Abstract
In the present study, 37 group A Streptococcus (GAS) strains belonging to 13 new emm sequence types identified among GAS strains randomly isolated in Brazil were characterized by using phenotypic and genotypic methods. The new types were designated st204, st211, st213, st809, st833, st854, st2904, st2911, st2917, st2926, st3757, st3765, and st6735. All isolates were susceptible to the antimicrobial agents tested, except to tetracycline. They all carried the speB gene, and 94.6% produced detectable SpeB. Most strains belonging to a given emm type had similar or highly related pulsed-field gel electrophoresis profiles that were distinct from profiles of strains of another type. The other characteristics were variable from isolate to isolate, although some associations were consistently found within some emm types. Unlike the other isolates, all type st213 isolates were speA positive and produced SpeA. Strains belonging to st3765 were T6 and opacity factor (OF) negative. Individual isolates within OF-positive emm types were associated with unique sof gene sequence types, while OF-negative isolates were sof negative by PCR. This report provides information on new emm sequence types first detected in GAS isolates from a geographic area not extensively surveyed. Such data can contribute to a better understanding of the local and global dynamics of GAS populations and of the epidemiological aspects of GAS infections occurring in tropical regions.
Streptococcus pyogenes, frequently referred to as group A Streptococcus (GAS), is a significant cause of human morbidity worldwide, and it is associated with a variety of mild and severe diseases that may occur either in areas where the diseases are endemic or as epidemics (10). Most of the knowledge that has been accumulated concerning GAS epidemiology is based upon serologic M and T typing. However, many GAS isolates are nontypeable due to the lack of appropriate type-specific antisera or possibly due to loss of antigen expression under cultivation. In recent years, DNA sequencing-based methods for characterizing GAS strains have been used, including sequence analysis of emm gene-specific PCR products (emm typing) (1, 2). This methodology has allowed the recognition of several previously unknown GAS types in different geographic areas, demonstrating the usefulness of emm typing for detecting genetic diversity among GAS isolates and for tracing GAS infections (1, 2, 5). A complementary molecular methodology (sof typing) is also based upon sequence analysis of a hypervariable virulence gene (3).
In the present study, we describe the characteristics of GAS isolates belonging to new emm types identified among strains recovered from patients living in Brazil. These strains were further characterized through phenotypic tests (T-protein typing, detection of opacity factor [OF], antimicrobial susceptibility, and production of streptococcal pyrogenic exotoxin A [SpeA] and SpeB) and additional genotypic tests (the presence of speA, speB, speC, and sof gene PCR products, sof sequence types, and analysis of genomic DNA restriction profiles).
MATERIALS AND METHODS
Bacterial strains.
A total of 37 GAS isolates belonging to new emm types were included in the present study. They were recognized among random GAS isolates recovered from sterile- and nonsterile-site clinical specimens obtained between 1995 and 1999 from patients living in the southeast region of Brazil. The isolates were identified on the basis of colony morphology and β-hemolysis on blood agar plates, gram stain characteristics, susceptibility to bacitracin, production of leucine aminopeptidase and pyrrolidonyl arylamidase, and reactivity with group A-specific antiserum.
Phenotypic characterization of the isolates.
Serologic T types and opacity factor (OF) reactions were determined as previously described (7, 8). Susceptibility to antimicrobial agents was tested by a broth microdilution assay using the CDC Strep panel (PML Microbiologicals, Wilsonville, Oreg.). The following 14 antimicrobial agents were tested: penicillin, amoxicillin, cefotaxime, cefuroxime, erythromycin, trimethoprim-sulfamethoxazole, clindamycin, chloramphenicol, tetracycline, vancomycin, trovafloxacin, rifampin, meropenem, and levofloxacin. The MIC results were interpreted according to NCCLS guidelines (9).
Detection of exotoxin production by immunoblotting assay.
Strains were tested for their ability to produce SpeA and SpeB in vitro by using procedures for exotoxin preparation and for exotoxin detection by a immunoblotting assay derived from those outlined by Talkington et al. (11).
Detection of streptococcal pyrogenic exotoxin genes by PCR.
PCRs were performed to detect the presence of the speA, speB, and speC genes. DNA extraction and PCR conditions were based on previously described methods (4, 11).
Analysis of chromosomal DNA restriction profiles by PFGE.
