Abstract
Simple Summary
The most common genera in the piglet microbiome were Lactobacillus, Escherichia-Shigella, Enterococcus, Bacteroides, and Fusobacterium. Bacteria of the Lactobacillus genus dominated in healthy piglets. An increased number of Escherichia-Shigella and Enterococcus was detected in diarrheal pigs. This indicates an important role of these bacteria in the pathogenesis of diarrhea. A decreased number of Bacteroides was detected in diarrheal pigs. According to the assessment of the microbiome composition in different sections of the intestine, bacteria of the Lactobacillus genus were the most common in the ileum, while Fusobacterium and Bacteroides were more common in the rectum. Our results show that the gut microbiome may make a significant contribution to the pathogenesis of diarrhea.
Abstract
Determining the taxonomic composition of microbial consortia of the piglet intestine is of great importance for pig production. However, knowledge on the variety of the intestinal microbiome in newborn piglets is limited. Piglet diarrhea is a serious gastrointestinal disease with a high morbidity and mortality that causes great economic damage to the pig industry. In this study, we investigated the microbiome of various sections of the piglet intestine and compared the microbiome composition of healthy and diarrheal piglets using high-throughput sequencing of the 16S rRNA gene. The results showed that bacteria of the Lactobacillus genus were the most common in the ileum, while Fusobacterium and Bacteroides dominated in the rectum. Comparing the microbiome composition of healthy and diarrheal piglets revealed a reduced number of Lactobacillus bacteria as a hallmark of diarrhea, as did an increased content of representatives of the Escherichia-Shigella genus and a reduced number of Bacteroides, which indicates the contribution of these bacteria to the development of diarrhea in piglets. The relative abundance of Enterococcus bacteria was higher in the diarrhea group. Although some bacteria of this genus are commensals, a small number of species may be associated with the development of diarrhea in piglets. Therefore, our results indicate that the gut microbiome may be an important factor in the development of diarrhea in piglets.
Keywords: piglet, microbiome, 16S rRNA, sequencing, diarrhea, intestines
1. Introduction
The formation of the gut microbiome at an early age is of particular importance for piglets’ health. Microbiome composition is a perspective predictive tool for health and disease assessment; however, it stays poorly described in terms of predisposition to diarrhea. Here, we aim to assess whether the composition of the gut microbiome is associated with differences in the susceptibility of pigs to diarrhea.
The gut microbiome has a complex organization, characterized by multiple options for inter-bacterial interaction, which greatly contributes to the health and immunity of mammals [1]. It is characterized by a large population diversity of microorganisms, the composition and abundance of which are influenced by both external and internal factors, such as environmental factors and the genetics of the host organism [2].
Populations of intestinal bacteria are essential in animal production. The development of high-throughput sequencing technologies has significantly advanced the study of the microbiomes of production animals [3,4].
The trend in recent years shows an exponential increase in the number of publications using the 16S rRNA gene sequencing approach to study the gut microbiome of pigs [5,6,7,8]. Interestingly, the composition of the pig gut microbiome correlates with such indicators as average body weight and daily weight gain [9,10], conversion factor, and food consumption [11,12]. At the same time, quite a small number of studies are devoted to a detailed analysis of microbiome profiles in different parts of the intestines of piglets [13,14,15]. In addition, only pigs over 120 days old or weaned pigs were analyzed in those studies. The dynamics of gut microbiome formation in newborn piglets are of particular importance as it affects the overall animal health and growth indicators [16].
It is known that from birth the digestive system of piglets is populated with facultative aerobic or anaerobic bacteria. They receive this compound together with colostrum and then from breast milk, which contains lactic acid bacteria such as Lactobacilli and Bifidobacterium as the main probiotic bacteria. The gut microbiome composition of piglets then changes and stabilizes, influenced by feeding and environmental characteristics [17,18]. The data show that the intestines of piglets are mainly colonized by the Clostdiaceae and Enterobacteriaceae families immediately after birth. One of the studies showed that the Streptococcaceae family can be observed in the gastrointestinal tract of piglets 6 h after birth, and in the period from 1 to 3 days it becomes the most numerous. Then, as a result of secondary colonization, they are gradually replaced by Lactobacillaceae and Clostridiaceae [19]. Other studies show the attendance of bacterial species such as Lactobacillus sobrius, Escherichia coli, Lactobacillus reuteri, Shigella flexneri, and Lactobacillus acidophilus in two-day-old pigs [17]. E. coli and Clostridium spp. were observed during the first 6 h after birth. Bacteroidetes spp. appear four days after birth and are therefore considered the latest representative of the emerging microbiota [20]. According to a study by Petri et al. [19], Lactobacillaceae were the most considerable part during the first 20 days of life, which contradicts the results, according to which representatives of this group are replaced by Clostridium spp. [20,21]. According to some studies, the microbiological composition of the digestive tract of piglets is quite stable during the lactation period [22,23,24,25]. Thus, the diversity of available data indicates the demand to investigate issues related to the bacterial composition of different parts of the intestine from the birth of piglets.
Pigs and piglets suffer from a variety of infections that can be caused by the environment, food, internal parasites, bacteria, viruses, and/or the simultaneous effect of these factors. These infections are a serious threat to agronomic health and ultimately to human health [26,27]. Hence, studying infectious conditions in pigs can help not only to improve the health of livestock but also to develop new treatments against bacterial infections. Diarrhea is considered to be one of the major microbial diseases in domestic piglets, and can have devastating consequences for animal health and therefore the food industry [28]. Post-weaning diarrhea is an economically important intestinal disease because of the financial losses that it causes [29]. It commonly occurs within 2 weeks after weaning and is characterized by profuse diarrhea, dehydration, high mortality (up to 20–30% within 1–2 months [29]), and weight loss in surviving pigs [30].
The main aim of this investigation was to assess the differences between the intestine microbiomes of piglets with diarrhea and healthy animals. We also studied the association between the composition of the intestinal bacterial community and the susceptibility of piglets to diarrhea. For this purpose, we studied the composition of the microbiome in four sections of the piglet intestine (ileum, cecum, colon, and rectum) using 16S rRNA gene sequencing on the Ion Torrent Personal Genome Machine (PGM) platform.
2. Materials and Methods
2.1. Samples
Twelve 3- to 4-day-old piglets (F1 hybrids) were obtained by crossing animals of the Large White and Landrace breeds. Six piglets with severe diarrhea and six healthy piglets were selected for the experiment and kept in a room at 30 ± 2 °C with a humidity of 55 ± 7%. Piglets with diarrhea were identified by the following clinical picture: shaky, unstable gait against the background of general dehydration, loose stools, fetid smell of fecal masses, and general contamination of piglets with feces. Molecular analysis of pathological material (feces, internal organs from piglets (thin and thick intestine, liver with gallbladder, kidney, and spleen)) did not detect viral diseases. Microscopy of smears-prints from the intestinal surface and in the feces did not detect pathogens of parasitic diseases. The piglets were euthanized and portions of their gastrointestinal tract were immediately removed. The contents of the intestinal lumen were collected from the ileum, cecum, colon, and rectum. In total, 48 samples were obtained (24 from sick and 24 from healthy animals), which were placed in microcentrifuge tubes and delivered to the laboratory on ice. However, in the course of sample preparation, seven samples of the gastrointestinal tract taken from the sick group of piglets were removed from the subsequent analysis, including one sample of the ileum, three of the cecum, one of the colon, and two of the rectum. Thus, a total of 41 samples of the gastrointestinal tract of pigs were sequenced: in the sick group, 17 samples were examined, of which five were of the ileum, three were of the cecum, five were of the colon, and four were of the rectum; in the healthy group, 24 samples were examined—six from each section of the intestine.
