Abstract
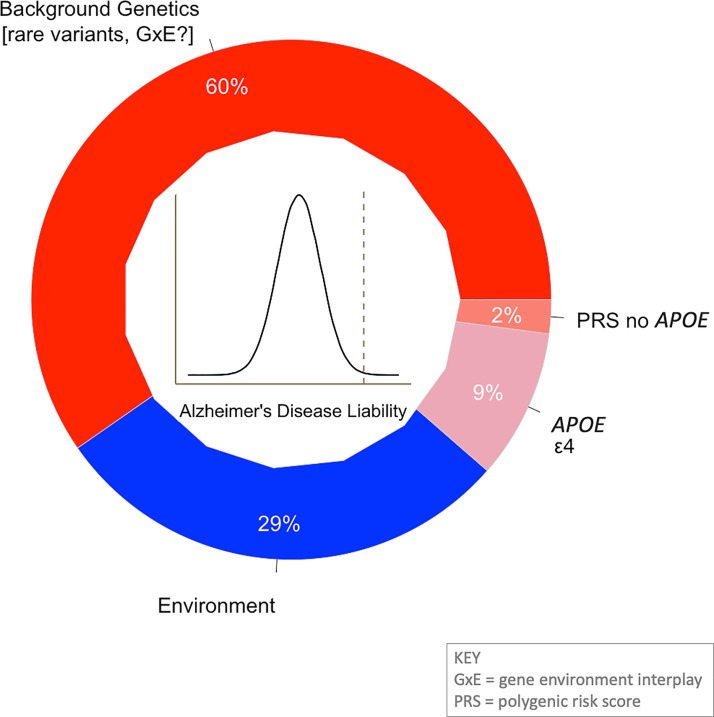
The heritability of Alzheimer’s disease estimated from twin studies is greater than the heritability derived from genome-based studies, for reasons that remain unclear. We apply both approaches to the same twin sample, considering both Alzheimer’s disease polygenic risk scores and heritability from twin models, to provide insight into the role of measured genetic variants and to quantify uncaptured genetic risk. A population-based heritability and polygenic association study of Alzheimer’s disease was conducted between 1986 and 2016 and is the first study to incorporate polygenic risk scores into biometrical twin models of Alzheimer’s disease. The sample included 1586 twins drawn from the Swedish Twin Registry which were nested within 1137 twin pairs (449 complete pairs and 688 incomplete pairs) with clinically based diagnoses and registry follow-up (Mage = 85.28, SD = 7.02; 44% male; 431 cases and 1155 controls). We report contributions of polygenic risk scores at P < 1 × 10−5, considering a full polygenic risk score (PRS), PRS without the APOE region (PRS.no.APOE) and PRS.no.APOE plus directly measured APOE alleles. Biometric twin models estimated the contribution of environmental influences and measured (PRS) and unmeasured genes to Alzheimer’s disease risk. The full PRS and PRS.no.APOE contributed 10.1 and 2.4% to Alzheimer’s disease risk, respectively. When APOE ɛ4 alleles were added to the model with the PRS.no.APOE, the total contribution was 11.4% to Alzheimer’s disease risk, where APOE ɛ4 explained 9.3% and PRS.no.APOE dropped from 2.4 to 2.1%. The total genetic contribution to Alzheimer’s disease risk, measured and unmeasured, was 71% while environmental influences unique to each twin accounted for 29% of the risk. The APOE region accounts for much of the measurable genetic contribution to Alzheimer’s disease, with a smaller contribution from other measured polygenic influences. Importantly, substantial background genetic influences remain to be understood.
Keywords: polygenic risk scores (PRSs), APOE, heritability, twins, Alzheimer’s disease
Karlsson et al. report that measured polygenic scores from genome-based studies, including an outsized role for APOE, explain only a fraction of the heritability indicated by twin models of Alzheimer’s disease, leaving most genetic risk for Alzheimer’s disease unexplained. Sensitive designs are needed to capture all the genetic influences.
Graphical Abstract
Introduction
Alzheimer’s disease is multifactorial with contributions of genetic and environmental influences. Twin studies leveraging the relative similarity of Alzheimer’s disease risk among identical or monozygotic (MZ) versus fraternal or dizygotic (DZ) twin pairs suggests an overall heritability of 0.58, with a maximum heritability of 0.79 if shared environmental influences are discounted.1 Thus, 58–79% of the liability to late-onset Alzheimer’s disease is heritable. By comparison, measured loci contributing to late-onset Alzheimer’s disease risk may capture up to 50% of the heritability.2 However, the comparability of estimates remains unclear as the estimation of polygenic contribution varies across study designs. We sought to provide insight into the role of APOE, which codes for apolipoprotein E, the major cholesterol transporter in the brain, and other measured genetic variants using polygenic risk scores (PRSs), as well as quantify uncaptured genetic risk in Alzheimer’s disease, within the same sample of twins.
