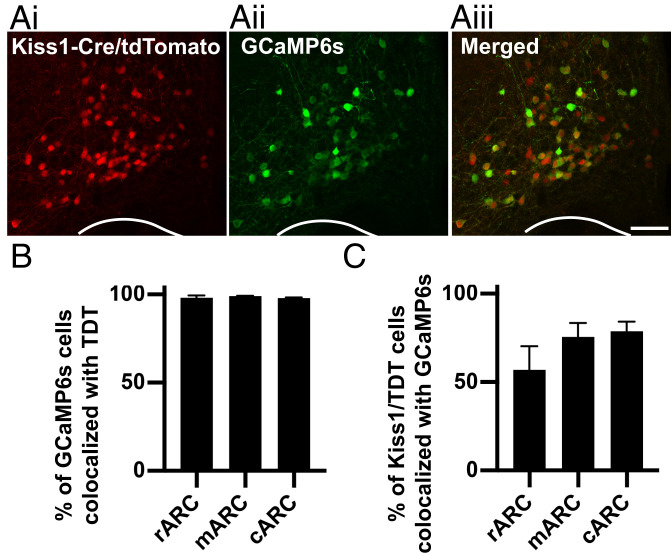
The viral vector used in this study (AAV9-CAG-Flex-GCaMP6s) contains the genetically encoded calcium sensor MP6s (GCaMP6s) and has been previously characterized as highly specific and selective to transfection of kisspeptin cells when injected into the ARC of Kiss1-Cre mice (22). The viral titer was diluted in this study to elongate the experimental timeline without toxicity to cells due to long-term transfection. To confirm selectivity of the viral vector to ARC kisspeptin cells, we unilaterally injected the Cre-dependent viral vector into the ARC of Kiss1-Cre/tdTomato reporter mice (Fig. 1A). We show here that 98.4 ± 0.5% of GCaMP6s-expressing cells were colocalized with tdTomato-expressing ARC kisspeptin cells, indicating, as in previous reports, the AAV (adenoassociated virus) is highly specific to Cre-positive cells (Fig. 1B, n = 3). Conversely, 70.5 ± 8.1% of tdTomato-expressing kisspeptin cells expressed GCaMP6s, showing over two-thirds of the KNDy population expresses the transgene (Fig. 1C, n = 3). No GCaMP6s-positive cells were detected in wild-type mice, consistent with Cre’s being required for transfection (n = 3).
Fig. 1.
Viral delivery of the calcium indicator GCaMP6s is highly specific to arcuate kisspeptin neurons. (A) Representative projected confocal images of the middle arcuate nucleus containing Kiss1-Cre cells expressing the tdTomato fluorophore (i, red), cells transfected with a Cre-dependent viral vector containing the calcium indicator GCaMP6s (ii, green), and merged images illustrating the high degree of colocalization between kisspeptin cells and GCaMP6s (iii). (B and C) Histograms depicting the percentage of GCaMP6-positive cells that are colocalized with Kiss1-Cre/tdTomato throughout the rostrocaudal extent of the ARC: in the rostral (r), middle (m), and caudal (c) portions (B) and the percentage of tdTomato-expressing kisspeptin cells that express GCaMP6s. (Scale bar, 100 µm.)
The KNDy Population at a Single-Cell Level Exhibits Synchronized Increases in Intracellular Calcium.
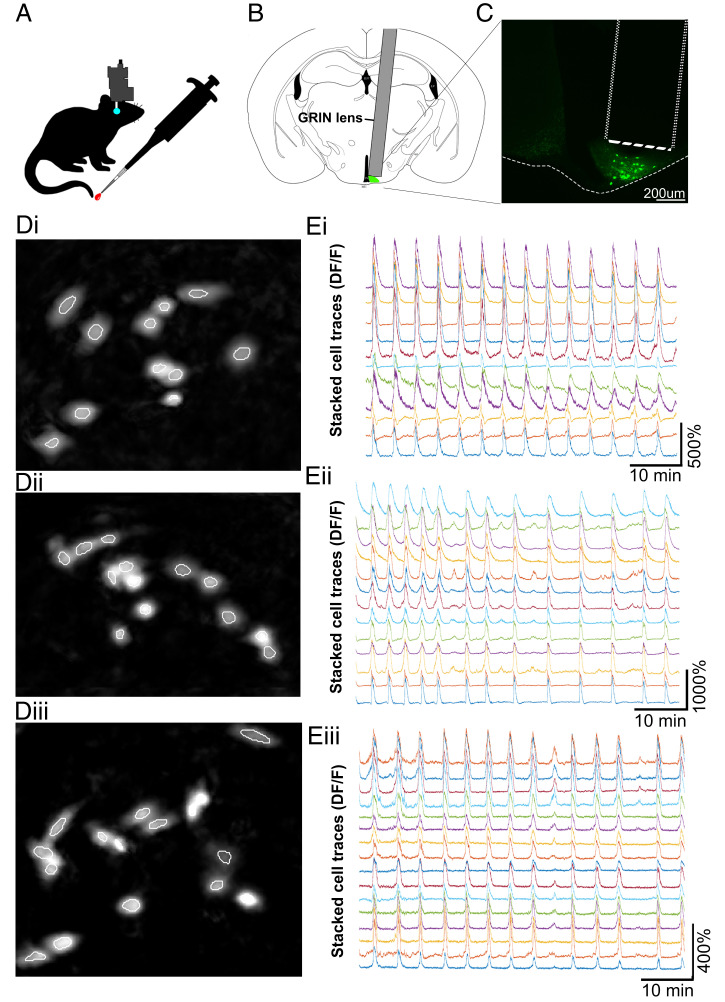
For in vivo calcium imaging experiments, Kiss1-Cre female mice were bilaterally ovariectomized (OVX) to remove inhibition of LH pulse generation by gonadal steroid hormone negative feedback and underwent Cre-dependent AAV transfection of ARC kisspeptin (KNDy) cells with GCaMP6s. To enable optical access to GCaMP6s-expressing KNDy cells, a 500-μm-diameter, 8.4-mm-gradient index (GRIN) lens was positioned directly above the ARC and secured via an integrated baseplate. After 5 wk of daily handling and habituation, unanesthetized and freely moving OVX Kiss1-GCaMP6s females underwent 60-min sessions in which calcium fluorescent signal was visualized and recorded at 10 Hz via attachment of a single-photon miniaturized microscope (nVoke; Inscopix Inc.) to the integrated baseplate, while tail-tip blood samples were collected every 3 to 4 min for LH pulse detection (n = 5, Movie S1A and Fig. 2 A–C). Histological examination at the completion of recording in perfusion-fixed tissue revealed that the majority of cells recorded were located within the middle region of the ARC. Fluorescent traces were extracted and analyzed from an average of 13 ± 1.6 cells per animal using the principal component analysis (PCA)–individual component analysis (ICA) cell detection algorithm (Movie S1 B and C and Fig. 2D). The average reduction in baseline fluorescence between the first and final 100 s of baseline activity was 1.8 ± 1.4%, suggesting little to no photobleaching of cells occurred within the 60-min recording period.
Fig. 2.
In vivo calcium imaging of KNDy cells in awake and freely moving mice reveals highly synchronized episodic activity at the single-cell level. (A) Ovariectomized female Kiss1-Cre mice with GCaMP6s expressed by ARC kisspeptin (KNDy) cells underwent 60 min of in vivo imaging of calcium activity in the arcuate nucleus using a microendoscope (Inscopix Inc.). Regular blood samples were collected from the tail tip for LH pulse analysis. Schematic representation (B) and fluorescent image (C) of GRIN lens placement above GCaMP6s-expressing KNDy cells. (D, i–iii) Images of KNDy cells from representative animals generated following segmentation by the PCA-ICA algorithm (white outlines). (E, i–iii) Representative traces of calcium activity extracted from individual KNDy cells identified using PCA-ICA in D, i–iii revealing synchronized and episodic changes in fluorescence (DF/F) in awake and freely moving mice.
Visualization of KNDy neurons at the single-cell level revealed episodes of elevated calcium fluorescence across the recording period with an interval of 4.1 ± 0.7 min (Fig. 2E). No observations of reduced KNDy cell activity were recorded. The average increase in fluorescent intensity relative to resting fluorescent intensity (DF/F) during episodes of activity in KNDy cells across animals was 263.98 ± 63.7% but ranged from 117.7 to 1211.9% between individual cells. To normalize for differences in DF/F between cells, analysis was henceforth conducted on z-scored cell traces, which was generated by calculating the SD of each recorded timepoint from the mean fluorescence of the cell.