Genomic DNA was prepared with agarose plugs, based on previously described recommendations (12). For lysis, plugs were treated with lysis solution containing 1 mg of lysozyme and 5 U of mutanolysin per ml. After digestion with SmaI, the fragments were resolved by pulsed-field gel electrophoresis (PFGE) in 1.3% agarose gels in 0.5× Tris-borate-EDTA buffer in a CHEF-DR III system (Bio-Rad Laboratories, Hercules, Calif.). The following parameters were used: running time, 25 h; temperature, 13°C; voltage gradient, 6 V/cm; initial pulse time, 5.3 s; final pulse time, 34.9 s. The DNA fragments in the gels were stained with ethidium bromide and photographed under UV light. Analysis of chromosomal DNA fragmentation profiles was performed by visual inspection of photographic registers, considering the criteria suggested by Tenover et al. (13). PFGE profiles were also analyzed and compared by using the Molecular Analyst Fingerprint Plus software package, version 1.12 (Bio-Rad). The percentages of similarities of the PFGE banding profiles were estimated with the Dice coefficient and clustered by the unweighted pair group method with arithmetic averages.
Detection of sof genes and sof sequence analysis
Amplification of 5′ sof gene fragments and sof sequence analysis were performed as previously described (3). GenBank accession numbers for sof gene sequences are as follows: sof213, AF 139743; sof833 (actually an allele of sof90; see reference 3), AF139741; sof6735 (also described in reference 3, with a 5′ sequence very similar in overlap to sof90).
Sequencing of emm gene-specific PCR products (emm typing).
Harvesting of the bacterial cells and the procedures for PCR and sequence analysis of emm-specific PCR products were essentially those already published (1, 2). DNA sequences were subjected to homology searches against all known emm sequences present in the GenBank and in the Centers for Disease Control and Prevention (CDC) database (http://www.cdc.gov/ncidod/biotech/infotech_hp.html) with Genetics Computer Group software, version 9, with the NCBI Blast programs. GenBank accession numbers are as follows: st204, AF056301; st211, AF 96179; st213, AF049855; st809, AF296180; st833, AF052425; st854, AF281048; st2904, AF192768; st2917, AF082864; st3757, AF074875; and st3765, AF074877.
Phylogram construction of M protein sequences.
Sequences with 103 residues (including 23 signal sequence residues and 80 amino acids of the mature N termini) were analyzed by sequential use of the Wisconsin Package (version 10.1) PileUp (gap creation and extension penalties of 8 and 2, respectively), Distances (uncorrected distance), and GrowTree (neighbor-joining) programs.
RESULTS AND DISCUSSION
The 37 GAS isolates selected for the present study were found to correspond to 13 previously undocumented emm sequence types. They comprised 12.3% of a total of 302 random GAS isolates recovered in Brazil, which are the subject of a study in progress. Phenotypic and genotypic characteristics of the 37 GAS isolates representing new emm types are shown in Table 1, in addition to their sources and city of isolation.