The application of the PRS approach, a weighted sum of single nucleotide polymorphism (SNP) variants based on the effect sizes from genome-wide association study (GWAS), leads to enhanced accuracy in the prediction of Alzheimer’s disease risk. For example, in case–control samples from the GERARD consortia, the best prediction accuracy using area under the curve (AUC) was 0.78 (0.77–0.80) based on a logistic regression model with measured apolipoprotein E (APOE) genotypes, a PRS comprising 20 SNPs from the Lambert et al.3 GWAS meta-analysis, sex and age.4 APOE ɛ4 alone achieves an AUC of about 0.685; however, when APOE ɛ4 carriers are excluded, the prediction accuracy of the PRS achieves an AUC of 0.65.5 That is, PRS prediction of risk is substantial even for those who do not carry the ɛ4 allele. Moreover, AUC model-based inferred heritability from maximum prediction models6 suggests that in neuropathologically confirmed cases and controls, heritability estimates can be inferred to lie between 27 and 55%7 based on common genome-wide SNPs contributing to liability and accounting for age-related increases in prevalence. This range is in line with other estimates of SNP-based heritability of 24–53%, with APOE ɛ4 accounting for approximately one-quarter of the genetic contributions to liability.8,9 Apart from APOE, other genes identified in recent GWAS are involved in amyloid precursor protein (APP) metabolism/β-amyloid (Aβ) formation and regulation of APP catabolic process, τ-protein binding, lipid metabolism and immune response.10,11
How much heritable variation a PRS captures for Alzheimer’s disease risk may be related to its genetic architecture. Recent work suggests that Alzheimer’s disease may be oligogenic, or influenced by a limited set of common genetic variants compared with other complex traits.12 However, the age distribution among Alzheimer’s disease cases versus controls, and thus differences in the prevalence of APOE ɛ2 versus ɛ4 allele frequencies can impact PRS prediction.13 In addition to Alzheimer’s disease risk, APOE is associated with longevity where the allele frequencies for ɛ2 become more prevalent in older samples and ɛ4 alleles become less prevalent, at least in samples of European and Asian ancestries.14–17 Moreover, the methods used to construct PRSs for Alzheimer’s disease can impact the composition of genetic variants included and hence prediction. A PRS constructed from a clumping and P-value threshold approach PRS(C + T) and related methods outperform or are comparable with other approaches (e.g. LDPRED and SBayesR).13 The best prediction was observed in a model combining directly measured APOE with the PRS excluding the APOE region at a threshold of P ≤ 0.10, whereas the prediction accuracy was attenuated at more relaxed thresholds despite increases in variants.13 Altogether, recent findings suggest that Alzheimer’s disease is polygenic and the age-related nature of the risk is essential to consider.13
The gap between heritability estimates from genome-based and twin-based studies is notable, although the upper range of genome or SNP-based heritability is at the cusp of heritability estimates observed in twin studies. That said, genome-based and twin-based estimates capture discrete components. While twin analyses typically model additive genetic effects, these estimates capture both additive and non-additive genetic variance shared among twins as well as gene–environment interplay, and contributions from both rare and common variants (and often is referred to as ‘broad-sense heritability’),18 whereas genome-based methods capture additive variance attributable to informative common genetic variants on genotyping arrays (known as ‘narrow-sense heritability’).8 In the current study, we implement two methods within the same twin samples and evaluate how Alzheimer’s disease PRS contributions to heritability vary and what Alzheimer’s disease PRS contributes beyond APOE.
Materials and methods
Participants
All participants were drawn from the Swedish Twin Registry (STR).19 The primary analysis sample included twins from four STR-based sub-studies: The Study of Dementia in Swedish Twins (HARMONY),20 the Swedish Adoption Twin Study of Aging (SATSA),21 Aging in Women and Men (GENDER)22 and Origins of Variance in the Oldest Old: Octogenarian Twins (OCTO-Twin),23 where informed consent was obtained from participants. Dementia was assessed using equivalent protocols that permits the combining of these data.24,25 SATSA, begun in 1984, followed 859 individuals aged 50 years and older from same-sex pairs across three decades with 10 in-person testing assessments commencing in 198621; the current analysis sample included 522 SATSA participants. OCTO-Twin, initiated in 1991, followed 351 same-sex twin pairs aged 80 years and older across 8 years with five biennial visits23; the current analysis sample included 66 OCTO-Twin participants. GENDER, initiated in 1995 includes three in-person follow-ups of 498 opposite-sex twin pairs aged 70 years and older22; the current analysis sample included 326 GENDER participants. HARMONY, commencing in 1998, screened 13 939 individuals from all STR individuals aged 65 years and older.20 Those who evidenced possible cognitive dysfunction were referred for a complete clinical work-up as well as their co-twin, plus a control sample, with a total clinical sample of 1557. A longitudinal follow-up after 2 years was done of those in the clinical work-up samples who showed possible dysfunction but did not meet the criteria for dementia. The current analysis sample included 666 HARMONY participants. Clinically based dementia and Alzheimer’s disease diagnoses were available from the in-person evaluations1 beginning in 1986 with additional follow-up through population-based registries up through 2016. Diagnoses available via registry sources are reliable.26
For individuals diagnosed with dementia, age at dementia diagnosis was used as the last follow-up. For controls, age at last follow-up was based on the age as on 31 December 2016 or death, whichever occurred first for those with register information as described below. The age at last follow-up, death or dementia onset was Mlastage = 85.28, SD = 7.02 years with 44% of the sample being male. Age distributions across cases and controls are similar although controls are on an average 2.32 years younger than the cases (see Supplementary Table 1). Age distributions within the sub-studies are generally similar among cases and controls overall, with average age differences between controls and cases ranging from −5.11 to 0.45 years, with the largest difference for SATSA.