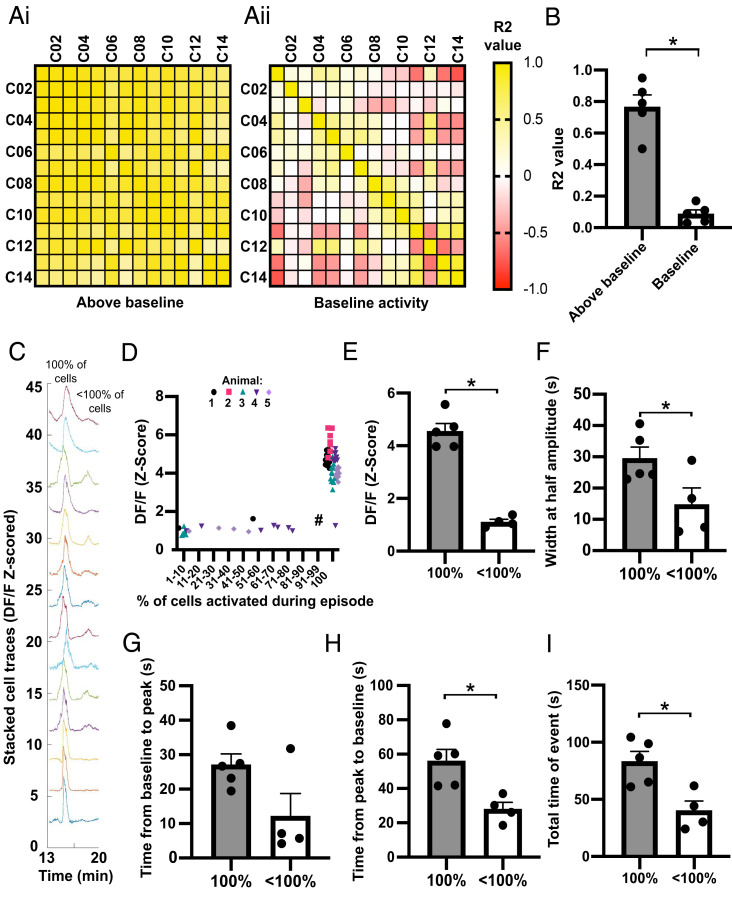
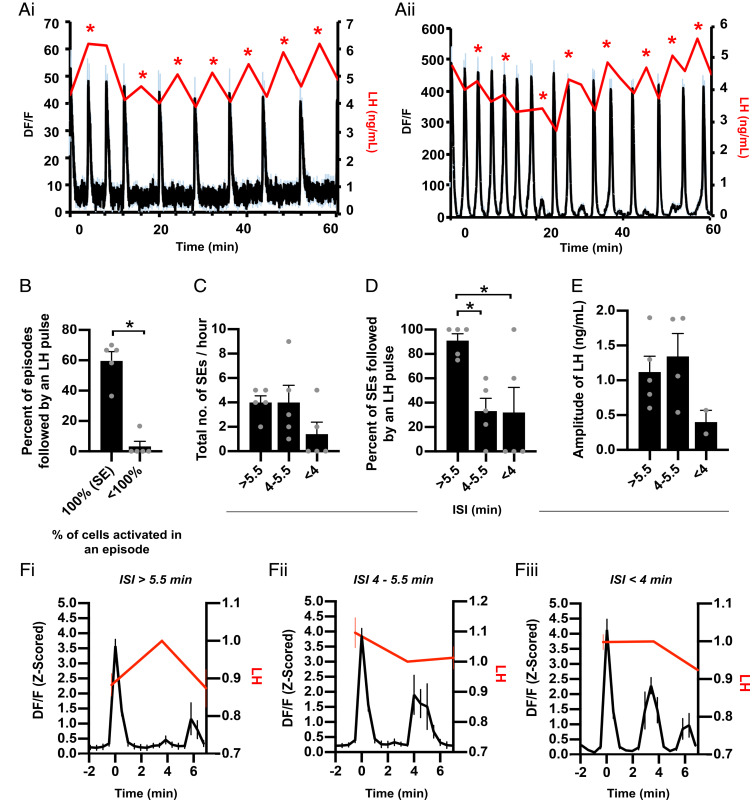
Correlation analysis of single-cell activity showed that although there is little to no correlation between individual cells at baseline activity there was a strong and significant increase in correlation between cells when the average activity of the population was above baseline (Fig. 3 A and B, P < 0.05). In support of this high correlation during episodic activity, we detected that the majority of episodes (75.2 ± 5.7%) were the result of an increase in calcium signal by all recorded KNDy cells, representative of synchronized activation of the KNDy neuronal network and referred to henceforth as synchronized episodes (SEs). In addition to SEs, episodes were also detected in which smaller percentages of the recorded KNDy cells (between 7.1 and 71.4%) showed episodes of elevated fluorescence, indicative of heterogenous KNDy cell activity (Fig. 3C). To compare whether full versus partial recruitment of the KNDy population affected the strength and duration of network activation, the amplitude at the peak of an episode, the width of the episode at half amplitude, and the time that calcium fluorescence was above baseline was compared between SEs and episodes with only subpopulations of cells displaying activation. The average amplitude at the peak of cell activation was dramatically and significantly higher in KNDy cells during an SE compared to episodes in which only partial activation of the KNDy neuronal population was achieved (Fig. 3 D and E, P < 0.05). Surprisingly, a positive linear relationship was not detected between the percentage of cells activated versus the amplitude at episode peak (R2 = 0.0251). Instead, except for one episode (marked # in Fig. 3D), the average amplitude of the episode peak remained low unless all cells were synchronously activated. The width (s) at half amplitude, a measure of the duration of the episode, was also significantly higher during SEs compared to episodes where subpopulations were activated (P < 0.05, Fig. 3F). The time for cells to reach peak amplitude from baseline was not significantly different between SEs and episodes with fewer than 100% of cells active (Fig. 3G). However, the time from peak amplitude to return to baseline was significantly higher during SEs compared to episodes with fewer than 100% of cells active (Fig. 3H, P < 0.05). Overall, the total time above baseline was significantly higher during SEs compared to episodes with fewer than 100% of the recorded cells showing activity (Fig. 3I, P < 0.05). Together, these data indicate that recruitment of all cells during an SE result in KNDy cell activity with both higher amplitude and longer duration.
Fig. 3.
The KNDy neuron population displays either large SEs of activation by all cells or smaller episodes of activation by subgroups of cells. (A) Matrices for a representative mouse showing the correlation of activity (R2) between individual cells (C01–C14) when the average calcium activity for the population is above (i) or at (ii) baseline. (B) The mean ± SEM. R2 based on all animals (n = 5) is significantly higher when activity is elevated above baseline. (C) Representative fluorescent traces from individual cells during an SE in which 100% of cells are robustly activated, followed by lower amplitude activity by less than 100% of cells. (D) Graph demonstrating the amplitude of episodes (Z-scored DF/F) remains low unless 100% of cells are activated. Each data point represents an episode in each of the animals (74 episodes from five animals). (E–I) Graphs demonstrating the mean ± SEM (n = 5) amplitude of episodes per animal (E) and the mean ± SEM width of an episode at half amplitude (F) is significantly higher when 100% of cells are activated compared to less than 100% of cells. The mean ± SEM time from baseline to peak is not significantly dependent on the percentage of cells activated (G); however, the time from peak to baseline (H) and the total time above baseline (I) is significantly higher when 100% of cells are activated in an episode compared to less than 100%. *P < 0.05.
KNDy Neurons Display a Temporal Order of Activation and Peak Amplitude during an SE.