TABLE 1.
Characteristics of 37 S. pyogenes strains belonging to 13 new emm gene sequence types isolated in Brazil
| Strain | Source | Origina | T typeb | OFc | sof sequence | emm sequence | spe gene | Sped | PFGE profile | Tete |
|---|---|---|---|---|---|---|---|---|---|---|
| CL-1459 | Wound | SP | NT | − | NDf | st204 | B, C | B | 204-A1 | R |
| CL-1463 | Skin | SP | 25/Imp 19 | − | st204 | B | B | 204-A2 | S | |
| CL-3785 | Throat | RJ | 2 | − | ND | st204 | B | B | 204-A2 | S |
| CL-2009 | Abscess | RJ | 25 | − | st204 | B | B | 204-A3 | S | |
| CL-3744 | Skin | RJ | 2/25 | − | st204 | B | B | 204-A3 | I | |
| CL-2187 | Wound | RJ | 2/6 | − | st204 | B | B | 204-A4 | S | |
| CL-1464 | Blood | SP | NT | − | st204 | A, B, C | B | 204-B | R | |
| CL-3745 | Skin | RJ | 2/25 | − | st204 | B, C | B | 204-C | S | |
| CL-4973 | Unknown | RJ | 2 | − | st204 | B | B | 204-D | R | |
| CL-1471 | Abscess | SP | 3/13 | − | st211 | B | B | 211-A | R | |
| CL-1472 | Abscess | SP | 3/13 | − | st211 | B | 211-A | R | ||
| CL-1473 | Abscess | SP | NT | + | sof213 | st213 | A, B | A, B | 213-A | S |
| CL-2190 | Wound | RJ | 4 | + | sof213 | st213 | A, B | A, B | 213-A | R |
| CL-2647 | Throat | RJ | 4 | + | sof213 | st213 | A, B | A, B | 213-A | R |
| CL-3095 | Abscess | RJ | NT | + | sof213 | st213 | A, B | A, B | 213-A | R |
| CL-3100 | Throat | RJ | NT | + | sof213 | st213 | A, B | A, B | 213-A | R |
| CL-4953 | Throat | RJ | 4 | + | sof213 | st213 | A, B | A, B | 213-A | R |
| CL-2588 | Blood | RJ | NT | − | st809 | B | B | 809-A | S | |
| CL-3832 | Unknown | RJ | 3/13/B | + | sof833 | st833 | B | B | 833-A | R |
| CL-2622 | Throat | RJ | 3/13 | + | sof833 | st833 | B | B | 833-B | R |
| CL-2652 | Sinus secretions | RJ | B3264 | − | st854 | B | B | 854-A | R | |
| CL-4907 | Throat | RJ | B3264 | − | st854 | B | B | 854-A | R | |
| CL-2177 | Throat | RJ | 3/13/B | + | sof2904 | st2904 | B | B | 2904-A | R |
| CL-2643 | Abscess | RJ | 3/13/B | − | st2911 | B | 2911-A | R | ||
| CL-3097 | Abscess | RJ | 3/B | − | st2917 | B | B | 2917-A1 | S | |
| CL-3747 | Skin | RJ | NT | − | st2917 | B | B | 2917-A2 | S | |
| CL-3780 | Throat | RJ | 3 | − | st2917 | B, C | B | 2917-B | S | |
| CL-3107 | Throat | RJ | 3/13/B | − | st2926 | B | B | 2926-A1 | R | |
| CL-3775 | Skin | RJ | 3/13/B | − | st2926 | B, C | B | 2926-A2 | R | |
| CL-3757 | Skin | RJ | 14 | − | st3757 | B, C | B | 3757-A | R | |
| CL-3779 | Throat | RJ | 14 | − | st3757 | B | B | 3757-B | R | |
| CL-3765 | Skin | RJ | 6 | − | st3765 | B | B | 3765-A1 | R | |
| CL-3822 | Abscess | RJ | 6 | − | st3765 | B | B | 3765-A2 | R | |
| CL-3994 | Abscess | RJ | 6 | − | st3765 | B | B | 3765-A2 | R | |
| CL-4988 | Throat | RJ | 6 | − | st3765 | B | B | 3765-A2 | R | |
| CL-4996 | Skin | RJ | 6 | − | st3765 | B | B | 3765-A2 | R | |
| CL-4987 | Throat | RJ | 11/12 | + | sof6735 | st6735 | B | B | 6735-A | R |
Locality of isolation: RJ, Rio de Janeiro; SP, São Paulo.
NT, T nontypeable.
+, OF positive; −, OF negative.
Production of SpeA and or SpeB as detected by immunoblotting assay.