Twins were selected for analyses where one or both members of the pair had information about a diagnosis consistent with Alzheimer’s disease or mixed Alzheimer’s disease and APOE genotyping. Exclusions included early-onset Alzheimer’s disease cases (aged <60 years, n = 3) and individuals with other forms of dementia (n = 382). Controls were excluded if they died before the age of 70 years (n = 38) or if they had possible cognitive impairment but did not meet the criteria for dementia (n = 110). Additional exclusionary criteria included no genome-wide genotyping (n = 76) or undetermined zygosity (n = 7). After these exclusions, a total of 1586 twins were available for the analytic samples (431 Alzheimer’s disease or mixed Alzheimer’s disease cases, 1155 controls). The 1586 twins were nested within 1137 twin pairs, with 898 individuals represented among 449 complete pairs and 688 individuals represented from 688 incomplete pairs.
Measures
Alzheimer’s disease assessment
A two-stage procedure identified dementia cases. First, cognitive screening by telephone was performed across the entire STR population by HARMONY or where twins missed a longitudinal assessment (SATSA, OCTO-Twin and GENDER), or where longitudinal performance declined markedly (e.g. mental status performance via a Mini-Mental Status Exam (MMSE)27 score <25 or a longitudinal drop by three points; low cognitive performance on verbal or spatial tasks in the bottom 10th percentile or dropping the equivalent of 1 SD from the prior assessment). Second, poor performance on the screening led to referral for in-person dementia diagnostic work-up for those twins, along with their cotwins.1 All studies also worked up samples of twin pairs who did not perform poorly on the cognitive screening. For individuals lost to follow-up due to the end of the parent study, or if a twin skipped an assessment wave, administrative sources were consulted, including the Swedish National Patient Register, the Cause of Death Register and the Prescribed Drug Register. The present study updated dementia status through 31 December 2016, using International Classification of Disease codes for Alzheimer’s disease and other dementias or Anatomical Therapeutic Chemical codes for Alzheimer’s disease medication (used as a proxy for an Alzheimer’s disease diagnosis).28
Genotyping
Direct APOE genotyping for two markers (rs7412 and rs429358) was available for all participants included in the analysis as described elsewhere.29 The distribution of APOE ɛ2/ɛ3/ɛ4 alleles in this analysis sample was 9.4/74.2/16.4% (taking all DZ twins and selecting one individual from each MZ pair). Genome-wide data were available from the Illumina PsychArray (N = 1451) or the Human OmniExpress array (N = 135) and imputed to 1000 Genomes Project phase1 version3.30 Initial exclusions of SNPs included those with a minor allele frequency of 0, >2% missing calls and those out of Hardy–Weinberg equilibrium (P < 1 × 10−6). Ancestral outliers (based on principal components) and individuals with >1% missing genotypes were excluded. PRSs were created in Plink 1.931 using summary statistics from the 2019 Alzheimer’s disease genetic meta-analysis.10 All non-ambiguous SNPs in the summary statistics were selected for PRS generation if they were also present in the study sample data with a minor allele frequency of 1% or higher and info score >0.8 (indicating good imputation quality) on both genotyping arrays. Using Plink 1.9,31 independent genetic variants were obtained through linkage disequilibrium (LD) clumping, setting the LD parameter r2 to 0.01. PRSs were then computed by summing up the number of risk alleles at each SNP, weighted by the effect size from the GWAS summary statistics.31 Eight different PRSs were computed based on significance level in the GWAS, at P ≤ 1, P ≤ 0.5, P ≤ 0.05, P ≤ 0.01, P ≤ 1 × 10−3, P ≤ 1 × 10−4, P ≤ 1 × 10−5 and P ≤ 5 × 10−8, with and without the APOE region. For 183 of the MZ twin pairs, only one twin was genotyped and the co-twin’s PRS imputed by taking the genotyped twin’s PRS.
Analysis
Regression analyses included both complete and incomplete pairs (N = 1586 individuals from 1137 twin pairs), whereas biometric models included complete pairs (N = 898 individuals, 449 pairs). PRSs were adjusted for the first four ancestry principal components and standardized within the SNP array.
PRS effects in a regression context were tested using the R package mixor32 (v.1.04) using a probit model as follows:
| (1) |
where AD reflects Alzheimer’s disease risk for the ith individual in the jth pair as predicted by an MZ twin type, Sex, LastAge (centered on 80 years, divided by 10), Array (Omni or Psych) and zPRS the residualized and standardized PRS scores. Random effects for MZ and DZ pairs were estimated at the pair level to account for sibling dependencies. Fit comparisons between a baseline model with covariates and adding the PRS or APOE alleles were made comparing deviances distributed as chi-square (Δχ2) with d.f. equal to the number of predictors added to the model. The probit model was prioritized as it underlies the biometrical model described below. However, a model assuming a logit link produced comparable estimates and is presented in Supplementary material for comparison with previously published work.