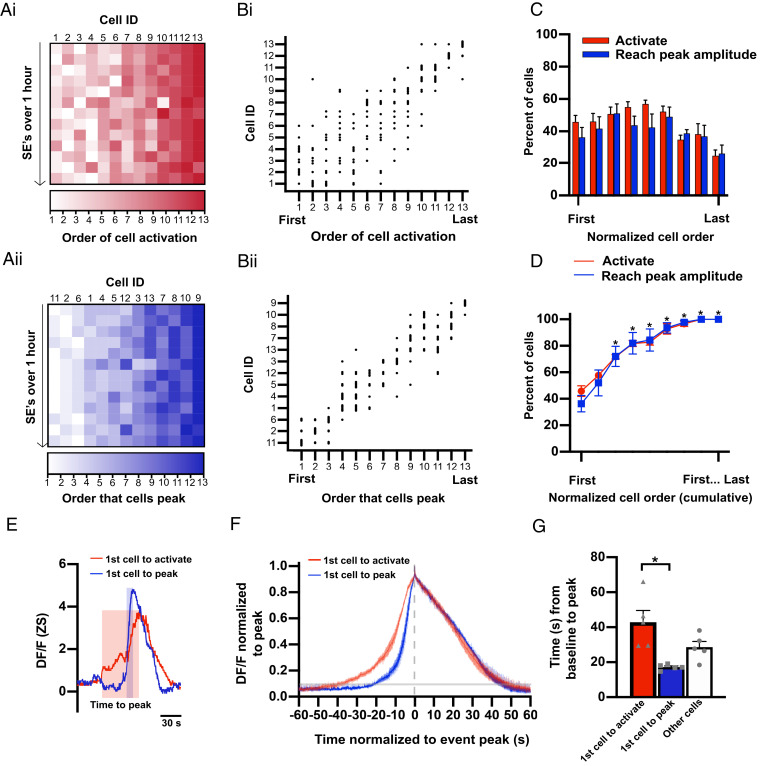
Although activity among KNDy cells was highly coordinated, the average time for all cells to activate (defined as when individual cell fluorescence elevates above baseline) in an episode was 31.28 ± 8.04 s. Similarly, the average interval between the first and last cell to reach peak amplitude was 23.51 ± 4.13 s. Within an SE, KNDy cells appeared to show sequential activity, with an average interval of 2.72 ± 0.65 s elapsing between the activation of each cell and 2.32 ± 0.63 s between the peak amplitude of each cell. Therefore, we next aimed to determine whether the sequential activity among KNDy cells occurred in a predictable temporal pattern. To achieve this, we determined the order that 1) cells were activated (Fig. 4 A, i and B, i) and 2) cells reached peak amplitude (Fig. 4 A, ii and B, ii) within each SE generated during the recording period of all animals. Across the average of 11.2 ± 0.9 SEs recorded in 60 min, 45.9 ± 4.1% of the recorded KNDy cells displayed at least one instance of activating first during an SE. Cells that were activated first did so on multiple SEs, on average 2.2 ± 0.3 times out of the 11.2 ± 0.9 SEs (or 19.8 ± 0.9% of SEs) (Fig. 4C). Likewise, we found that 36.6 ± 6.1% of recorded KNDy cells displayed at least one instance of reaching peak amplitude first. Cells that peaked first would do so on an average of 3.1 ± 0.5 out of 11.2 ± 0.9 episodes (or 26.72 ± 3.2% of SEs) (Fig. 4C). These data indicate the presence of a potential “leader cell” subpopulation, in which cells take turns to repeatedly activate or reach peak amplitude. Next, we aimed to assess the order cells in the leader subgroup activate and reach peak amplitude when not leading the generation of an SE. To achieve this, we quantified the cumulative percentage of the cell population that was recruited (e.g., the percentage of KNDy cells that activated or peaked first or second; first, second, or third; etc.) (Fig. 4D). Up until the point that 33.3% of the total KNDy cell population had activated or reached peak amplitude, the size of the KNDy neuron population recruited was not significantly different from the size of the cell population that activated or peaked first. This indicates that when cells with a recorded instance of activating or peaking first are not leading an episode they will still activate or peak early within an SE prior to two-thirds of the remaining population. An example of this can be seen in Fig. 4 B, ii, where KNDy cells that peak first will, on alternate episodes, reach peak amplitude either second, third, or fourth. These data indicate that KNDy cells are segregated into “leader” cells that are temporally restricted to exhibit activity early within an SE, and “follower” cells that are temporally restricted to activity later within an SE. Consistent with this, only 24.9 ± 3.6% and 26.2 ± 5.3% of the recorded KNDy population exhibited at least one instance where they were last to activate or peak during the hour-long recording window, respectively (Fig. 4C). Therefore, cells that were last to activate or peak in an SE did so 5.1 ± 0.9 and 5.4 ± 1.3 times out of 11.2 ± 0.9 episodes during the 60-min recording. Only 5.0 ± 5.6% and 2.5 ± 2.8% of cells that activated or peaked first at least once during the recording period also displayed an instance of activating or peaking last, supporting that leader cells that begin an event and follower cells that terminate an event represent distinct subpopulations.
Fig. 4.
KNDy cells activate and peak in a predictable temporal order during an SE. (A) Heat map from a representative animal depicting the order that fluorescence in individual cells (Cell ID, x axis) elevates above baseline (i, activation) and reaches peak amplitude (ii) during SEs (y axis, 13 SEs in 60 min). The cell IDs in i correspond with the cell IDs in ii. (B) Scatterplot mapping the order in which individual cells activate (i) and reach peak amplitude (ii) from the animal in A. (C and D) Graphs demonstrating the mean ± SEM percentage (C) and the mean ± SEM cumulative percentage (D) of KNDy cells per animal (n = 5) that activate or peak in order of first to last over multiple SEs in a 60-min recording. The asterisk in D indicates a significant increase compared to the percent of cells that activate or peak first. (E) Calcium traces from representative cells that activate first (red) and peak first (blue) during a single SE. The shaded boxes depict the time from baseline to peak for the cell that activates first (red box) and the cell that peak first (blue box). (F) The mean ± SEM calcium fluorescence (normalized to peak) per animal (n = 5) of cells that activate first versus cells that peak first, graphed with time 0 as the peak of fluorescent traces (gray dotted line). The shaded line on the y axis depicts the average point at which cell activity is above baseline. (G) Graph illustrating the mean ± SEM time from baseline to peak amplitude per animal (n = 5) for cells that activate first in an SE is significantly longer compared to cells that reach peak amplitude first. *P < 0.05.
Finally, we examined whether the temporal order of cell activity was consistent between SEs generated by the same leader cell. In 61.4 ± 10.5% of SEs in which the same cell was the first to activate, the cell to reach peak amplitude first was also the same. Similarly, in 63.0 ± 6.9% of SEs in which the same cell was first to reach peak amplitude, the cell next in order to reach peak amplitude was identical. These data indicate that in the majority of, but not all, episodes a set order of cell activity occurs when an SE is generated by a common leader cell. The probability that the temporal order in which cells reached peak amplitude within the nonleader, or putative “follower,” population during the maintenance phase (cells not active first or last in an episode) appeared weaker, with an average of 53.0 ± 2.0% of SEs showing the same cell-to-cell order of activity. However, in 83.2 ± 9.3% of SEs, the temporal order at the termination of the episode was identical, indicating that the cell that reaches peak amplitude at the termination of an event is highly predictable. We also aimed to determine whether the interval of time between cell activity was consistent within SEs generated by the same leader cell. We quantified that, despite the common temporal order, there was no significant difference in the range or SD of variation (STDEV) between the interval of SEs with a common cell-to-cell order (range = 1.99 ± 0.27 s, STDEV = 0.73 ± 0.15 s) versus all SEs (range = 2.76 ± 0.12 s, STDEV = 0.95 ± 0.26) in the recording period (P > 0.05). Together, these data indicate that KNDy cells are temporally restricted to activity either early in an SE (leader cells) or during the maintenance or termination phase of an SE (follower cells). Within these subpopulations, some variation in the precise temporal order and time of signal transmission occurs between episodes.
SEs Are Initiated by a Slowly Activating Population of Leader Cells.
Although the percentage of KNDy cells capable of activating first was not significantly different from the population of cells that peak first (Fig. 4 C and D, P > 0.05), only 9.1 ± 2.6% of SEs exhibited a cell that both activated first and reached peak amplitude first. Visual inspection of cell traces noted two patterns of activity that occurred between the baseline and peak of episodes. The first pattern would exhibit a rapid increase in fluorescence from baseline to peak, followed by a slower reduction in fluorescence to baseline (Fig. 4 E and F, blue line). The second pattern would exhibit a slower increase in fluorescence before reaching the peak of the episode (Fig. 4 E and F, red line). These two patterns were not observed within distinct cell subpopulations. Instead, cells could exhibit either pattern in different SEs. In line with these observations, cells that were first to activate during an SE took significantly longer, by 6.9 ± 1.5 s, to reach peak amplitude compared to the cells that reached peak amplitude first (Fig. 4G, P < 0.05).