2.2. Isolation of DNA
DNA was extracted from each sample using a ZymoBiomics DNA Miniprep Kit (Zymo Research, Los Angeles, CA, USA) according to the manufacturer’s instructions. The isolated DNA was quantified using a Qubit 2.0 benchtop fluorometer (Invitrogen, San Diego, CA, USA).
During DNA isolation from samples, we added the sample that served as a negative control for data analysis and contained only the Milli-Q water which is used in the laboratory. This sample underwent sample preparation completely identically to the test samples to exclude the contamination of the test samples in the laboratory. During the bioinformatics analysis of the obtained data we used the decontam R package to identify and subsequently remove any laboratory contaminants. This additional step makes it possible to obtain more accurate data on the composition of the studied bacterial communities, which is based on the analysis of the V3 hypervariable region of the 16S rRNA marker gene and metagenomic data.
2.3. Amplification of the 16S rRNA Gene
In our work, to study the gut microbiome of piglets using sequencing on the Ion Torrent PGM platform we selected the hypervariable region V3 of the 16S rRNA gene. For the amplification of bacterial DNA we used universal primers 337F and 518R (Table 1).
Table 1.
Primers used in the study.
| Primer | Sequence |
|---|---|
| 337F | 5′-GACTCCTACGGGAGGCWGCAG-3′ |
| 518R | 5′-GTATTACCGCGGCTGCTGG-3′ |
The amplification was performed with a 5 × ScreenMix-HS Master Mix kit (Evrogen, Moscow, Russia) in the following regime: 94 °C for 4 min; 37 cycles of 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 30 s; and final elongation at 72 °C for 5 min.
2.4. Ion Torrent PGM Sequencing
PCR products were purified using AMPureXP magnetic particles (Beckman Coulter, Brea, CA, USA). The preparation of libraries for sequencing was performed using a NEBNext Fast DNA Library Prep kit (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocol. After that, the obtained libraries were mixed in equimolar amounts for emulsion PCR on a OneTouch 2 System (Thermo Fisher Scientific, Madison, WI, USA). Sequencing was performed with an Ion PGM Hi-Q View Sequencing Kit (Thermo Fisher Scientific, Madison, WI, USA) using an Ion Torrent PGM system.
2.5. Statistical Data Analysis
The sets of sequences for each sample were obtained in a BAM format. Next, the files were converted into a FastQ format using SAMtools v.1.2 software and analyzed using the R programming language in the RStudio environment. The raw reads were filtered by length and the quality was controlled by using the functions of VSEARCH v.2.8.2 software. The samples were pooled and unique sequences were identified before searching for operational taxonomic units (OTUs). To find the OTUs, we used the UNOISE2 algorithm, which reduces noise by correcting errors. To generate the OTUs and make the OTU table, we combined all the reads for all the samples. After that, the processed reads were consistent with the reference readings in the SILVA v.123 databases (https://www.arb-silva.de, accessed on 26 October 2021) using the DADA2 package. The DADA2 package provides a native implementation of the naive Bayesian classifier method for this purpose.
The statistical analysis was performed using GraphPad Prism 9 software (GraphPad, Sand Diego, CA, USA). The differences in the bacterial composition of the microbiomes from the intestine sections were analyzed using two-way analysis of variance (ANOVA). The results are expressed as mean ± standard error of the mean (SEM).
3. Results
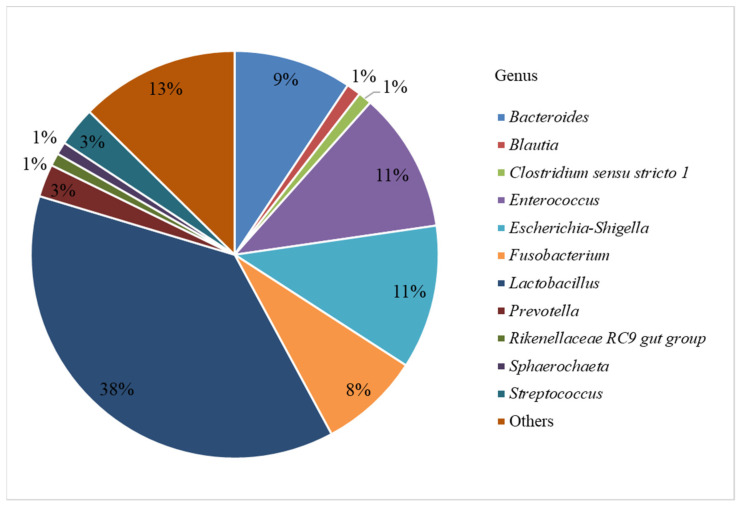
In this study, we investigated the bacterial profiles (at the level of genera) of the ileum, cecum, colon, and rectum from twelve piglets. After filtering the reads obtained by sequencing of 41 studied samples, a total of 307,977 unique sequences were identified, which corresponded to 166 genera (99% identity) (Figure 1).
Figure 1.
Relative abundance of identified bacterial genera.
Figure 1 shows that 38% of the sequences belonged to members of the Lactobacillus genus. The next in numbers were Escherichia-Shigella and Enterococcus (11% each), Bacteroides (9%), Fusobacterium (8%), Streptococcus and Prevotella (3% each), and Blautia, Clostridium sensu stricto 1, Rikenellaceae RC9 gut group, and Sphaerochaeta (1% each). The genera with the content below 1% were combined into the “Others” group.
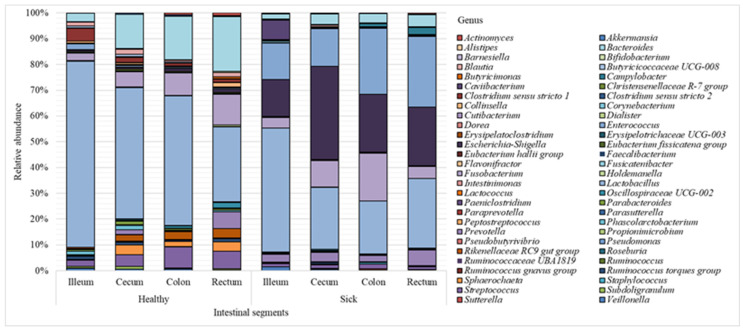
The results revealed the differences between the microbial profiles of different sections of piglet gastrointestinal tract, as well as between sick and healthy piglets (the 40 most common genera identified for each intestinal section are shown in Figure 2). The bacterial composition of individual samples can be found in Supplementary Material Figure S1.