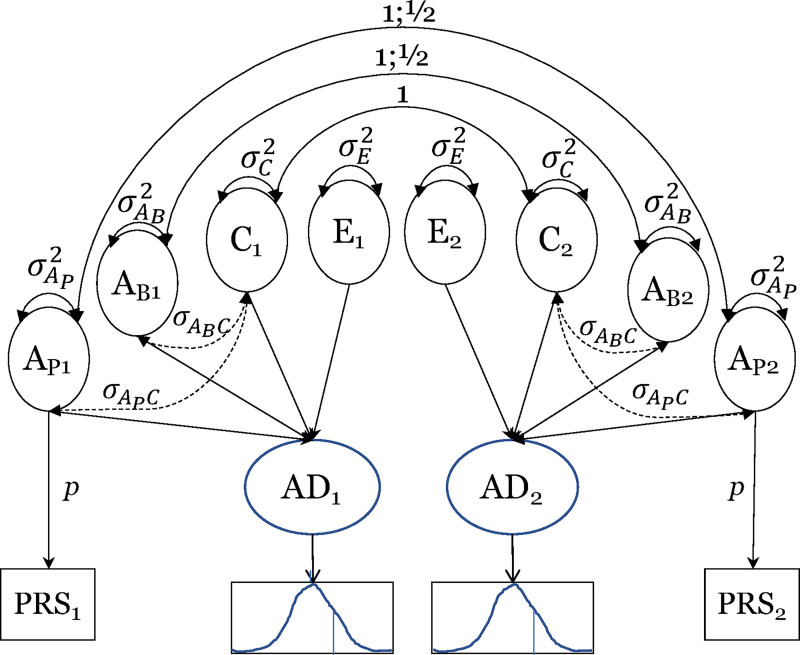
PRS contributions in the context of a biometric model were tested using the R package OpenMx33 (v. 2.18.1), assuming a latent-liability probit model with maximum-likelihood estimation. We fitted an extended ACE biometric twin model34 (see Fig. 1), decomposing underlying liability to Alzheimer’s disease into total additive genetic (A) influences, common (C) and non-shared or person-specific environmental (E) influences, and covariance between A and C (covAC). Notably, E also includes any measurement error and stochastic factors. Additive genetic influences include the unmeasured background genetic (AB) component and a latent polygenic risk score (AP) that was perfectly defined by the measured PRS and its observed variance scaled by the parameter p (i.e., σ2PRS = p2 x σ2Aᴩ). An identifying constraint included no covariance between AB and AP (σAᴩ,Aʙ = 0). The sum of variance components was constrained such that
| (2) |
Hence, σ2Aᴩ represents the proportion of variance in Alzheimer’s disease liability explained by the measured PRS and σ2Aᴩ + σ2Aʙ represents the proportion of variance due to all genetic influences. In addition, the total covariance between A and C (covAC) was constrained as:
| (3) |
Figure 1.
Biometrical ACE model with Alzheimer’s disease PRS. AD, Alzheimer’s disease liability; PRS, polygenic risk score. AP, additive genetic influences due to the PRS which are correlated at 1.0 among MZ twin pair members and 1/2 for DZ twins pair members; AB, background additive genetic influences which are correlated at 1 among MZ and 1/2 for DZ twin pairs; C, common environmental influences that are perfectly correlated among both MZ and DZ pairs; E, non-shared environmental influences. Subscripts of 1 refer to Twin 1 and subscripts of 2 refer to Twin 2. Total A = AP + AB + 2covAC, where covAC is the total covariance of A and C.
Hence, the expected correlations among MZ twins who share 100% of their genes while DZ twins on average share 50% of their segregating alleles were:
| (4) |
| (5) |
The models freely estimated variance components without boundary constraints to allow for unbiased fit statistics and correct Type I error rates.35 We fixed the Alzheimer’s disease liability threshold to 0 and estimated its mean for ease in analysis given that the mean estimation was already specified for the PRSs, and is a statistically equivalent approach to estimating the threshold and fixing the mean to 0.36 95% confidence intervals were estimated.
Data availability
Raw data were generated at the Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden. The derived data supporting the findings of this study are available from the corresponding author on request.
Results
In comparing the IGAP2 summary statistics10,11 with GWAS of our current samples, the β coefficients suggested similar effect sizes for those included in the PRS at P < 1 × 10−5 (rGWAS = 0.54, P < 6.1 × 10−8; nSNPs = 89) and P < 1 × 10−4 (rGWAS = 0.36, P < 2.1 × 10−8; nSNPs = 233). The best regression predictions were observed at P < 1 × 10−5 and P < 1 × 10−4 thresholds (Nagelkerke R2 = 0.062 for both) and improved over P < 5 × 10−8 (Nagelkerke R2 = 0.058), whereas predictions fell off at P < 1 × 10−3 (Nagelkerke R2 = 0.053) (see Supplemental Table 2 for all thresholds). The comparable predictions based on the PRS without the APOE region were Nagelkerke R2 values of 0.011 and 0.012 at the P < 1 × 10−5 and P < 1 × 10−4 thresholds, respectively. We present findings for the P < 1 × 10−5 threshold (based on the rGWAS and comparable Nagelkerke R2), evaluating a full PRS, a PRS without the APOE region (PRS.no.APOE) and the latter with directly measured APOE alleles (PRS.no.APOE + ɛ2 + ɛ4 alleles).