LH Pulses Are Preceded by SEs of KNDy Cell Activity.
Serial blood samples were collected from mice throughout the 60-min recording periods to correlate KNDy population activity with LH pulsatile release (Fig. 5A). As expected in OVX female mice, a rapid frequency of 7.4 ± 0.9 LH pulses per hour was recorded. All LH pulses were preceded by KNDy neuron population activation, with an average interval of 3.1 ± 0.3 min lapsing between the peak amplitude of an LH pulse and the peak of the immediately preceding fluorescent episode. Although LH pulses faithfully followed KNDy neuron activation, only 49 ± 7.8% of all detected KNDy neuron activation episodes were followed by an LH pulse. Notably, LH pulses primarily followed the SEs, as 59.6 ± 6.1% of SEs (with 100% of cells activated) were followed by an LH pulse, while only one episode in which less than 100% of cells were activated (3.3 ± 3.3% of subpopulation episodes) preceeded LH pulsatile release (Fig. 5B). In addition, although the average interval of time between the peak of SEs (inter-SE interval, ISI) across animals was 5.3 ± 0.4 min, we compared LH output within a variety of ISIs. The majority of SEs had an ISI of over 4 min, although two animals exhibited episodes in which the ISI was more rapid (Fig. 5C). When analyzing LH pulsatile release after KNDy SEs at varying ISIs, we found that most SEs with an ISI of over 5.5 min elicited an LH pulse (Fig. 5D). When the ISI dropped beneath 5.5 min, the incidence of LH pulse generation was significantly reduced, indicating that the detection of an LH pulse depends on the ISI (Fig. 5 D and F, i–iii). Effects of ISI on LH pulse amplitude were also noted. The amplitude of LH release following SEs with an ISI of 4 to 5.5min was not significantly different compared to episodes with an ISI of over 5.5 min (Fig. 5E, P > 0.05). However, in the two animals that exhibited SEs with an ISI of under 4 min and some LH pulses were detected (Fig. 5D) the amplitude of the LH pulses was markedly lower compared to pulses following SEs with a larger ISI (Fig. 5E). Finally, no significant difference was detected in the amplitude of SEs that were immediately followed by an LH pulse versus the amplitude of SEs that were not followed by an LH pulse (4.31 ± 0.2 versus 4.48 ± 0.4 Z-scored DF/F, P = 0.7). Together, these data demonstrate that synchronized activation of the KNDy population precedes LH pulsatile release, but this relationship is not perfectly correlated when KNDy SEs are generated at high frequency.
Fig. 5.
LH pulses are generated by synchronized KNDy population activity with an interval of over 5 min. (A, i and ii) Representative examples from two animals of the mean (black line) ± SEM (blue error bars) change in fluorescence (DF/F) within the KNDy neuron population over a 60-min recording coupled with LH pulsatile release (red line). The asterisk depicts an LH pulse. (B) The percentage of episodes with KNDy cell activity followed by an LH pulse reveal episodes with less than 100% of cells recruited rarely elicit an LH pulse. (C) The number of SEs over a 60-min recording period that have an interval of over 5.5 min, 4 to 5.5 min, and under 4 min. (D) Graph summarizing that SEs with an ISI of over 5.5 min are followed by an LH pulse, whereas an ISI of under 5.5 min significantly reduces LH pulsatile release. (E) Graph illustrating that the amplitude of LH release is lower when a pulse is generated following a KNDy SE with an ISI of less than 4 min. (F) Fluorescent traces (peak at time 0) with an ISI of over 5.5 min (i), between 4 and 5.5 min (ii), and under 4 min (iii) plotted against LH that has normalized to the sample collected after the KNDy SE peak. Data in B–F are depicted as averages per animal and expressed as mean ± SEM; n = 5; *P < 0.05.
Discussion
Using in vivo calcium imaging at single-cell resolution in freely behaving female mice, we demonstrate here that ARC KNDy cells, proposed as the GnRH pulse generator, show synchronous activation of all recorded cells prior to pulsatile LH release. Strikingly, we identified that the order in which cells fire during an SE is temporally organized, revealing subsets of “leaders” and “followers” that may be critical for the initiation, maintenance, and termination of GnRH pulse generation. Furthermore, although past studies have provided evidence for presumed in vivo synchronization within episodic KNDy cell activity (22) our study provides visualization in freely behaving animals of KNDy cell synchronization at the level of individual neurons, and demonstration that their synchronized pattern of activity occurs before each LH pulse. Together with previous studies (22, 25), these data provide a critical additional piece of evidence supporting the role of KNDy cells as the GnRH/LH pulse generator and evidence for functional subsets of KNDy neurons that may dictate the initiation of activity within this reciprocally interconnected population.
Sixty-minute recordings of GCaMP6s fluorescent signal from the KNDy population at single-cell resolution revealed that activation of all recorded cells occurred in ∼75% of episodic events. These in vivo data validate anatomical (14, 16, 26) and in vitro electrophysiological data (25) which suggest KNDy cells form an interconnected network capable of synchronization. It further extends these observations by demonstrating that highly correlated synchronized activity occurs among all KNDy cells detected during an episode and always precedes an LH pulse. In most animals, we also detected episodes in which less than 100% of the cell population displayed activity. In these instances, the mean amplitude of the calcium signal across cells was significantly lower and not followed by LH release. Although this may indicate that complete synchronization of the KNDy population is necessary to drive LH secretion, this is not necessarily supported by previous reports. For instance, bilateral lesioning of KNDy cells can disrupt LH release (27–29), but the unilateral suppression of KNDy neurons using inhibitory optogenetics is insufficient to suppress LH pulsatile release from ovariectomized mice (22). Similarly, the knockdown of ∼30% of the KNDy cell population does not have an appreciable effect on LH pulse generation besides a slight reduction in LH pulse frequency (30). Combined with our results, this suggests that, in situ, all KNDy cells may participate in pulse generation, but not all cells may necessarily be required. Potentially, the larger amplitude recorded during endogenous SEs may be a critical factor for LH release. Currently, the role of smaller-amplitude episodic events from subsets of the KNDy population is unknown. It is possible that these episodes represent failed attempts to release the necessary signal, such as NKB, believed to be a start signal for each pulse (9, 12, 21, 31), that drives a full SE and LH release.
Another consideration is that our data were collected from cells primarily in the middle division of the ARC. It is possible that KNDy cells in more rostral or more caudal divisions may not show the same synchronized activity before an LH pulse. For instance, previous attempts to suppress LH pulsatile release using inhibitory optogenetics was successful in the middle and caudal ARC but not the rostral portion of the nucleus (22). Further, projections from the rostral half of the arcuate differ to projections from more caudal neurons (32). These data suggest that although we recorded from cells displaying synchronized activity consistent with pulse generation, future studies may identify anatomically and functionally distinct subpopulations in other ARC regions that do not demonstrate the episodic activity prior to LH release and instead are related to other functions of KNDy cells, such as their role in the modulation of metabolic responses and/or stress (33–36).
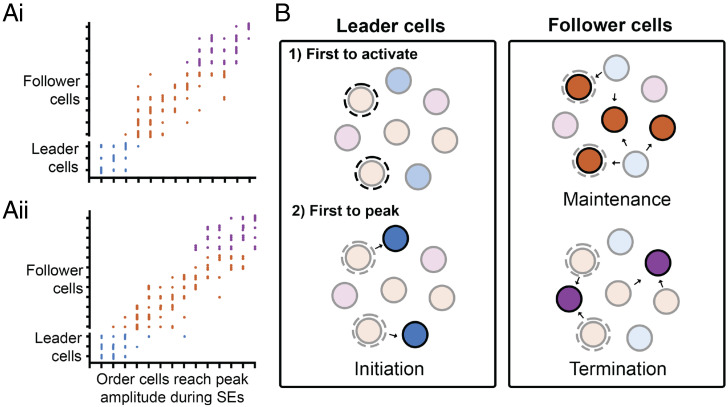
Intriguingly, we found KNDy cells display a temporal order of firing within each SE. Analysis across multiple SEs within 60-min recording periods from each animal revealed that a subpopulation of the recorded cells peaked first on multiple occasions. When not first, this subpopulation was still temporally restricted to reach peak amplitude early within an episode. Similarly, cells that peaked during either the midway or endpoint of the event were restricted to this time point across all SEs studied. This suggests the possibility that the KNDy population contain a subset of “leader” cells capable of driving the initiation phase of an episode and “follower” cells that only fire during the maintenance and termination phase. Analysis of SEs with the same leader cell revealed that the temporal order of activity was repeated in the majority of episodes, but not all. This suggests that although there is a hierarchy that divides KNDy cells into leader and follower cells, some plasticity within the precise coordination of synchronization may occur between episodes within these subpopulations. Currently, in vivo calcium imaging and therefore the analysis of a temporal order within the KNDy population is restricted to the imaging field of a GRIN lens. Therefore, KNDy cells not recorded in our analysis, including reciprocally connected contralateral KNDy cells (25), may be interspersed in the order we report here and contribute toward the later activation of follower cells. Recruitment of unrecorded cells during an SE may also explain the variation in the interval of time that we report between cells that have an identical order of activity.