Figure 2.
Microbiome composition (bacterial genera) of piglet intestine.
Figure 2 shows that the Lactobacillus genus prevailed over other genera in all sections of the intestine; however, a greater number of these bacteria was found in healthy piglets (51% on average) vs. animals with diarrhea (30% on average). Bacteria of the Escherichia-Shigella genus were also found in large numbers (24%) in sick piglets compared to 1% in healthy animals. Representatives of the Bacteroides genus were more common in the healthy group (14%) vs. 4% in sick piglets. The abundance of Streptococcus in the healthy animals was 6%, while in piglets with diarrhea, the content of these bacteria was approximately 1%. In the healthy group, the amount of Sphaerochaeta was 2%; in the sick group, these bacteria were absent. The relative content of representatives of the Enterococcus genus in the healthy and sick groups was 1% and 20%, respectively.
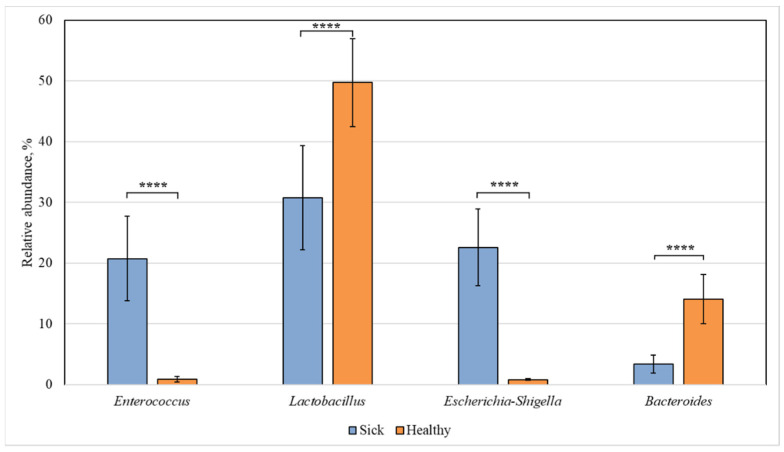
Figure 3 shows the bacterial genera, the intestinal content of which differed statistically in the healthy and diseased groups.
Figure 3.
Statistically significant differences in the content of bacterial genera in the intestine of healthy and diseased piglets. Results are expressed as mean ± SEM (**** p ≤ 0.0001).
Among all identified bacterial genera, statistically significant differences were found for the representatives of Enterococcus, Lactobacillus, Escherichia-Shigella, and Bacteroides. The content of Enterococcus and Escherichia-Shigella bacteria in the sick group increased (to 21 and 23%, respectively) compared to the healthy animals, in which their relative abundance was 1% for both genera. At the same time, in sick piglets, the content of Lactobacillus (31%) and Bacteroides (3%) was reduced compared to 50% and 14%, respectively, in the healthy group.
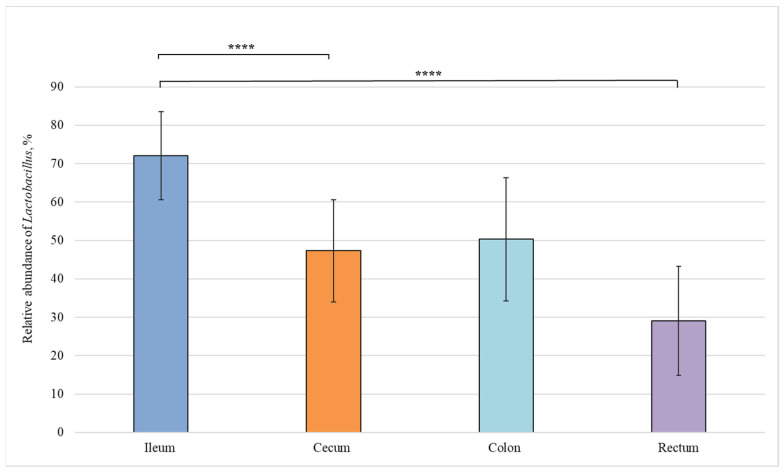
We also carried out a comparative analysis of the bacterial composition in different sections of the intestine from healthy piglets and revealed statistically significant differences in the relative abundance of bacteria belonging to the Lactobacillus, Fusobacterium, and Bacteroides genera (Figure 4, Figure 5 and Figure 6).
Figure 4.
Relative abundance of Lactobacillus genus in the intestinal sections of healthy piglets. Results are expressed as mean ± SEM (**** p ≤ 0.0001).
Figure 5.
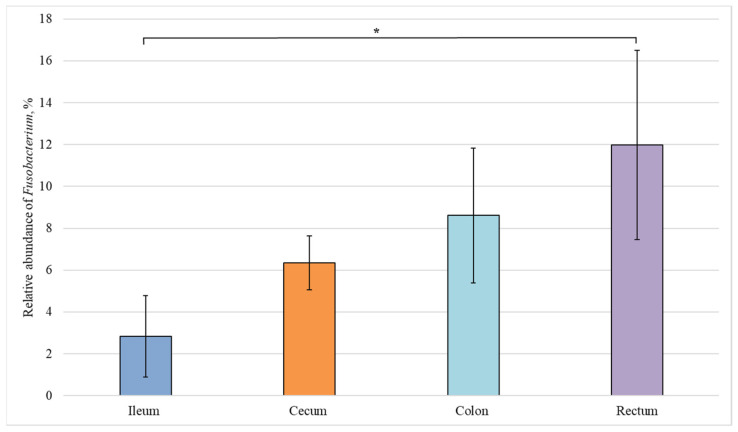
Relative abundance of the Fusobacterium genus in the intestinal sections of healthy piglets. Results are expressed as mean ± SEM (* p ≤ 0.05).
Figure 6.
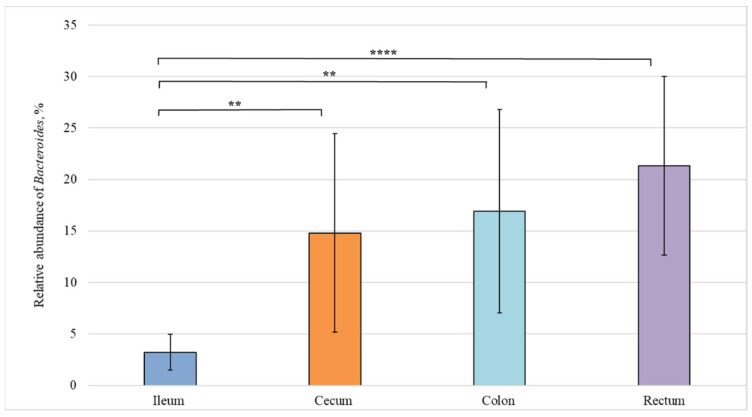
Relative abundance of the Bacteroides genus in the intestinal sections of healthy piglets. Results are expressed as mean ± SEM (** p ≤ 0.01, **** p ≤ 0.0001).
Figure 4 shows that the amount of Lactobacillus bacteria in the ileum (72%) was significantly higher than in the cecum (50%) and rectum (29%).