Probit regression models
Entering the full PRS at P < 1 × 10−5 to the baseline model with covariates led to a significant increase in fit [Δχ2(d.f. = 1) = 65.82, P < 4.93 × 10−16; Nagelkerke R2 = 0.062] (Table 1). Entering PRS.no.APOE also led to a significant increase in fit [Δχ2(d.f. = 1) = 11.27, P < 7.86 × 10−4; Nagelkerke R2 = 0.011] (Table 1). When adding directly genotyped APOE ɛ2 alleles and ɛ4 alleles (PRS.no.APOE + ɛ2 + ɛ4 alleles), the resulting gain in prediction was evident [Δχ2(d.f. = 2) = 81.29, P < 2.23 × 10−18] with a Nagelkerke R2 of 0.076, driven by APOE ɛ4 (P = 2.05 × 10−12) and with a non-significant reduction in risk by the number of ɛ2 alleles (P = 1.38 × 10−1) (Table 1). The AUC values across all models were high ranging from 0.97 to 0.98, suggesting that background characteristics perform well in distinguishing cases from non-cases. Logistic regression models produced similar results (see Supplementary Table 3). Sensitivity analyses using only complete twin pairs produced consistent results as the full sample analysis (see Supplementary Table 4). Finally, analyses adding in adjustment for sub-study resulted in slight differences: the Nagelkerke R2 dropped from 0.062 to 0.055 = 0.007 for PRS with APOE and from 0.011 to 0.009 for PRS.no.APOE (see Supplementary Table 5). Overall, the best genetic prediction was observed for directly measured APOE ɛ2 and ɛ4 plus PRS.no.APOE (Table 1).
Table 1.
Probit regression analyses (N = 1586): Alzheimer’s disease PRS at P < 1 × 10−5
| Parameters | Baseline | PRS P < 1 × 10−5 | PRS.no.APOE P < 1 × 10−5 | PRS.no.APOE + APOE alleles | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SE | P(>|z|) | B | SE | P(>|z|) | B | SE | P(>|z|) | B | SE | P(>|z|) | |
| Intercept | −1.56 | 0.20 | 4.89E−15 | −1.67 | 0.21 | 8.88E−16 | −1.58 | 0.20 | 2.89E−15 | −1.82 | 0.22 | <2.20E−16 |
| MZ | 0.04 | 0.16 | 7.92E−1 | 0.06 | 0.15 | 6.79E−1 | 0.08 | 0.16 | 6.15E−1 | 0.07 | 0.15 | 6.08E−1 |
| Sex (0 = M, 1 = F) | 0.44 | 0.09 | 3.83E−6 | 0.44 | 0.10 | 4.15E−6 | 0.44 | 0.09 | 3.02E−6 | 0.41 | 0.09 | 1.40E−5 |
| LastAge | 0.97 | 0.12 | 8.88E−15 | 0.97 | 0.13 | 2.75E−14 | 0.95 | 0.13 | 4.31E−14 | 0.96 | 0.13 | 6.51E−14 |
| LastAge2 | −0.55 | 0.09 | 5.12E−10 | −0.50 | 0.09 | 1.29E−8 | −0.53 | 0.09 | 1.78E−9 | −0.47 | 0.09 | 4.27E−8 |
| Array | 0.40 | 0.16 | 1.55E−2 | 0.44 | 0.17 | 9.83E−3 | 0.40 | 0.17 | 1.69E−2 | 0.38 | 0.17 | 2.88E−2 |
| PRS | — | — | — | 0.38 | 0.06 | 7.53E−12 | 0.16 | 0.05 | 1.41E−3 | 0.16 | 0.05 | 1.06E−3 |
| APOE ɛ2 alleles | — | — | — | — | — | — | — | — | — | −0.18 | 0.12 | 1.38E−1 |
| APOE ɛ4 alleles | — | — | — | — | — | — | — | — | — | 0.75 | 0.11 | 2.05E−12 |
| Random·MZ | 2.23 | 0.76 | 3.37E−3 | 1.77 | 0.62 | 4.49E−3 | 2.06 | 0.73 | 4.53E−3 | 1.61 | 0.58 | 5.60E−3 |
| Random·DZ | 0.37 | 0.23 | 1.04E−1 | 0.39 | 0.24 | 1.02E−1 | 0.39 | 0.24 | 1.02E−1 | 0.36 | 0.24 | 1.40E−1 |
| Fit statistics | ||||||||||||
| Deviance | 1703.37 | 1637.55 | 1692.10 | 1610.81 | ||||||||
| AIC | −859.69 | −827.78 | −855.05 | −816.40 | ||||||||
| SBC | −879.83 | −850.44 | −877.71 | −844.10 | ||||||||
| ICC·MZ | 0.691 | 0.638 | 0.673 | 0.616 | ||||||||
| ICC·DZ | 0.269 | 0.282 | 0.280 | 0.263 | ||||||||
| AUC | 0.976 | 0.972 | 0.977 | 0.970 | ||||||||
| R 2 Nagelkerke | 0.084 | 0.062 | 0.011 | 0.076 | ||||||||
Regression analyses estimating varying intraclass correlations (ICCs) with clustered twin data were adapted from code in Archer et al.32 using mixor. MZ, monozygotic twin; DZ, dizygotic twin; LastAge, age at last follow-up, death or dementia onset, centred on age 80 years and divided by 10; Array, Human OmniExpress = 0, Illumina PsychArray = 1; PRS, polygenic risk score at P < 1 × 10−5 residualized for four PCs and standardized within array type; PRS.no.APOE, PRS without APOE region; Random, random effect; Deviance, –2ln(Likelihood); AIC, Akaike Information Criteria; SBC, Schwarz Bayesian Criterion; ICCs measured as Random·MZ/(1 + Random·MZ) and Random·DZ/(1 + Random·DZ)32; AUC, area under the curve.