Results of antimicrobial susceptibility tests for tetracycline: R, resistant; S, susceptible; I, intermediate.
ND, not determined.
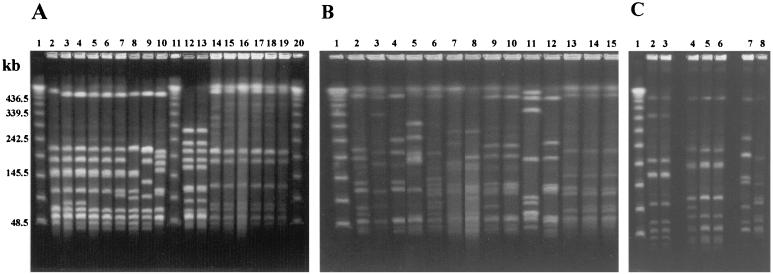
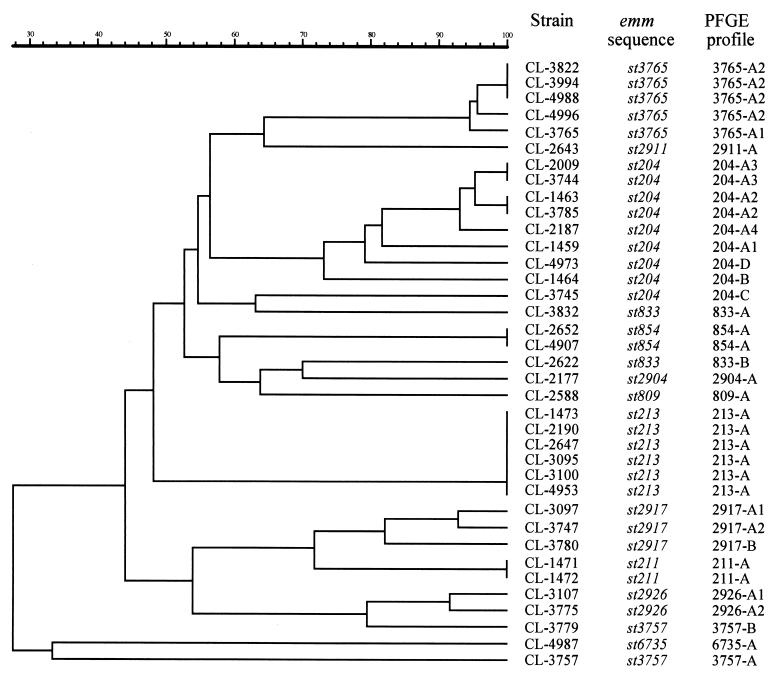
All strains were susceptible to most of the antimicrobial agents tested. The exception was tetracycline, to which 26 strains were resistant and 1 was intermediate. The speB gene was detected in all isolates, while speA and speC genes were found in seven and six isolates, respectively. Most isolates (94.6%) produced SpeB. PFGE profiles of isolates belonging to a given emm type were usually similar or highly related. In some instances, PFGE profiles within a given emm type were not related, but they were always distinct from profiles of strains of another emm type (Fig. 1 and 2). This data indicated that the majority of isolates within each individual emm type shared a high degree of genetic relatedness. This high degree of relatedness within certain emm types might hold true only within this particular geographical area, since emm sequence type 854 (st854) isolates recovered in Egypt as this study was being completed had PFGE profiles clearly distinct from those of the Brazilian st854 isolates (Fig. 1C). It should also be noted that after its initial discovery in 1997 in Brazilian isolates (this study), the 549-base emm st854 sequence was found to be identical over its overlap with material recently assigned GenBank accession number AF018179 (225 bases), derived from an impetigo isolate in Northern Australia (6).
FIG. 1.
Representative PFGE profiles of SmaI-digested genomic DNA of Streptococcus pyogenes belonging to new emm sequence types isolated in Brazil. Lanes 1 (A through C), 11 (B), and 20 (A), molecular mass markers expressed in kilobases (lambda DNA concatemers ranging from 48.5 to 1,018.5 kb). (A) Lanes: 2 to 10, st204 strains (lane 2, CL-1459; lane 3, CL-1463; lane 4, CL-3785; lane 5, CL-2009; lane 6, CL-3744; lane 7, CL-2187; lane 8, CL-4973; lane 9, CL-1464; lane 10, CL-3745); 12 and 13, st211 strains (lane 12, CL-1471; lane 13, CL-1472); 14 to 19, st213 strains (lane 14, CL-1473; lane 15, CL-2190; lane 16, CL-2647; lane 17, CL-3095; lane 18, CL-3100; lane 19, CL-4953). (B) Lanes: 2, st809 strain CL-2588; 3 and 4, st833 strains (lane 3, CL-3832; lane14, CL-2622); 5, st2904 strain CL-2177; 6, st2911 strain CL-2643; 7 and 8, st2917 strains (lane 7, CL-3097; lane 8, CL-3780); 9 and 10, st2926 strains (lane 9, CL-3107; lane 10, CL-3775); 11 and 12, st3757 strains (lane 11, CL-3757; lane 12, CL-3779); 13 to 15, st3765 strains (lane 13, CL-3765; lane 14, CL-3822; lane 15, CL-3994). (C) Lanes: 2 and 3, st854 strains from Brazil (lane 2, CL-2652; lane 3, CL-4907); 4 to 6, st854 strains from Egypt; 7, st6735 strain CL-4987; 8, st3765 strain CL-4996.
FIG. 2.
Dendrogram resulting from a computer-assisted analysis of the PFGE profiles of SmaI-digested genomic DNA of S. pyogenes belonging to new emm sequence types isolated in Brazil. The scale represents average percentages of similarity.