The relative number of representatives of the Fusobacterium genus increases in the direction from the small to the large intestine (Figure 5). However, a statistically significant difference was observed only between the content of Fusobacterium bacteria in the ileum and rectum (3% and 12%, respectively), i.e., in the most distant sections.
Figure 6 demonstrates a statistically significant difference in the content of Bacteroides genus between the ileum (3%) and cecum (15%), as well as between the ileum (3%) and colon (21%).
Next, we compared the bacterial composition of each intestinal section in the healthy and sick groups (Figure 7, Figure 8, Figure 9 and Figure 10).
Figure 7.
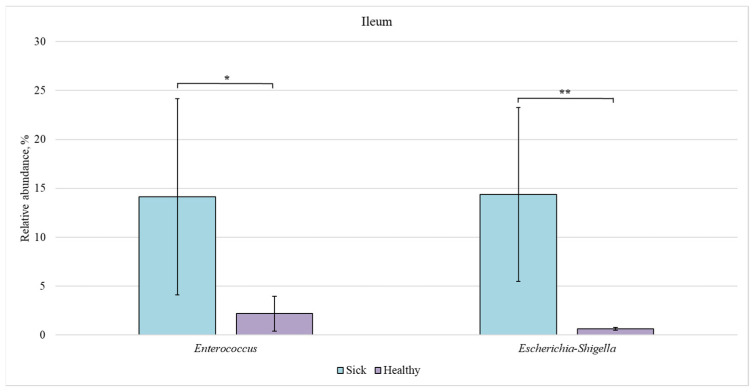
Relative abundance of Enterococcus and Escherichia-Shigella bacteria in the ileum of sick and healthy piglets. Results are expressed as mean ± SEM (* p ≤ 0.05, ** p ≤ 0.01).
Figure 8.
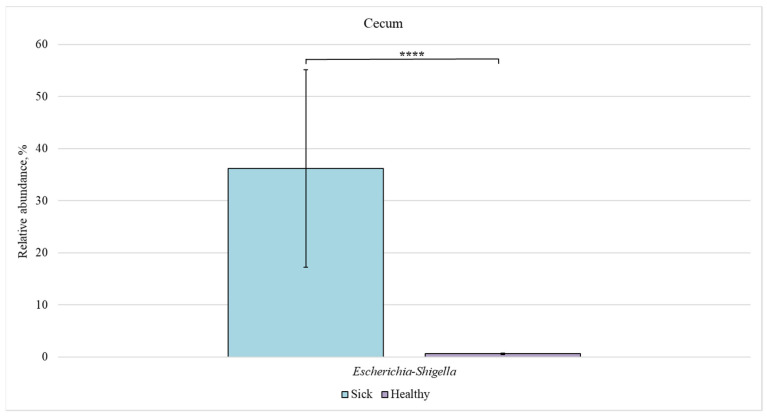
Relative abundance of Escherichia-Shigella bacteria in the cecum of sick and healthy piglets. Results are expressed as mean ± SEM (**** p ≤ 0.0001).
Figure 9.
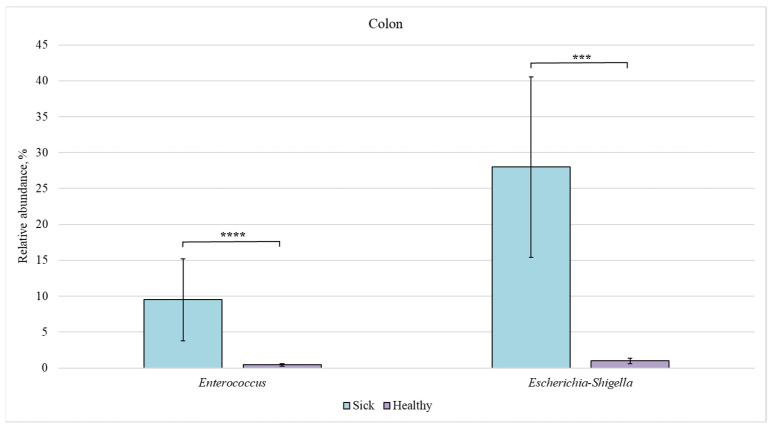
Relative abundance of Enterococcus and Escherichia-Shigella bacteria in the colon of sick and healthy piglets. Results are expressed as mean ± SEM (*** p ≤ 0.001, **** p ≤ 0.0001).
Figure 10.
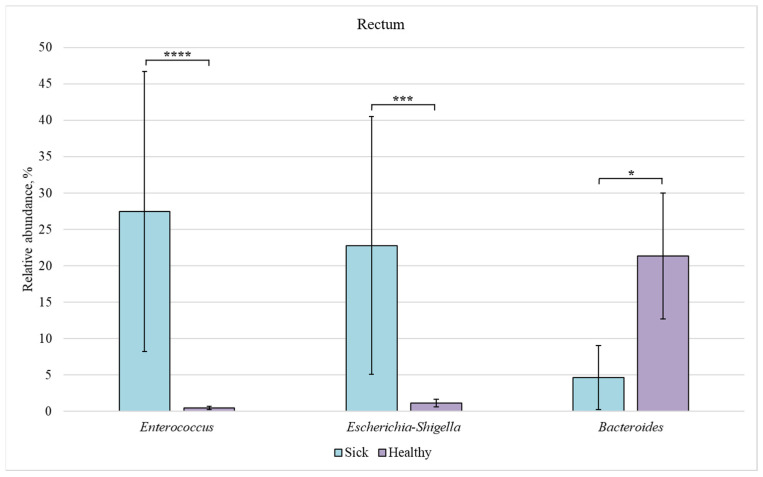
Relative abundance of Enterococcus, Escherichia-Shigella, and Bacteroides bacteria in the rectum of sick and healthy piglets. Results are expressed as mean ± SEM (* p ≤ 0.05, *** p ≤ 0.001, **** p ≤ 0.0001).
In the ileum of sick piglets, the content of Enterococcus and Escherichia-Shigella bacteria (14% each) was approximately 7 and 20 times higher than in the healthy animals (2 and 0.7%, respectively).
In the cecum, a significant difference in the relative content was found only for the Escherichia-Shigella genus (36% in sick animals vs. 0.6% in healthy piglets).
The relative content of Enterococcus (9.5%) and Escherichia-Shigella (28%) bacteria was increased in the colon and ileum of sick piglets (compared to 0.5% and 1%, respectively, in the healthy group).
In the rectum, a significant difference in the relative abundance was observed for the three genera. The content of Enterococcus and Escherichia-Shigella bacteria was increased in the sick group as compared to the healthy animals (27% and 23% vs. 0.5% and 1%, respectively). However, the number of representatives of the Bacteroides genus was reduced in sick piglets (5%) in comparison with the healthy ones (21%).
4. Discussion
In this study, we discovered that Lactobacillus was one of the major genera of the pig gastrointestinal tract (Figure 1). Lactobacillus representatives are common in both proximal and distal sections of pig digestive tract, which they colonize shortly after birth [31].