Surprisingly, cells which were first to exhibit an increase in activity above baseline (activate) at the beginning of an episode were not the first to reach peak amplitude. Analysis of these two potential subpopulations of leader cells, cells first to activate and cells first to peak, confirmed that cells first to activate exhibited a slower “shoulder” period between baseline and peak amplitude, whereas cells that peaked first demonstrated a more rapid increase in calcium activity until peak amplitude was reached. Potentially, these distinct patterns of activity may indicate a model in which “activating” leader cells undergo a slow increase in activity toward a threshold that elicits the robust depolarization of a neighboring KNDy cell, and this activity is then propagated from cell to cell, as illustrated in Fig. 6. As the interval between the activation of KNDy cells during an SE was on the order of seconds, this supports in vitro observations that slow, peptidergic synaptic signaling by a neuropeptide such as NKB underlies synchronization of the population (25), as opposed to fast amino acid synaptic transmission which would have an expected time lag of milliseconds. The initial source of excitation for activating leader cells is currently unknown.
Fig. 6.
Predicted model for the temporal ordering of KNDy neuron activation during SEs. (A) Scatterplots mapping the order of cell activity during SEs from 60-min recordings from two representative animals. Each dot represents the order in which a cell reached peak amplitude during an SE. KNDy neurons have been divided into “leader” and “follower cells” depending on their order of activation across multiple SEs and color-coded into the categories shown in B. (B) Schematic depicting the predicted temporal activation of KNDy neurons that drives LH pulsatile release. First, a population of leader cells first initiate an LH pulse (1) by exhibiting an increase in activity toward threshold and driving activation of a subpopulation of leader cells that peak first (2, blue cells), which activates reciprocally connected follower cells. Follower cells are further divided into cells that peak during the maintenance (orange cells) and termination (purple cells) phase of the KNDy/GnRH/LH pulse.
The identification of leader cells within synchronized cell populations that drive episodic pituitary hormone output has remained a major unresolved issue in multiple neuroendocrine systems. For instance, oxytocin neurons of the hypothalamus display synchronized bursts of firing that lead to pulses of oxytocin release from the posterior pituitary gland (37, 38). Attempts using techniques such as paired electrophysiological recordings have been unable to determine leader and follower subpopulations (39, 40). However, a recent study exploring oscillatory beta cell calcium dynamics in the pancreas, thought to underlie the pulsatile secretion of insulin, using an ex vivo zebrafish preparation and in vivo engrafted mouse islets revealed temporally defined subpopulations of “leader” cells which, similar to what we have shown here, are capable of repeatedly leading calcium responses within a highly connected cell population (24). The demonstration of leader cells within peripheral endocrine organs together with our results showing a strikingly similar phenomena in KNDy neurons suggests leader populations may be a common feature within peripheral and central endocrine cell populations. Further, the ablation of leader cells in the zebrafish model prevented ongoing pan-islet signaling, whereas the ablation of follower cells had no effect. These data suggest that leader cell populations may be a necessary feature for the generation of pulsatile hormone secretion in oscillatory endocrine systems.
Using serial blood sampling from the tail tip during imaging, we found SEs with full activation of the KNDy cell population always preceded individual LH pulses. Surprisingly, and unlike reports using fiber photometry in male mice and intact female mice (22, 41, 42), we also detected SEs that were not followed by an LH pulse. Further analysis revealed that the vast majority of SEs with an interval between the peaks of SEs (ISIs) of over 5.5 min generated a detectable LH pulse, whereas SEs with a shorter ISI weakened this relationship. Prior fiber photometry recordings detected near-perfect correlation between LH pulsatile release and preceding KNDy population activity in gonadectomized males with an interval ranging between 4.5 and 20.7 min (22). However, our observation that rapid SEs were not followed by LH release may have two explanations. First, an LH pulse with an ISI below 5.5 min will be difficult to capture using a blood sampling interval of 3 min. Here, in ovariectomized female mice, SEs displayed an average ISI of 5.3 min and a range between 3 and 9 min. The surprisingly fast pace of the GnRH pulse generator in this study may result from long-term ovariectomy which, in these studies, was typically a 6- to 8-wk period before recording. Currently, we are unable to test whether a brief pulse of LH follows SEs with an interval of under 4 min, as collection of mouse tail blood samples more frequently than every 3 min in freely moving mice is currently not technically feasible over the length of time required for multiple SEs to occur. Alternatively, high-frequency GnRH pulses may desensitize the pituitary to GnRH, leading to reduced or absent LH secretion. This is supported by historical pulse generation studies in OVX rhesus monkeys which demonstrated that using exogenous delivery of GnRH to increase the physiological frequency of GnRH stimulation of the pituitary gland from one pulse an hour to two, three and five pulses an hour resulted in a gradual decline in LH output (43). Similar results have been observed in perifused sheep pituitary cells, in which exogenous delivery of GnRH pulses more rapidly than every 16 min also reduced LH release (44).
A number of interesting questions arise from the results reported here. What are cellular and/or molecular features that distinguish “leader” from “follower” KNDy cells? Which signals are responsible for synchronization of “leader” cells that are first to activate versus those which are first to peak? Do manipulations of either “leader” or “follower” cells result in altered profiles of GnRH release and downstream effects on pituitary gonadotropin secretion? As the current studies were conducted in ovariectomized mice, it will also be of interest to investigate whether steroid hormone replacement exerts negative feedback upon GnRH/LH pulsatile secretion by reducing the activity of KNDy cells. Further, does the identity of “leader” cells remain consistent under different steroid hormone feedback conditions, and is the subsequent recruitment of cells during SEs altered by the level of steroid hormone feedback? Given the fundamental role that the GnRH pulse generator plays in control of reproduction, identification of the mechanisms responsible for temporal ordering of KNDy cell synchronization during a pulse may have important translational relevance for understanding reproductive disease as well as normal function. In the clinic, LH pulse frequency outside the physiological norm leads to significant deficits in reproductive capacity in both male and female patients, manifesting in disorders such as hypothalamic amenorrhea, in which LH pulses are less frequent or absent (45), and polycystic ovary syndrome (PCOS), in which LH pulse frequency is increased due to impaired steroid hormone feedback at afferent GnRH populations (46). In both of these disorders, changes in LH pulse frequency disrupt follicular maturation and appropriate steroid hormone secretion, leading to infertility (46). Impairments to kisspeptin and KNDy cells have been investigated as an underlying cause for altered LH pulse secretion in preclinical models (47, 48), and KNDy peptide antagonists have already been intensely studied as therapeutic tools in the treatment of these disorders (49–51). Further characterization of leader versus follower cells, and the signaling pathways controlling their synchronization, may provide a basis for development of new therapeutics to either trigger or inhibit KNDy pulses and novel approaches to the control of reproduction in fertility disorders such as PCOS.
Materials and Methods
Animals.
All mice were bred and housed in the Kent State University animal facility on a 12-h light/dark cycle and given access to food and water for ad libitum intake prior to calcium imaging experiments. Experimental procedures in mice were conducted from 50 d of age. All experimental protocols and procedures were approved by Kent State University Institutional Animal Care and Use Committee under protocol 484 LC 19-08 and conform to guidelines outlined by the United States National Institutes of Health for animal research. Heterozygous Kiss1-Cre mice, in which Cre-recombinase expression is driven by Kiss1 regulatory elements (52) (breeding pairs kindly donated by Carol Elias, University of Michigan, Ann Arbor, MI, JAX mice, stock 023426), were crossed with either C57BL/6J mice (JAX mice, stock 000664) to generate hemizygote and wild-type Kiss1-Cre mice or with B6.Cg-Gt-(ROSA)26Sortm9(CAG-tdTomato)Hze/J floxed-stop reporter mice (JAX mice, stock 007907) to generate heterozygous Kiss1-Cre/tdTomato female mice.