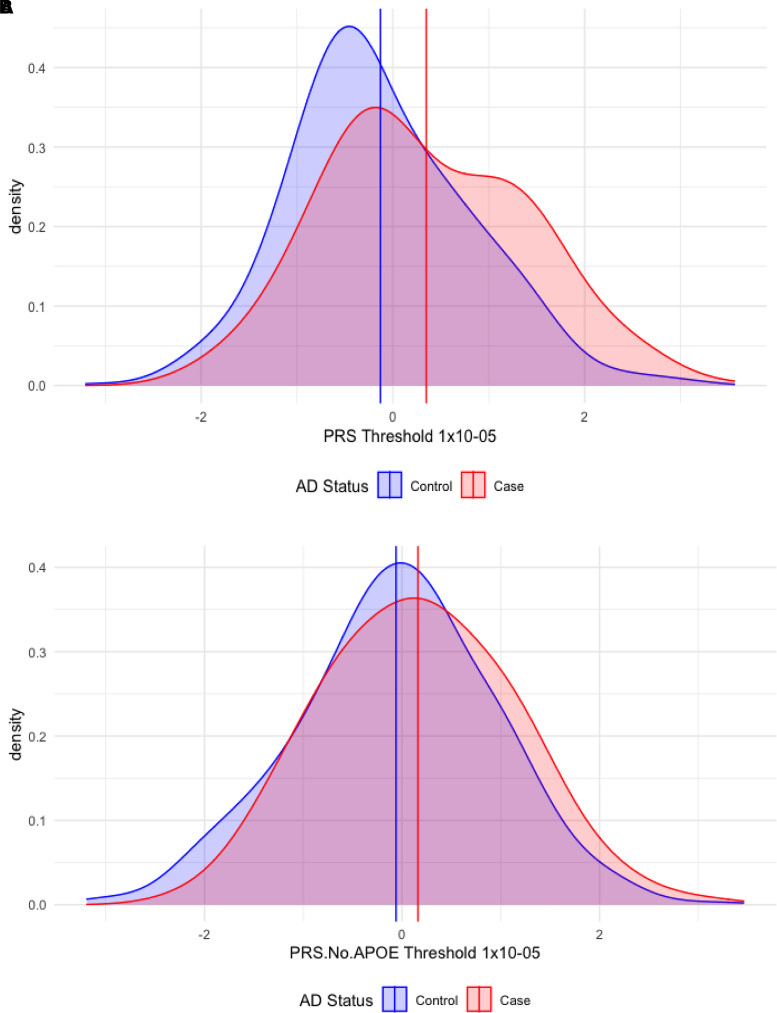
The standardized PRS distribution at P < 1 × 10−5, by Alzheimer’s disease status, is shown in Fig. 2A, adjusted for the first four ancestry PCs and array type. The mean PRS for controls was −0.13 (SD = 0.95) versus cases at 0.35 (SD = 1.06), an effect size difference of z = 0.48. The standardized PRS distribution for PRS.no.APOE at P < 1 × 10−5, by Alzheimer’s disease status, is shown in Fig. 2B, adjusted for the first four ancestry PCs and array type. The mean PRS.no.APOE for controls was −0.06 (SD = 1.00) versus cases at 0.16 (SD = 0.97), an effect size difference of z = 0.22. Hence, the offset in the PRS distributions between cases and controls is over 2-fold for the full PRS containing the APOE region compared with the distribution of PRS.no.APOE.
Figure 2.
Density plots of Alzheimer’s disease PRSs at the P < 1 × 10−5 threshold in cases and controls. (A) PRS. (B) PRS.no.APOE (PRS without APOE region). PRS, polygenic risk score. Vertical lines indicate the mean PRS values for Alzheimer’s disease (AD) cases (red line) and controls (blue line). PRSs are based on independent genetic variants reaching a significance threshold of P < 1 × 10−5 in the GWAS.