Lactobacillus bacteria possess several probiotic properties necessary for resistance to infections and diseases of the gastrointestinal tract. They exhibit antipathogenic activity [32,33] and antioxidant activity [34,35] and are involved in the regulation of immune system [36]. All these traits affect gut microbial populations, such that propagation of opportunistic pathogens, such as Salmonella, Clostridia, and Enterobacteriaceae, is controlled, resulting in the prevention of infections and intestinal disorders [37,38,39]. It has been shown that Lactobacillus species colonize piglet intestine shortly after birth and are stable members of the gut microbiome throughout the entire intestinal tract [40]. The low abundance of Lactobacillus representatives, which are considered beneficial, may be an indicator of gastrointestinal problems in pigs. In our study (Figure 3), we observed a reduced amount of Lactobacillus bacteria in piglets with diarrhea, which confirms the probiotic properties of the Lactobacillus genus.
Physiological variations along the length of the intestine include chemical and nutrient gradients, as well as compartmentalized host immune activity. All these factors influence bacterial community composition [41]. It is well known that piglets experience extreme stress when they are weaned from the sow. This can lead to the development of intestinal dysfunction and immune system disorders, which ultimately leads to a deterioration in the health of piglets, especially during the first week after weaning. We would like to point out that only suckling piglets were involved in this study. This made it possible to exclude the influence of stress endured during weaning on changes in the microbiome and, as a consequence, the development of diarrhea [42].
According to the results of our study, Lactobacillus dominated in the distal part of the small intestine (ileum) (Figure 4). Lactobacillus species grow in an oxygen-free or low-oxygen atmosphere. Oxygen present in the small intestine is gradually depleted by aerobic bacteria, and only a small amount of it remains in the ileum, where most Lactobacillus bacteria are found. Some Lactobacillus species produce acetic acid, which has a fairly strong antipathogenic effect [43].
In our study, the content of Escherichia-Shigella bacteria was much higher in piglets with diarrhea (Figure 3). Several Escherichia-Shigella species are believed to play an important role in the development of diarrhea in piglets and to have a serious effect on the barrier function of the animal intestine [44]. An increased number of Escherichia-Shigella bacteria in sick piglets vs. healthy ones was observed in all sections of the intestine (Figure 7, Figure 8, Figure 9 and Figure 10). However, we found no differences in the number of these bacteria in the intestine sections in the healthy group. This suggests that Escherichia-Shigella bacteria are distributed approximately evenly in the investigated sections of the intestine.
Members of the Bacteroides genus are often found in the digestive tract of mammals. They are early colonizers of the intestines of healthy piglets [19]. Bacteroides species play an important role in health promotion by producing butyrate, which activates T cell-mediated immune response, thus limiting colonization of the digestive tract by potentially pathogenic bacteria [45]. In the healthy group, bacteria of this genus were more common in the rectum (Figure 6). The number of Bacteroides representatives in the rectum of piglets with diarrhea was lower than in the control group (Figure 10).
We also found an increased number of Enterococcus bacteria in sick piglets (Figure 3). Information on the enterococcal flora of healthy newborn piglets is scanty, but a small number of representatives of this genus can be a part of the normal gut microbiota [46,47]. However, Enterococcus is sometimes associated with piglet diarrhea [48,49]. Although many members of the Enterococcus genus are believed to be commensals of the intestinal tract, some representatives cause diarrhea in suckling animals of various species [50,51,52,53,54,55].
We also found an increased number of Enterococcus bacteria in the ileum (Figure 7), colon (Figure 9), and rectum (Figure 10) of piglets with diarrhea compared to healthy animals. This may indicate that pathogenic Enterococcus representatives are prevalent in the intestinal microbiome, which may be associated with the diarrhea of newborns piglets.
Fusobacterium species are anaerobic, Gram-negative, non-spore-forming, non-motile, rod-shaped bacteria. The main metabolite produced by these bacteria is butyric acid [56]. According to numerous data, Fusobacterium is considered to be a normal representative of the oropharyngeal, gastrointestinal and genital microbiota. At the same time, this genus is the second most commonly isolated anaerobic microbial group from clinical samples of both humans and animals, especially in the case of purulent-necrotic infections [57]. Fusobacterium is involved in various clinical anaerobic infections and can cause intestinal inflammation [58]. Even though many studies have shown increased levels of Fusobacterium in piglets with various intestinal disorders compared to the healthy ones [59,60,61,62,63], we did not find statistically significant differences in the content of this genus in the studied groups. However, we found an increased content of Fusobacterium members in the rectum compared to the ileum of healthy pigs. The content of Fusobacterium bacteria in newborn piglets requires further research.
We studied the bacterial composition inhabiting the intestines of healthy newborn piglets, as well as piglets with diarrhea. Thus, our study, using high-throughput sequencing, showed that microbial communities in the studied samples included many commensal and opportunistic microorganisms. Compared to classical microbiological and immunological approaches, as well as PCR, the advantage of this method is the ability to identify all bacteria contained in the test sample, including non-culturable microorganisms [64]. We used the universal primer pair 337F/518R (see Section 2.3. Amplification of the 16S rRNA gene) to amplify a fragment of the 16S rRNA gene that includes the V3 region, which is considered one of the most effective hypervariable regions for phylogenetic analysis and taxonomic classification of bacterial species [65,66].
The results of this study can be used to predict bacterial taxa indicative of healthy development of gut microbiome in suckling piglets, as well as to identify taxa that can be used as probiotics to prevent post-weaning diarrhea.
5. Conclusions
The most common genera in the piglet microbiome were Lactobacillus, Escherichia-Shigella, Enterococcus, Bacteroides, and Fusobacterium. Bacteria of the Lactobacillus genus dominated in healthy piglets, which once again proves their probiotic effect. An increased number of Escherichia-Shigella representatives in diarrheal pigs indicates the contribution of these bacteria to the development of diarrhea. A decreased number of Bacteroides may also indicate the development of diarrhea. The content of Enterococcus bacteria was higher in sick piglets. According to the assessment of the microbiome composition in different sections of the intestine, bacteria of the Lactobacillus genus were most common in the ileum, while Fusobacterium and Bacteroides were more common in the rectum.
Further studies are needed to understand the protective mechanisms of the gut microbial community and to develop clinical interventions to improve gut health in piglets.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani12030320/s1, Figure S1: The abundance of microorganisms for the samples.
Author Contributions
Conceptualization, M.Y.S., E.V.M. and V.N.P.; methodology, N.A.S., M.Y.S. and E.V.M.; software, M.V.G., M.Y.S. and Y.D.D.; validation, M.V.G., M.Y.S. and Y.D.D.; formal analysis, M.V.G. and Y.D.D.; investigation, N.A.S., M.V.G. and Y.D.D.; resources, M.Y.S., E.V.M. and V.N.P.; data curation, M.V.G. and Y.D.D.; writing—original draft preparation, M.V.G., S.V.S., and M.Y.S.; writing—review and editing, M.Y.S. and V.N.P.; visualization, M.V.G. and Y.D.D.; supervision, P.A.P., S.V.S. and M.Y.S.; project administration, V.N.P.; funding acquisition, V.N.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Ministry of Science and Higher Education of the Russian Federation in the framework of the national project “Science” (project FZGW-2020-0001, unique number of the register of State tasks 075001X39782002).