Viral Vectors.
The AAV pAAV2/9.CAG.FLEX.GCaMP6s.WPRE.SV40 was purchased from Addgene (Addgene viral prep 100842-AAV9; www.addgene.org/100842/; RRID: Addgene_100842). Viral titers were reduced to 2.5 × 1012 with 0.1 M phosphate-buffered salilne (PBS) before use. Previous studies using cell-attached recordings of ARC kisspeptin neurons in acute brain slices support that kisspeptin neuron activity is faithfully reported by changes in GCaMP6s fluorescence following transfection with the current viral vector (22).
Surgical Procedures.
Stereotaxic viral injection and ovariectomy.
Kiss1-Cre+/−, Kiss1-Cre+/−/tdTomato+/−, or Kiss1-Cre−/−/tdTomato+/− mice were anesthetized with isoflurane (2%) and placed in a stereotaxic frame (Stoelting Co.). Using a Drill and Microinjection Robot (Neurostar), a small hole was drilled into the skull 1 mm posterior to bregma and 0.92 mm lateral to midline. A 29-gauge cannula attached to a 2.5-µL Hamilton syringe was loaded with 500 nL of diluted AAV, angled at 6° toward midline and slowly lowered 5.88 mm ventral to dura into the unilateral ARC at a rate of 150 µm/min. The needle was left in situ for 10 min before the viral vector was injected at a rate of 50 nL/min. Following injection, syringes were left in situ for a further 10 min before the needle was slowly removed at a rate of 150 µm/min. During the viral injection procedure, mice were bilaterally ovariectomized to generate a state of absent steroid hormone inhibition of KNDy cell activity. Mice were returned to clean home cages and pair-housed until Kiss1-Cre mice were implanted with GRIN lenses 1 to 3 wk after viral injection for calcium imaging. Kiss1-Cre/tdTomato mice were transcardially perfused with 4% paraformaldehyde (PFA) in 0.1 M PBS 3 wk after viral injection for immunohistochemical analysis of viral specificity with Cre-expressing arcuate kisspeptin cells.
GRIN lens placement.
For calcium imaging, Kiss1-Cre mice injected with Cre-dependent GCAMP6s AAVs were implanted with Proview Integrated Lenses that combine a 500-µm diameter, 8.4-mm-length GRIN lens with a baseplate (1050-004611, 0.5-mm diameter, 8.4-mm length; Inscopix Inc.) designed for attachment of a miniaturized, single-photon fluorescent microscope (nVoke; Inscopix Inc.). First, a 600-µm-diameter optical fiber (FT600UMT; Thorlabs Inc.) with a sharpened tip was inserted into polyimide tubing (624-µm diameter, cut to 8.4-mm length, 141-0159; Nordson Medical). The polyimide-optical fiber pairing was attached to the stereotaxic frame and implanted 200 to 300µm above the arcuate nucleus. The implant was angled at 6° toward midline to reduce motion artifacts from proximity to the third ventricle. The polyimide tubing was secured to the skull using C&B metabond dental cement (375-0407; Parkell, Inc.) and, once dry, the optical fiber was retracted. The integrated GRIN lens was inserted through the polyimide tubing until 200 µm above the ARC and the baseplate was secured to the skull using C&B dental cement. After surgery, mice were housed individually and allowed to recover for at least 1 wk before receiving daily habituation for microscope attachment and serial blood sampling. No gross behavioral abnormalities were observed in animals that received surgery.
Habituation to Microscope Attachment and Serial Blood Sampling.
One week following surgery, mice were habituated daily for 3 to 5 wk to serial blood sampling protocols as reported previously (53). Briefly, mice were trained to enter the hand of the investigator from inside the home cage and the tail was stroked to mimic the blood collection protocol. Mice would be placed back in their home cage when samples were not being collected. In addition, mice were scruffed during handling for habituation to the restraint necessary for microscope attachment and removal.
In Vivo Imaging and Blood Sampling.
Images were acquired using a single-photon epifluorescence microscope (nVoke miniaturized microscope; Inscopix) and Inscopix Data Acquisition Software. Images were captured at 10 frames per second (10 Hz) with 0.8 to 1 mW of LED (light emitting diode) power and a gain of 4. Images were collected for a total of 60 min per mouse. To assess LH pulsatile secretion, serial 3-µL blood samples were collected from the tail tip of freely moving mice every 3 or 4 min over the 60-min imaging period and diluted in 57 µL of 0.1M PBS with 0.02% Tween 20. Blood samples diluted in PBS-Tween 20 were immediately frozen on dry ice and then stored at −20 C until measurement of LH levels using enzyme-linked immunosorbent assay (ELISA).
Postprocessing of Images.
All calcium imaging data were processed using IDPS software. First, data were cropped to areas containing GCaMP6s fluorescence. Second, background was subtracted from the images using a spatial band-passing Gaussian filter. Each video was corrected for motion using the mean image of the entire video as a reference and the change in fluorescence (DF) relative to the resting F (DF/F) was generated. Within DF/F videos PCA-ICA was used to extract GCaMP6s fluorescent responses associated with individual neurons (54). It should be noted that while the use of automated segmentation programs to extract data from individual cells avoids bias and subjectivity in cell identification for in vivo calcium imaging studies, it does not perfectly detect all fluorescent cells within a recording. PCA-ICA data were manually inspected for signal from nonsomatic compartments, signal detected over multiple cells, or for cells with movement that is unable to be corrected or distortion due to location on the edge of the imaging plane. Currently, there are two main segmentation programs used for microendoscope data: PCA-ICA and constrained nonnegative matrix factorization for microendoscope imaging (CNMFe). Although CNMFe reduces the contribution of fluorescent signal from cells above and below the imaging plane when compared to PCA-ICA, it also has lower precision and identifies false positive cells (55). In our hands, we found CNMFe had poor recognition for the somatic compartment of cells and therefore opted to use PCA-ICA data for analysis of calcium traces. Individual traces were exported for analysis in Excel and MATLAB (The MathWorks Inc.).
Analysis.
Analysis of cell activation.
For all imaging experiments, potential for bleaching effects over the 60-min imaging period were checked by comparing the baseline signal in raw traces at the beginning and end of experiments. In MATLAB, data were normalized to z-scores for quantitative analysis. Baseline fluorescence was identified using the asymmetric least square (ALS) function and was set to 0 for analysis of fluctuations in calcium fluorescence. Peaks in calcium fluorescence within individual cells were identified across the 60-min imaging period using the Minpeak and Mindistance function, which detects peaks once signal surpasses 0.75 SDs above baseline (Z-score of 0.75 from 0) with a minimum duration of 100 s between peaks. The time and amplitude of peak fluorescence and the width (s) at half amplitude was identified. Activity above baseline was defined for each cell as the point when fluorescence reached three times the SD of the baseline of the cell. Time to peak and time to return to baseline for each episode was calculated for each cell, in addition to the interval of time between when cells activated (fluorescence rose above baseline) and the interval of time between the peak amplitude of cells.
Correlation matrix construction.
To assess the correlation of activity between individual cells, correlation matrix construction was conducted using PRISM software. Z-scored traces were separated into data points at baseline activity and data points when fluorescence was above baseline, as defined above. For both datasets, Pearson correlation was used to measure the correlation coefficient between individual cells over the 60-min imaging period. R2 values from each cell-to-cell correlation were averaged per animal. R2 values of zero indicate a random association between neurons or no correlated activity, whereas values close to 1 or −1 indicate a high degree of positive or negative correlation, respectively.
Order of cell activation.
To determine whether KNDy neurons fire in a predictable pattern, the order that cell activity rose above baseline (cells activated) and the order that cells reached peak amplitude was determined for each episode recorded over a 60-min period from each animal. From this data, we calculated the percentage of the recorded cell population that displayed at least one instance of activating for each order point over the 60-min recording session. As the number of episodes and cells recorded differed between animals, we normalized the number of cells and episodes. To determine whether the order of cell activation and peak amplitude was predicted by the identity of the preceding cell, we determined the percentage of SEs in which the same cell activated in the same temporal order. In addition, we calculated the interval of time between when cells activated or when cells reached peaked amplitude when in the same temporal order and compared the range (seconds) and the SD of variation (STDEV) of the interval to that of all SEs across the recorded period. Finally, starting from the percentage of cells that displayed at least one instance of activating or peaking first over 60 min of recording, we calculated the cumulative percentage of the population when including cells that activated or peaked later during the episode.