Several characteristics were variable from strain to strain, although some associations were found within emm types. Six of nine isolates representing the predominant new type st204 had related PFGE profiles, despite differences in susceptibility to tetracycline and T-typing results. T types and OF phenotypes can vary extensively even between different passages of the same strain, possibly due to the effects of laboratory growth conditions on gene expression (unpublished observations). All six st213 isolates contained both speA and speB genes and, in contrast to all of the other isolates, produced both SpeA and SpeB. All six st213 isolates were OF positive and contained the unique sof213 sequence. Three st213 isolates were T type 4, while the other three were T nontypeable. The five st3765 isolates were T type 6 and OF negative. All three st2917 isolates were susceptible to tetracycline and were OF negative. Two st2917 isolates typed with components of the T 3/13/B complex while one was T nontypeable.
Characteristics varied among isolates with the other new emm types represented by one or two isolates (st211, st809, st833, st854, st2904, st2911, st2926, st3757, and st6735). Most of these isolates typed with components of the T 3/13/B complex, except for st3757 isolates (T type 14) and st6735 isolates (T type 11/12).
Representative isolates from 4 of the 13 new emm types shown in Table 1 were found to be sof PCR positive. The partial sof213 sequence from the st213 isolate CL-1473 was unique from all other sof sequences analyzed to date (at least 80 at present time [3]), with only 79% sequence identity to its closest match. Similarly, the best match to sof2904 was only 80% identical over 547 bases. It was interesting to find that the 5′ sof sequences associated with both st833 and st6735 types were nearly identical to sof90 from an emm type M serotype 90 isolate (3). While the st833 sequence is very similar to emm90, it differs from emm90 by the deletion of the codons encoding mature M residues 2 to 5 and 17 to 21 and does not confer the M90 serotype. Nonetheless, a st833 sof833 isolate was found to be anti-OF type 90 (3). The similarity between this pair of chromosomally unlinked genes with these genes from the emm90 reference strain, together with related T-agglutination patterns (either T 3/13 or T 3/13/B3264) suggests that sequence types st833, sof833, and emm90 sof90 strains are highly related genomically (see http://www.cdc.gov/ncidod/biotech/infotech_hp.html for information concerning emm90 and other known emm types). In contrast, while sof6735 and sof90 (or sof833) 5′ sequences were highly similar (3), the st6735 sequence and T types were found to be quite divergent from these markers in the emm90 reference strain. The PFGE results clearly indicate that CL-4987 (st6735) and CL-2622 (st833) had distinct chromosomal restriction profiles (Table 1; Fig. 1 and 2).
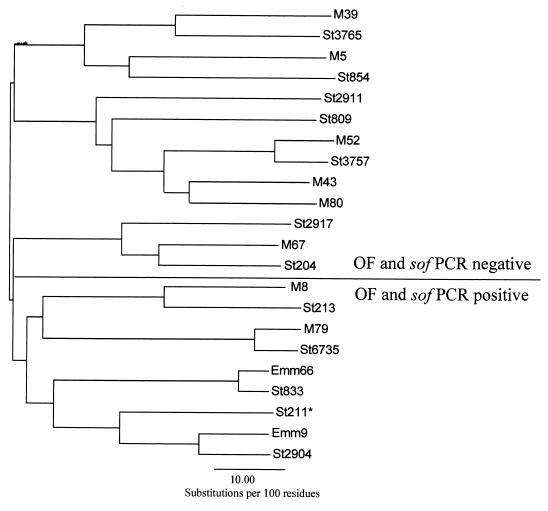
Figure 3 shows a dendrogram based upon sequence comparisons of 114 residue N-terminal portions of the various deduced M proteins representing the new sequence types first encountered in Brazil. These 114 residues include 19 signal sequence amino acids and the 95 mature N terminal residues. For reference, the corresponding sequence from the closest “classical” M protein sequence match was included in the analysis. It is striking that all of the 17 sequences, representing OF- and sof-negative strains, were neatly segregated from the 4 sequences derived from OF- and sof-positive strains, with the single exception of emm type st211, which was obtained from an OF- and sof PCR-negative isolate. This relationship, where the majority of OF-positive emm types are segregated from the majority of OF-negative emm types, holds true with a similar analysis representing the majority of known deduced M sequences (14; www.cdc.gov/ncidod/biotech/strep/strepindex.html). These results support the notion that OF-negative and -positive strains represent two distinct GAS lineages between which little horizontal exchange occurs. As also shown in Fig. 3, the branch point for emm type st211 is much deeper than the other branch points in the OF-positive section of the dendrogram, which indicates that this sequence is the most divergent sequence included in this analysis.
FIG. 3.