Biometric twin models
A simple baseline ACE model fitted to complete twin pairs (190 MZ and 259 DZ) suggested a significant additive genetic contribution (A), a non-significant common environmental variance (C), and a significant non-shared or person-specific environmental variance (E) (see Table 2, Full Model 0). A reduced model dropping common environmental variance (C) [Δχ2(d.f. = 1) = 0.81, P = 3.69 × 10−1] was the best-fitting and suggested that 70.7% of the liability for Alzheimer’s disease was attributable to additive genetic influences (σ2A = 0.707, CI95 = 0.542, 0.832) and the remainder attributable to non-shared environment (Table 2, Reduced Model 0). Model fit comparisons between the full baseline ACE and all sub-models testing individual variance components are shown in Supplementary Table 6.
Table 2.
Biometric twin model results: Alzheimer’s disease PRS at P < 1 × 10−5
| Model | VC | Full model | Reduced model | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Est | SE | LL | UL | Est | SE | LL | UL | ||
| 0. ACE | A | 0.960 | 0.294 | 0.391 | 1.546 | 0.707 | 0.074 | 0.542 | 0.832 |
| C | −0.232 | 0.265 | −0.774 | 0.258 | — | — | — | — | |
| E | 0.272 | 0.073 | 0.153 | 0.440 | 0.293 | 0.074 | 0.168 | 0.458 | |
| −2LL | 888.71 | 889.52 | |||||||
| AIC | 902.71 | 901.52 | |||||||
| BIC | 931.46 | 926.16 | |||||||
| 1. PRS | A P | 0.136 | 0.040 | 0.058 | 0.342 | 0.101 | 0.029 | 0.051 | 0.164 |
| A B | 0.882 | 0.083 | 0.343 | 1.410 | 0.614 | 0.075 | 0.452 | 0.744 | |
| C | 0.003 | 4.26E−4 | −9.31E−7 | 0.668 | — | — | — | — | |
| E | 0.263 | 0.069 | 0.149 | 0.425 | 0.285 | 0.072 | 0.164 | 0.446 | |
| covAC | −0.142 | 0.008 | −0.618 | 0.098 | — | — | — | — | |
| −2LL | 2746.32 | 2747.52 | |||||||
| AIC | 2768.32 | 2765.52 | |||||||
| BIC | 2813.49 | 2802.48 | |||||||
| 2. PRS.no.APOE | A P | 0.030 | 0.020 | 0.012 | 0.139 | 0.024 | 0.016 | 0.004 | 0.065 |
| A B | 0.924 | 0.080 | 0.367 | 1.498 | 0.685 | 0.075 | 0.519 | 0.811 | |
| C | 4.29E−4 | 3.07E−4 | −1.75E−6 | 1.101 | — | — | — | — | |
| E | 0.271 | 0.071 | 0.152 | 0.439 | 0.291 | 0.074 | 0.167 | 0.456 | |
| covAC | −0.113 | 0.005 | −0.781 | 0.115 | — | — | — | — | |
| −2LL | 2824.69 | 2825.45 | |||||||
| AIC | 2846.69 | 2843.45 | |||||||
| BIC | 2891.87 | 2880.42 | |||||||
| 3. PRS.no.APOE + ɛ4 alleles | A P | 0.027 | 0.013 | 0.025 | 0.087 | 0.021 | 0.015 | 0.005 | 0.059 |
| A e4 | 0.118 | 0.024 | 0.049 | 0.242 | 0.093 | 0.027 | 0.046 | 0.152 | |
| A B | 0.825 | 0.077 | 0.318 | 1.351 | 0.596 | 0.075 | 0.434 | 0.728 | |
| C | 4.17E−4 | 4.94E−4 | −5.419E−11 | 0.474 | — | — | — | — | |
| E | 0.268 | 0.069 | 0.153 | 0.433 | 0.289 | 0.073 | 0.167 | 0.451 | |
| covAC | −0.120 | 0.007 | −0.491 | 0.115 | — | — | — | — | |
| −2LL | 3783.69 | 3784.47 | |||||||
| AIC | 3811.69 | 3808.47 | |||||||
| BIC | 3869.19 | 3857.76 | |||||||
Biometrical analyses of Alzheimer’s disease risk with entry of a PRS were fitted adapting code in Dolan et al.34 using OpenMx.33 VC, variance component; Est, Estimate; SE, standard error; LL, lower 95% confidence interval; UL, upper 95% confidence interval; PRS, polygenic risk score at P < 1 × 10−5 residualized for four PCs and standardized within array type; PRS.no.APOE, PRS without APOE region; A, additive genetic influences; C, common environmental influences; E, non-shared environmental influences; AP, genetic influences due to the PRS; Aɛ4, genetic influences due to APOE ɛ4 alleles; AB, background additive genetic influences; total A = AP + AB + 2covAC. The Reduced Model dropped C (common environmental variance) and covAC. All models adjusted for Sex, LastAge and LastAge2. 95% confidence intervals are shown.