LH ELISA and pulse detection.
Detection of LH in serial blood samples was achieved using an ultrasensitive sandwich ELISA, as previously reported (56). The assay sensitivity was 0.04 ng/mL and the intra- and interassay coefficient of variation was 6.8% and 7.9%, respectively. In serial blood samples from OVX female mice, an LH pulse was identified when LH rose >10% from the previous one or two samples and lowered >20% in the following one or two samples, as previously described (22). The percentage of LH pulses that were preceded by KNDy neuron activation and, conversely, the percentage of KNDy population episodes followed by an LH pulse were calculated. Additionally, the time from peak of LH to the peak of the preceding episode was measured. Finally, we measured the time between peaks of adjacent SEs (ISI) and determined whether LH pulsatile output occurred within the interval. From this, we calculated the percentage of SEs that were followed by an LH pulse as determined by an ISI of over 5.5 min, between 4 and 5.5 min and under 4 min.
Tissue collection and analysis of GCaMP6s specificity and expression.
Following the completion of in vivo imaging experiments, mice were transcardially perfused with ice-cold 4% PFA in 0.1M PBS. The brain was removed from the skull and the integrated GRIN lens with cannula was carefully removed. The brain was postfixed in 4% PFA for 1 h at room temperature before switching to a 20% sucrose in 0.1 M PBS solution until sunk. Brains were then cut into three parallel series of coronal sections at 30-µm thickness using a freezing microtome. Tissue sections underwent 10-min washes in 0.1 M PB for 3 h and were mounted onto superfrost charged slides, air-dried, and coverslipped using an aqueous mounting medium in order to image endogenous fluorescence [Gelvotol (57), containing the anti-fade agent 1,4-diazabicylo(2,2)octane (Sigma-Aldrich; 50 mg/mL)].
To assess the specificity of the viral vector with kisspeptin neurons, the colocalization of GCaMP6s with tdTomato cells in Kiss1-Cre/tdTomato mice was calculated. Two sections containing the rostral, middle, and caudal arcuate nucleus were imaged per animal (n = 3) using an Olympus FV3000 confocal microscope. A 20× objective was used to enable imaging of the entire ARC area. Optical sections with a 1.25-µm step size were acquired using 488-nm and 550-nm channels. The number of GCaMP6s-positive cells, tdTomato cells, and dual-labeled cells were quantified and the percentage of tdTomato-positive cells colocalized with GCaMP6s and the percentage of GCaMP6s-positive cells colocalized with tdTomato was calculated.
To assess viral GCaMP expression and GRINS lens placement in Kiss1-Cre mice that underwent in vivo Ca2+ imaging, tissue sections were imaged using epifluorescent microscopy (DM500B; Lecia Microsystems) and a digital camera (Microfire A/R; Optronics) paired with MicroBrightField Neurolucida Software (Williston) and a 20× objective. Only mice with accurate lens placement above the ARC with viral expression of GCAMP6s displayed visible calcium signal and were included in this study. Approximately 50% of animals exhibited correct lens placement, viral infection, and fluctuations in calcium signal, and these mice were used for subsequent analysis (n = 5).
Statistics.
All statistical analyses were made using Prism8 (GraphPad Software Inc.) using the methods described below. All data, unless otherwise stated, are represented as the mean ± SEM per group (n = 3 animals in Fig. 1, n = 5 animals in Figs. 2–6) and a P value of <0.05 was accepted as statistically significant. First, statistical comparisons of R2 values generated using Pearson’s correlation coefficient to determine the interaction between individual cells when at baseline activity compared to above baseline were made using two-tailed unpaired Student’s t test (Fig. 3). Comparison of SEs in which 100% of cells were activated versus episodes where less than 100% of cells were activated were conducted using two-tailed unpaired Student’s t test (Fig. 3). Comparison of the time lag between cells with a common cell-to-cell order versus the time-lag between all cells was conducted using two-tailed unpaired Student’s t test. To compare the size of the recorded KNDy population that activated or peaked in order of first to last, we compared data from each order point using two-way ANOVA with post hoc Bonferroni tests (Fig. 4 C and D). The time between cell activation and peak amplitude for leader cells that activated first and peaked first and nonleader cells was compared using one-way ANOVA with Bonferroni posttest (Fig. 4G). Finally, the occurrence of LH pulsatile release following episodes in which all cells versus some cells were active was compared using a two-tailed Student’s t test (Fig. 6B), and LH pulse generation following SEs with different ISIs were compared using one-way ANOVA with Bonferroni posttest (Fig. 5 C–E).
Supplementary Material
Acknowledgments
We thank Dayanara B. Lohr for her excellent technical assistance during this research. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the NIH under award numbers K99HD096120 to A.M.M. and R01HD039916 to M.N.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117767119/-/DCSupplemental.
Data Availability
Raw data have been deposited in Open Science Framework (https://osf.io/m5dxc/) (58).
References
- 1.Nakai Y., Plant T. M., Hess D. L., Keogh E. J., Knobil E., On the sites of the negative and positive feedback actions of estradiol in the control of gonadotropin secretion in the rhesus monkey. Endocrinology 102, 1008–1014 (1978). [DOI] [PubMed] [Google Scholar]
- 2.Belchetz P. E., Plant T. M., Nakai Y., Keogh E. J., Knobil E., Hypophysial responses to continuous and intermittent delivery of hypopthalamic gonadotropin-releasing hormone. Science 202, 631–633 (1978). [DOI] [PubMed] [Google Scholar]
- 3.de Roux N., et al. , Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc. Natl. Acad. Sci. U.S.A. 100, 10972–10976 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seminara S. B., et al. , The GPR54 gene as a regulator of puberty. N. Engl. J. Med. 349, 1614–1627 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Irwig M. S., et al. , Kisspeptin activation of gonadotropin releasing hormone neurons and regulation of KiSS-1 mRNA in the male rat. Neuroendocrinology 80, 264–272 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Messager S., et al. , Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc. Natl. Acad. Sci. U.S.A. 102, 1761–1766 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman R. L., et al. , Kisspeptin neurons in the arcuate nucleus of the ewe express both dynorphin A and neurokinin B. Endocrinology 148, 5752–5760 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Ramaswamy S., et al. , Neurokinin B stimulates GnRH release in the male monkey (Macaca mulatta) and is colocalized with kisspeptin in the arcuate nucleus. Endocrinology 151, 4494–4503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navarro V. M., et al. , Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J. Neurosci. 29, 11859–11866 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.True C., Kirigiti M., Ciofi P., Grove K. L., Smith M. S., Characterisation of arcuate nucleus kisspeptin/neurokinin B neuronal projections and regulation during lactation in the rat. J. Neuroendocrinol. 23, 52–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassaneen A., et al. , Immunohistochemical characterization of the arcuate kisspeptin/neurokinin B/dynorphin (KNDy) and preoptic kisspeptin neuronal populations in the hypothalamus during the estrous cycle in heifers. J. Reprod. Dev. 62, 471–477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakabayashi Y., et al. , Neurokinin B and dynorphin A in kisspeptin neurons of the arcuate nucleus participate in generation of periodic oscillation of neural activity driving pulsatile gonadotropin-releasing hormone secretion in the goat. J. Neurosci. 30, 3124–3132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore A. M., Coolen L. M., Porter D. T., Goodman R. L., Lehman M. N., KNDy cells revisited. Endocrinology 159, 3219–3234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke M. C., Letts P. A., Krajewski S. J., Rance N. E., Coexpression of dynorphin and neurokinin B immunoreactivity in the rat hypothalamus: Morphologic evidence of interrelated function within the arcuate nucleus. J. Comp. Neurol. 498, 712–726 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Weems P. W., et al. , κ-opioid receptor is colocalized in GnRH and KNDy cells in the female ovine and rat brain. Endocrinology 157, 2367–2379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foradori C. D., Amstalden M., Goodman R. L., Lehman M. N., Colocalisation of dynorphin A and neurokinin B immunoreactivity in the arcuate nucleus and median eminence of the sheep. J. Neuroendocrinol. 