Institutional Review Board Statement
The experiments on animals were approved by the Ethics Commission of FSBSI “All-Russian Veterinary Research Institute of Pathology, Pharmacology and Therapy” (FSBSI “ARVRIPP&T”) (Protocol No. 01/12-2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw sequencing data are available in the NCBI BioProject database (BioProject ID: PRJNA794980).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cho I., Blaser M.J. The Human Microbiome: At the Interface of Health and Disease. Nat. Rev. Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu D., Tiezzi F., Schillebeeckx C., McNulty N.P., Schwab C., Shull C., Maltecca C. Host Contributes to Longitudinal Diversity of Fecal Microbiota in Swine Selected for Lean Growth. Microbiome. 2018;6:4. doi: 10.1186/s40168-017-0384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeoman C.J., White B.A. Gastrointestinal Tract Microbiota and Probiotics in Production Animals. Annu. Rev. Anim. Biosci. 2014;2:469–486. doi: 10.1146/annurev-animal-022513-114149. [DOI] [PubMed] [Google Scholar]
- 4.De Rodas B., Youmans B.P., Danzeisen J.L., Tran H., Johnson T.J. Microbiome Profiling of Commercial Pigs from Farrow to Finish. J. Anim. Sci. 2018;96:1798–1994. doi: 10.1093/jas/sky109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivarsson E., Roos S., Liu H.Y., Lindberg J.E. Fermentable Non-Starch Polysaccharides Increases the Abundance of Bacteroides-Prevotella-Porphyromonas in Ileal Microbial Community of Growing Pigs. Anim. Int. J. Anim. Biosci. 2014;8:1777–1787. doi: 10.1017/S1751731114001827. [DOI] [PubMed] [Google Scholar]
- 6.Kraler M., Ghanbari M., Domig K.J., Schedle K., Kneifel W. The Intestinal Microbiota of Piglets Fed with Wheat Bran Variants as Characterised by 16S RRNA Next-Generation Amplicon Sequencing. Arch. Anim. Nutr. 2016;70:173–189. doi: 10.1080/1745039X.2016.1160534. [DOI] [PubMed] [Google Scholar]
- 7.Mach N., Berri M., Estellé J., Levenez F., Lemonnier G., Denis C., Leplat J.J., Chevaleyre C., Billon Y., Doré J., et al. Early-Life Establishment of the Swine Gut Microbiome and Impact on Host Phenotypes. Environ. Microbiol. Rep. 2015;7:554–569. doi: 10.1111/1758-2229.12285. [DOI] [PubMed] [Google Scholar]
- 8.Slifierz M.J., Friendship R.M., Weese J.S. Longitudinal Study of the Early-Life Fecal and Nasal Microbiotas of the Domestic Pig. BMC Microbiol. 2015;15:184. doi: 10.1186/s12866-015-0512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramayo-Caldas Y., Mach N., Lepage P., Levenez F., Denis C., Lemonnier G., Leplat J.J., Billon Y., Berri M., Doré J., et al. Phylogenetic Network Analysis Applied to Pig Gut Microbiota Identifies an Ecosystem Structure Linked with Growth Traits. ISME J. 2016;10:2973–2977. doi: 10.1038/ismej.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsiamis G., Ntougias S., Sabree Z.L., Chen C., He M., Fang S., Huang X., Zhao Y., Ke S., Yang H., et al. Evaluating the Contribution of Gut Microbiota to the Variation of Porcine Fatness with the Cecum and Fecal Samples. Front. Microbiol. 2016;7:2108. doi: 10.3389/fmicb.2016.02108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camarinha-Silva A., Maushammer M., Wellmann R., Vital M., Preuss S., Bennewitz J. Host Genome Influence on Gut Microbial Composition and Microbial Prediction of Complex Traits in Pigs. Genetics. 2017;206:1637–1644. doi: 10.1534/genetics.117.200782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormack U.M., Curião T., Buzoianu S.G., Prieto M.L., Ryan T., Varley P., Crispie F., Magowan E., Metzler-Zebeli B.U., Berry D., et al. Exploring a Possible Link between the Intestinal Microbiota and Feed Efficiency in Pigs. Appl. Environ. Microbiol. 2017;83:1–42. doi: 10.1128/AEM.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crespo-Piazuelo D., Estellé J., Revilla M., Criado-Mesas L., Ramayo-Caldas Y., Óvilo C., Fernández A.I., Ballester M., Folch J.M. Characterization of Bacterial Microbiota Compositions along the Intestinal Tract in Pigs and Their Interactions and Functions. Sci. Rep. 2018;8:12727. doi: 10.1038/s41598-018-30932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly J., Daly K., Moran A.W., Ryan S., Bravo D., Shirazi-Beechey S.P. Composition and Diversity of Mucosa-Associated Microbiota along the Entire Length of the Pig Gastrointestinal Tract; Dietary Influences. Environ. Microbiol. 2017;19:1425–1438. doi: 10.1111/1462-2920.13619. [DOI] [PubMed] [Google Scholar]
- 15.Yang H., Huang X., Fang S., Xin W., Huang L., Chen C. Uncovering the Composition of Microbial Community Structure and Metagenomics among Three Gut Locations in Pigs with Distinct Fatness. Sci. Rep. 2016;6:27427. doi: 10.1038/srep27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guevarra R.B., Lee J.H., Lee S.H., Seok M.J., Kim D.W., Kang B.N., Johnson T.J., Isaacson R.E., Kim H.B. Piglet Gut Microbial Shifts Early in Life: Causes and Effects. J. Anim. Sci. Biotechnol. 2019;10:1. doi: 10.1186/s40104-018-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konstantinov S.R., Awati A.A., Williams B.A., Miller B.G., Jones P., Stokes C.R., Akkermans A.D.L., Smidt H., de Vos W.M. Post-Natal Development of the Porcine Microbiota Composition and Activities. Environ. Microbiol. 2006;8:1191–1199. doi: 10.1111/j.1462-2920.2006.01009.x. [DOI] [PubMed] [Google Scholar]
- 18.Naito S., Hayashidani H., Kaneko K., Ogawa M., Benno Y. Development of Intestinal Lactobacilli in Normal Piglets. J. Appl. Bacteriol. 1995;79:230–236. doi: 10.1111/j.1365-2672.1995.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 19.Petri D., Hill J.E., van Kessel A.G. Microbial Succession in the Gastrointestinal Tract (GIT) of the Preweaned Pig. Livest. Sci. 2010;133:107–109. doi: 10.1016/j.livsci.2010.06.037. [DOI] [Google Scholar]
- 20.Swords W.E., Wu C.C., Champlin F.R., Buddington R.K. Postnatal Changes in Selected Bacterial Groups of the Pig Colonic Microflora. Biol. Neonate. 1993;63:191–200. doi: 10.1159/000243931. [DOI] [PubMed] [Google Scholar]