Dendrogram based upon sequence comparisons of 114 residue N-terminal portions of the various deduced M proteins representing new emm sequence types first encountered among S. pyogenes isolated in Brazil (*St211 was an exception within this group of M N-terminal sequences in that the strain harboring st211 was OF and sof PCR negative. Note that the branch point for st211 is much deeper than the other branch points in the OF-positive section of the dendrogram).
This report presents the phenotypic and genotypic characteristics of GAS isolates belonging to 13 new emm sequence types that were first detected among randomly collected Brazilian isolates. The data indicate the existence of a significant proportion of newly recognized emm types with a variety of characteristics, suggesting that strains circulating in Brazil, an area not previously extensively surveyed, may represent a large pool of M serotypes and clonal groups different from those known to circulate in other geographic areas. Such information can contribute to a better understanding of the local and global dynamics of GAS populations, and of the epidemiological aspects of GAS infections occurring in tropical regions.
ACKNOWLEDGMENTS
This study was supported in part by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Financiadora de Estudos e Projetos (FINEP), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and Ministério da Ciência e Tecnologia (MCT/PRONEX), Brazil.
We thank Theresa Hoenes, Raji Viswanathan, and Zhongya Li for their excellent technical assistance in the CDC Streptococcal Genetics Laboratory. We also thank the CDC Biotechnology Core Facility Branch computing group, most notably Scott Sammons, Elizabeth Neuhaus, and Sarah McKneally, for assistance in constructing and maintaining the web site http://www.cdc.gov/ncidod/biotech/infotech_hp.html.
REFERENCES
- 1.Beall B, Facklam R, Hoenes T, Schwartz B. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J Clin Microbiol. 1997;35:1231–1235. doi: 10.1128/jcm.35.5.1231-1235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall B, Gherardi G, Lovgren M, Facklam R R, Forwick B A, Tyrrell G J. emm and sof gene sequence variation in relation to serological typing of opacity-factor-positive group A streptococci. Microbiology. 2000;146:1195–1209. doi: 10.1099/00221287-146-5-1195. [DOI] [PubMed] [Google Scholar]
- 4.Black C M, Talkington D F, Messmer T O, Facklam R R, Hornes E, Olsvik O. Detection of streptococcal pyrogenic exotoxin genes by a nested polymerase chain reaction. Mol Cell Probes. 1993;7:255–259. doi: 10.1006/mcpr.1993.1038. [DOI] [PubMed] [Google Scholar]
- 5.Facklam R, Beall B, Efstratiou A, Fischetti V, Johnson D, Kaplan E, Kriz P, Lovgren M, Martin D, Schwartz B, Totolian A, Bessen A D, Hollingshead S, Rubin F, Scott J, Tyrrell G. emm typing and validation of provisional M types for group streptococci. Emerg Infect Dis. 1999;5:247–253. doi: 10.3201/eid0502.990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardiner D L, Goodfellow A M, Martin D R, Sriprakash K S. Group A streptococcal Vir types are M-protein gene (emm) sequence type specific. J Clin Microbiol. 1998;36:902–907. doi: 10.1128/jcm.36.4.902-907.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maxted W R, Widdowson J P, Fraser C A M, Ball L C, Bassett D C J. The use of serum opacity reaction in the typing of group A streptococci. J Med Microbiol. 1973;68:83–90. doi: 10.1099/00222615-6-1-83. [DOI] [PubMed] [Google Scholar]
- 8.Moody M D, Padula J, Lizana D, Hall C T. Epidemiologic characterization of group A streptococci by T-agglutination and M-precipitation tests in the public health laboratory. Health Lab Sci. 1965;2:149–162. [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement, M100–S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 10.Stevens D L. Group A beta-hemolytic streptococci: virulence factors, pathogenesis, and spectrum of clinical infections. In: Stevens D L, Kaplan E L, editors. Streptococcal infections; clinical aspects, microbiology, and molecular pathogenesis. New York, N.Y: Oxford University Press; 2000. pp. 19–36. [Google Scholar]
- 11.Talkington D F, Schwartz B, Black C M, Todd J K, Elliott J, Breiman R F, Facklam R R. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect Immun. 1993;61:3369–3374. doi: 10.1128/iai.61.8.3369-3374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teixeira L M, Carvalho M G S, Merquior V L C, Steigerwalt A G, Brenner D J, Facklam R R. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J Clin Microbiol. 1997;35:2778–2781. doi: 10.1128/jcm.35.11.2778-2781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenover F C, Arbeit R D, Georing R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whatmore A M, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Noncongruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]