18, 534–541 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Lehman M. N., Coolen L. M., Goodman R. L., Minireview: Kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: A central node in the control of gonadotropin-releasing hormone secretion. Endocrinology 151, 3479–3489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbison A. E., The gonadotropin-releasing hormone pulse generator. Endocrinology 159, 3723–3736 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Okamura H., et al. , Kisspeptin and GnRH pulse generation. Adv. Exp. Med. Biol. 784, 297–323 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Clarke H., Dhillo W. S., Jayasena C. N., Comprehensive review on kisspeptin and its role in reproductive disorders. Endocrinol. Metab. (Seoul) 30, 124–141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman R. L., et al. , Kisspeptin, neurokinin B, and dynorphin act in the arcuate nucleus to control activity of the GnRH pulse generator in ewes. Endocrinology 154, 4259–4269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarkson J., et al. , Definition of the hypothalamic GnRH pulse generator in mice. Proc. Natl. Acad. Sci. U.S.A. 114, E10216–E10223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han S. Y., McLennan T., Czieselsky K., Herbison A. E., Selective optogenetic activation of arcuate kisspeptin neurons generates pulsatile luteinizing hormone secretion. Proc. Natl. Acad. Sci. U.S.A. 112, 13109–13114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salem V., et al. , Leader β-cells coordinate Ca2+ dynamics across pancreatic islets in vivo. Nat. Metab. 1, 615–629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu J., et al. , High-frequency stimulation-induced peptide release synchronizes arcuate kisspeptin neurons and excites GnRH neurons. eLife 5, e16246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krajewski S. J., Burke M. C., Anderson M. J., McMullen N. T., Rance N. E., Forebrain projections of arcuate neurokinin B neurons demonstrated by anterograde tract-tracing and monosodium glutamate lesions in the rat. Neuroscience 166, 680–697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beale K. E., et al. , The physiological role of arcuate kisspeptin neurons in the control of reproductive function in female rats. Endocrinology 155, 1091–1098 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittelman-Smith M. A., et al. , Arcuate kisspeptin/neurokinin B/dynorphin (KNDy) neurons mediate the estrogen suppression of gonadotropin secretion and body weight. Endocrinology 153, 2800–2812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittelman-Smith M. A., Krajewski-Hall S. J., McMullen N. T., Rance N. E., Ablation of KNDy neurons results in hypogonadotropic hypogonadism and amplifies the steroid-induced LH surge in female rats. Endocrinology 157, 2015–2027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu M. H., et al. , Relative importance of the arcuate and anteroventral periventricular kisspeptin neurons in control of puberty and reproductive function in female rats. Endocrinology 156, 2619–2631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamura T., Wakabayashi Y., Ohkura S., Navarro V. M., Okamura H., Effects of intravenous administration of neurokinin receptor subtype-selective agonists on gonadotropin-releasing hormone pulse generator activity and luteinizing hormone secretion in goats. J. Reprod. Dev. 61, 20–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo S.-H., Herbison A. E., Projections of arcuate nucleus and rostral periventricular kisspeptin neurons in the adult female mouse brain. Endocrinology 152, 2387–2399 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Roa J., Tena-Sempere M., Connecting metabolism and reproduction: Roles of central energy sensors and key molecular mediators. Mol. Cell. Endocrinol. 397, 4–14 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Nestor C. C., Kelly M. J., Rønnekleiv O. K., Cross-talk between reproduction and energy homeostasis: Central impact of estrogens, leptin and kisspeptin signaling. Horm. Mol. Biol. Clin. Investig. 17, 109–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grachev P., et al. , Neurokinin B signaling in the female rat: A novel link between stress and reproduction. Endocrinology 155, 2589–2601 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Yang J. A., et al. , Acute psychosocial stress inhibits LH pulsatility and kiss1 neuronal activation in female mice. Endocrinology 158, 3716–3723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otsuki Y., Yamaji K., Fujita M., Takagi T., Tanizawa O., Serial plasma oxytocin levels during pregnancy and labor. Acta Obstet. Gynecol. Scand. 62, 15–18 (1983). [DOI] [PubMed] [Google Scholar]
- 38.Perkinson M. R., Kim J. S., Iremonger K. J., Brown C. H., Visualising oxytocin neurone activity in vivo: The key to unlocking central regulation of parturition and lactation. J. Neuroendocrinol. 33, e13012 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Belin V., Moos F., Paired recordings from supraoptic and paraventricular oxytocin cells in suckled rats: Recruitment and synchronization. J. Physiol. 377, 369–390 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belin V., Moos F., Richard P., Synchronization of oxytocin cells in the hypothalamic paraventricular and supraoptic nuclei in suckled rats: Direct proof with paired extracellular recordings. Exp. Brain Res. 57, 201–203 (1984). [DOI] [PubMed] [Google Scholar]
- 41.McQuillan H. J., Han S. Y., Cheong I., Herbison A. E., GnRH pulse generator activity across the estrous cycle of female mice. Endocrinology 160, 1480–1491 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Han S. Y., Cheong I., McLennan T., Herbison A. E., Neural determinants of pulsatile luteinizing hormone secretion in male mice. Endocrinology 161, bqz045 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Wildt L., et al. , Frequency and amplitude of gonadotropin-releasing hormone stimulation and gonadotropin secretion in the rhesus monkey. Endocrinology 109, 376–385 (1981). [DOI] [PubMed] [Google Scholar]
- 44.McIntosh R. P., McIntosh J. E., Influence of the characteristics of pulses of gonadotrophin releasing hormone on the dynamics of luteinizing hormone release from perifused sheep pituitary cells. J. Endocrinol. 98, 411–421 (1983). [DOI] [PubMed] [Google Scholar]
- 45.Perkins R. B., Hall J. E., Martin K. A., Aetiology, previous menstrual function and patterns of neuro-endocrine disturbance as prognostic indicators in hypothalamic amenorrhoea. Hum. Reprod. 16, 2198–2205 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Coyle C., Campbell R. E., Pathological pulses in PCOS. Mol. Cell. Endocrinol. 498, 110561 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Coutinho E. A., Kauffman A. S., The role of the brain in the pathogenesis and physiology of polycystic ovary syndrome (PCOS). Med. Sci. (Basel) 7, E84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore A. M., Lohr D. B., Coolen L. M., Lehman M. N., Prenatal androgen exposure alters KNDy neurons and their afferent network in a model of polycystic ovarian syndrome. Endocrinology 162, bqab158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.George J. T., et al. , Neurokinin B receptor antagonism in women with polycystic ovary syndrome: A randomized, placebo-controlled trial. J. Clin. Endocrinol. Metab. 101, 4313–4321 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Prague J. K., et al. , Neurokinin 3 receptor antagonism as a novel treatment for menopausal hot flushes: A phase 2, randomised, double-blind, placebo-controlled trial. Lancet 389, 1809–1820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jayasena C. N., et al. , Subcutaneous injection of kisspeptin-54 acutely stimulates gonadotropin secretion in women with hypothalamic amenorrhea, but chronic administration causes tachyphylaxis. J. Clin. Endocrinol. Metab. 94, 4315–4323 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Cravo R. M., et al. , Characterization of Kiss1 neurons using transgenic mouse models. Neuroscience 173, 37–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCosh R. B., Kreisman M. J., Breen K. M., Frequent tail-tip blood sampling in mice for the assessment of pulsatile luteinizing hormone secretion. J. Vis. Exp., 2018, 57894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukamel E. A., Nimmerjahn A., Schnitzer M. J., Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamatakis A. M., et al. , Miniature microscopes for manipulating and recording in vivo brain activity. Microscopy (Oxf) 70, 399–414 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steyn F. J., et al. , Development of a methodology for and assessment of pulsatile luteinizing hormone secretion in juvenile and adult male mice. Endocrinology 154, 4939–4945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuiper L. B., Beloate L. N., Dupuy B. M., Coolen L. M., Drug-taking in a socio-sexual context enhances vulnerability for addiction in male rats. Neuropsychopharmacology 44, 503–513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. M. Moore, L. M. Coolen, M. N. Lehman, In vivo imaging of the GnRH pulse generator reveals a temporal order of neuronal activation and synchronization during each pulse. Open Science Framework. https://osf.io/m5dxc/. Deposited 11 January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data have been deposited in Open Science Framework (https://osf.io/m5dxc/) (58).