- 21.Inoue R., Tsukahara T., Nakanishi N., Ushida K. Development of the Intestinal Microbiota in the Piglet. J. Gen. Appl. Microbiol. 2005;51:257–265. doi: 10.2323/jgam.51.257. [DOI] [PubMed] [Google Scholar]
- 22.Konstantinov S.R., Awati A., Smidt H., Williams B.A., Akkermans A.D.L., de Vos W.M. Specific Response of a Novel and Abundant Lactobacillus amylovorus-Like Phylotype to Dietary Prebiotics in the Guts of Weaning Piglets. Appl. Environ. Microbiol. 2004;70:3821–3830. doi: 10.1128/AEM.70.7.3821-3830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M., Gong J., Cottrill M., Yu H., de Lange C., Burton J., Topp E. Evaluation of QIAamp DNA Stool Mini Kit for Ecological Studies of Gut Microbiota. J. Microbiol. Methods. 2003;54:13–20. doi: 10.1016/S0167-7012(02)00260-9. [DOI] [PubMed] [Google Scholar]
- 24.Hill J.E., Hemmingsen S.M., Goldade B.G., Dumonceaux T.J., Klassen J., Zijlstra R.T., Swee H.G., van Kessel A.G. Comparison of Ileum Microflora of Pigs Fed Corn-, Wheat-, or Barley-Based Diets by Chaperonin-60 Sequencing and Quantitative PCR. Appl. Environ. Microbiol. 2005;71:867–875. doi: 10.1128/AEM.71.2.867-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knecht D., Cholewińska P., Jankowska-Mąkosa A., Czyż K. Development of Swine’s Digestive Tract Microbiota and Its Relation to Production Indices—A Review. Animals. 2020;10:527. doi: 10.3390/ani10030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng X.J., Lindsay D.S. Wild Boars as Sources for Infectious Diseases in Livestock and Humans. Philos. Trans. R. Soc. B Biol. Sci. 2009;364:2697–2707. doi: 10.1098/rstb.2009.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers K.P., Olsen C.W., Gray G.C. Cases of Swine Influenza in Humans: A Review of the Literature. Clin. Infect. Dis. 2007;44:1084–1088. doi: 10.1086/512813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue S., Li Z., Hu F., Picimbon J.F. Curing Piglets from Diarrhea and Preparation of a Healthy Microbiome with Bacillus Treatment for Industrial Animal Breeding. Sci. Rep. 2020;10:19476. doi: 10.1038/s41598-020-75207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amezcua R., Friendship R.M., Dewey C.E., Gyles C., Fairbrother J.M. Presentation of Postweaning Escherichia coli Diarrhea in Southern Ontario, Prevalence of Hemolytic E. coli Serogroups Involved, and Their Antimicrobial Resistance Patterns. Can. J. Vet. Res. 2002;66:73–78. [PMC free article] [PubMed] [Google Scholar]
- 30.Fairbrother J.M., Nadeau É., Gyles C.L. Escherichia coli in Postweaning Diarrhea in Pigs: An Update on Bacterial Types, Pathogenesis, and Prevention Strategies. Anim. Health Res. Rev. 2005;6:17–39. doi: 10.1079/AHR2005105. [DOI] [PubMed] [Google Scholar]
- 31.Delia E., Tafaj M., Männer K. Probiotic in Animals. IntechOpen; London, UK: 2012. Efficiency of Probiotics in Farm Animals. [DOI] [Google Scholar]
- 32.Bomba A., Nemcová R., Gancarčíková S., Herich R., Kaštel R. Potentiation of the Effectiveness of Lactobacillus casei in the Prevention of E. coli Induced Diarrhea in Conventional and Gnotobiotic Pigs. Adv. Exp. Med. Biol. 2000;473:185–190. doi: 10.1007/978-1-4615-4143-1_18. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y., Zhao X., Le M.H.A., Zijlstra R.T., Gänzle M.G. Reutericyclin Producing Lactobacillus reuteri Modulates Development of Fecal Microbiota in Weanling Pigs. Front. Microbiol. 2015;6:762. doi: 10.3389/fmicb.2015.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang A., Yu H., Gao X., Li X., Qiao S. Influence of Lactobacillus fermentum I5007 on the Intestinal and Systemic Immune Responses of Healthy and E. coli Challenged Piglets. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2009;96:89–98. doi: 10.1007/s10482-009-9339-2. [DOI] [PubMed] [Google Scholar]
- 35.Wang A.N., Cai C.J., Zeng X.F., Zhang F.R., Zhang G.L., Thacker P.A., Wang J.J., Qiao S.Y. Dietary Supplementation with Lactobacillus fermentum I5007 Improves the Anti-Oxidative Activity of Weanling Piglets Challenged with Diquat. J. Appl. Microbiol. 2013;114:1582–1591. doi: 10.1111/jam.12188. [DOI] [PubMed] [Google Scholar]
- 36.Suda Y., Villena J., Takahashi Y., Hosoya S., Tomosada Y., Tsukida K., Shimazu T., Aso H., Tohno M., Ishida M., et al. Immunobiotic Lactobacillus jensenii as Immune-Health Promoting Factor to Improve Growth Performance and Productivity in Post-Weaning Pigs. BMC Immunol. 2014;15:24. doi: 10.1186/1471-2172-15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemcová R., Bomba A., Gancarčiková S., Herich R., Guba P. Study of the Effect of Lactobacillus paracasei and Fructooligosaccharides on the Faecal Microflora in Weanling Piglets. Berl. Munch. Tierarztl. Wochenschr. 1999;112:225–228. doi: 10.1080/09540100220145000e. [DOI] [PubMed] [Google Scholar]
- 38.Gebru E., Lee J.S., Son J.C., Yang S.Y., Shin S.A., Kim B., Kim M.K., Park S.C. Effect of Probiotic-, Bacteriophage-, or Organic Acid-Supplemented Feeds or Fermented Soybean Meal on the Growth Performance, Acute-Phase Response, and Bacterial Shedding of Grower Pigs Challenged with Salmonella enterica Serotype Typhimurium. J. Anim. Sci. 2010;88:3880–3886. doi: 10.2527/jas.2010-2939. [DOI] [PubMed] [Google Scholar]
- 39.Liu H., Zhang J., Zhang S., Yang F., Thacker P.A., Zhang G., Qiao S., Ma X. Oral Administration of Lactobacillus fermentum I5007 Favors Intestinal Development and Alters the Intestinal Microbiota in Formula-Fed Piglets. J. Agric. Food Chem. 2014;62:860–866. doi: 10.1021/jf403288r. [DOI] [PubMed] [Google Scholar]
- 40.Kenworthy R., Crabb W.E. The intestinal flora of young pigs, with reference to early weaning, Escherichia coli and scours. J. Comp. Pathol. 1963;73:215–228. doi: 10.1016/S0368-1742(63)80025-9. [DOI] [PubMed] [Google Scholar]
- 41.Donaldson G.P., Lee S.M., Mazmanian S.K. Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell J.M., Crenshaw J.D., Polo J. The Biological Stress of Early Weaned Piglets. J. Anim. Sci. Biotechnol. 2013;4:19. doi: 10.1186/2049-1891-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandis-Heep A. Encyclopedia of Food Microbiology. Academic Press; San Diego, CA, USA: 2000. [Google Scholar]
- 44.Chen L., Xu Y., Chen X., Fang C., Zhao L., Chen F. The Maturing Development of Gut Microbiota in Commercial Piglets during the Weaning Transition. Front. Microbiol. 2017;8:1688. doi: 10.3389/fmicb.2017.01688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W., Ma C., Xie P., Zhu Q., Wang X., Yin Y., Kong X. Gut Microbiota of Newborn Piglets with Intrauterine Growth Restriction Have Lower Diversity and Different Taxonomic Abundances. J. Appl. Microbiol. 2019;127:354–369. doi: 10.1111/jam.14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devriese L.A., Hommez J., Pot B., Haesebrouck F. Identification and Composition of the Streptococcal and Enterococcal Flora of Tonsils, Intestines and Faeces of Pigs. J. Appl. Bacteriol. 1994;77:31–36. doi: 10.1111/j.1365-2672.1994.tb03040.x. [DOI] [PubMed] [Google Scholar]
- 47.Leser T.D., Amenuvor J.Z., Jensen T.K., Lindecrona R.H., Boye M., Moøller K. Culture-Independent Analysis of Gut Bacteria: The Pig Gastrointestinal Tract Microbiota Revisited. Appl. Environ. Microbiol. 2002;68:673–690. doi: 10.1128/AEM.68.2.673-690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coudert P. Les Principales Maladies Du Porc. Actual. Pharm. 2018;57:50. doi: 10.1016/j.actpha.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheon D.S., Chae C. Outbreak of Diarrhea Associated with Enterococcus durans in Piglets. J. Vet. Diagn. Investig. 1996;8:123–124. doi: 10.1177/104063879600800123. [DOI] [PubMed] [Google Scholar]
- 50.Tzipori S., Hayes J., Sims L., Withers M. Streptococcus Durans: An Unexpected Enteropathogen of Foals. J. Infect. Dis. 1984;150:589–593. doi: 10.1093/infdis/150.4.589. [DOI] [PubMed] [Google Scholar]
- 51.Collins J.E., Bergeland M.E., Lindeman C.J., Duimstra J.R. Enterococcus (Streptococcus) durans Adherence in the Small Intestine of a Diarrheic Pup. Vet. Pathol. 1988;25:396–398. doi: 10.1177/030098588802500513. [DOI] [PubMed] [Google Scholar]
- 52.Etheridge M.E., Vonderfecht S.L. Diarrhea Caused by a Slow-Growing Enterococcus-like Agent in Neonatal Rats. Lab. Anim. Sci. 1992;42:548–550. [PubMed] [Google Scholar]
- 53.Rogers D.G., Zeman D.H., Erickson E.D. Diarrhea Associated with Enterococcus durans in Calves. J. Vet. Diagn. Investig. 1992;4:471–472. doi: 10.1177/104063879200400423. [DOI] [PubMed] [Google Scholar]
- 54.Nicklas J.L., Moisan P., Stone M.R., Gookin J.L. In Situ Molecular Diagnosis and Histopathological Characterization of Enteroadherent Enterococcus hirae Infection in Pre-Weaning-Age Kittens. J. Clin. Microbiol. 2010;48:2814–2820. doi: 10.1128/JCM.00916-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vela A.I., Fernández A., Moreno B., Casamayor A., Chacón G., Villa A., Comenge J., Fernández-Garayzábal J.F. Isolation of Enterococcus hirae from Suckling Rabbits with Diarrhoea. Vet. Rec. 2010;167:345–346. doi: 10.1136/vr.c4460. [DOI] [PubMed] [Google Scholar]
- 56.Aladame N. Bergey’s Manual of Systematic Bacteriology. Ann. Inst. Pasteur Microbiol. 1987;138:1–46. doi: 10.1016/0769-2609(87)90099-8. [DOI] [Google Scholar]
- 57.Rowland M.D., Del Bene V.E., Lewis J.W. Factors Affecting Antimicrobial Susceptibility of Fusobacterium Species. J. Clin. Microbiol. 1987;25:476–479. doi: 10.1128/jcm.25.3.476-479.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan Z., Dong W., Ding Y., Ding X., Zhang Q., Jiang L. Changes in Cecal Microbiota Community of Suckling Piglets Infected with Porcine Epidemic Diarrhea Virus. PLoS ONE. 2019;14:e0219868. doi: 10.1371/journal.pone.0219868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swidsinski A., Dörffel Y., Loening-Baucke V., Theissig F., Rückert J.C., Ismail M., Rau W.A., Gaschler D., Weizenegger M., Kühn S., et al. Acute Appendicitis Is Characterised by Local Invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 60.Yoneda S., Loeser B., Feng J., Dmytryk J., Qi F., Merritt J. Ubiquitous Sialometabolism Present among Oral Fusobacteria. PLoS ONE. 2014;9:e99263. doi: 10.1371/journal.pone.0099263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jia Y.P., Wang K., Zhang Z.J., Tong Y.N., Han D., Hu C.Y., Li Q., Xiang Y., Mao X.H., Tang B. TLR2/TLR4 Activation Induces Tregs and Suppresses Intestinal Inflammation Caused by Fusobacterium nucleatum in Vivo. PLoS ONE. 2017;12:e0186179. doi: 10.1371/journal.pone.0186179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flemer B., Warren R.D., Barrett M.P., Cisek K., Das A., Jeffery I.B., Hurley E., O’Riordain M., Shanahan F., O’Toole P.W. The Oral Microbiota in Colorectal Cancer Is Distinctive and Predictive. Gut. 2018;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang A., Cai R., Wang Q., Shi L., Li C., Yan H. Dynamic Change of Gut Microbiota during Porcine Epidemic Diarrhea Virus Infection in Suckling Piglets. Front. Microbiol. 2019;10:322. doi: 10.3389/fmicb.2019.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta S., Mortensen M.S., Schjørring S., Trivedi U., Vestergaard G., Stokholm J., Bisgaard H., Krogfelt K.A., Sørensen S.J. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun. Biol. 2019;2:291. doi: 10.1038/s42003-019-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bukin Y., Galachyants Y., Morozov I., Bukin S.V., Zakharenko A.S., Zemskaya T.I. The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci. Data. 2019;6:190007. doi: 10.1038/sdata.2019.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sirichoat A., Sankuntaw N., Engchanil C., Buppasiri P., Faksri K., Namwat W., Chantratita W., Lulitanond V. Comparison of different hypervariable regions of 16S rRNA for taxonomic profiling of vaginal microbiota using next-generation sequencing. Arch. Microbiol. 2021;203:1159–1166. doi: 10.1007/s00203-020-02114-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available in the NCBI BioProject database (BioProject ID: PRJNA794980).