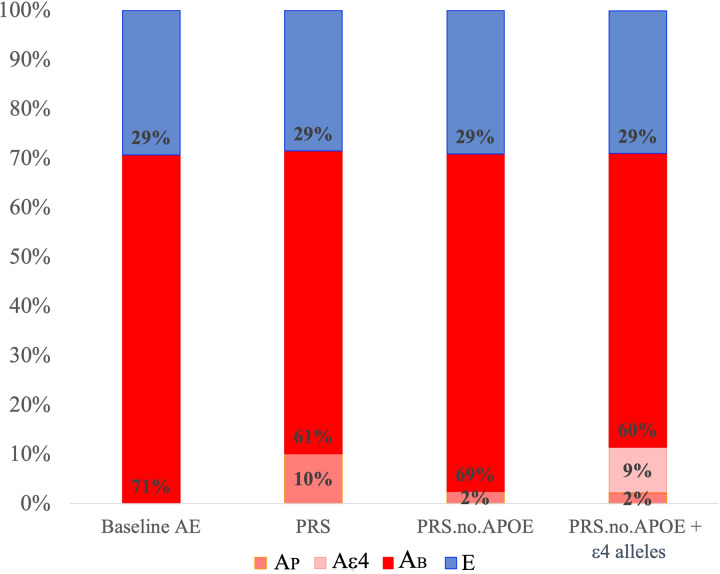
Next, we expanded the full baseline ACE model to consider the PRS at P < 1 × 10−5 as the measured polygenic risk (AP), remaining background additive genetic (AB) variance as well as common environmental variance (C), the covariance of A and C (covAC) and E. Both C and covAC could be dropped (P ≥ 5.48 × 10−1) (see Supplementary Table 6). In Reduced Model 1, AP for the full PRS accounted for 10.1% (σ2Aᴩ = 0.101, CI95 = 0.051, 0.164) of variation contributing to Alzheimer’s disease risk (see Table 2), whereas, in Reduced Model 2, AP for the PRS.no.APOE accounted for 2.4% (σ2Aᴩ-APOE = 0.024, CI95 = 0.004, 0.065) (see Table 2). Notably, when APOE ɛ4 alleles were added to the model with PRS.no.APOE, the total measured prediction (PRS.no.APOE + ɛ4 alleles) was 11.4% (σ2Aᴩ-APOE = 0.021, CI95 = 0.005, 0.059; σ2Aɛ4 = 0.093, CI95 = 0.046, 0.152) and the remaining genetic background variance was 59.6% (σ2A = 0.596, CI95 = 0.434, 0.728) (see Fig. 3). Overall, in the context of twin biometrical models, the best measured genetic prediction was observed for directly measured APOE ɛ4 alleles + PRS.no.APOE, but substantial background genetic contributions remain that are not captured by these measured sources.
Figure 3.
Biometrical AE model results including Alzheimer’s disease PRSs at the P < 1 × 10−5 threshold. E, non-shared environmental influences; A, additive genetic influences; AB, background additive genetic influences; AP, genetic influences due to a polygenic risk score (PRS); Aɛ4, genetic influences due to APOE ɛ4 alleles. Total A = AP + Aɛ4 + AB (values from Table 2, Reduced Model). PRSs are based on independent genetic variants reaching a significance threshold of P < 1 × 10−5 in the GWAS.
Our observed power for our given estimate of A of 0.71 was 0.80.37 Our observed power for evaluating PRS.no.APOE and APOE ɛ4 alleles based on the Reduced Model 3 was 0.77 for PRS.no.APOE and approached 1.00 for APOE ɛ4 alleles.
Discussion
There are many ways to evaluate the importance of genetic influences on Alzheimer’s disease. To date, twin-based models and contributions of PRS have been considered independently. In bringing these approaches together for the first time in the same twin sample, we observed that much of the genetic variance contributing to Alzheimer’s disease liability is not explained by directly measured APOE or common genetic influences currently captured by GWAS contributing to a polygenic score. The Alzheimer’s disease PRS contribution to Alzheimer’s disease risk was as high as 0.101, or 10.1% in the twin biometric model. The APOE region accounts for much of the measurable contribution to Alzheimer’s disease, with smaller polygenic contribution from other measured common genetic influences. Considering the best biometrical model, directly measured APOE ɛ4 explained 9.3% and PRS.no.APOE an additional 2.1% of Alzheimer’s disease risk, leaving much of the genetic risk uncaptured (i.e. 71.1% total minus 11.4% measured).
Our estimates of measured contributions of the PRS to background heritability for Alzheimer’s disease risk, in the same sample, are smaller than the SNP-heritability estimates as well as that for APOE ɛ4.7–9 While the small contribution of the PRS in this study can potentially be explained by the fact that it is based on the most significant SNPs (N = 89),7 we note that including PRSs at more relaxed P-value thresholds did not pick up more heritability than SNPs with P < 1 × 10−5. As the GWAS of Alzheimer’s disease is still of comparatively small sample size, based on 21 982 cases and 41 944 controls, this may indicate that substantial genetic variation will be discovered as GWAS sample size increases.
PRS methods rely on the power of GWAS, whereas other genome-wide heritability methods, such as GCTA, are less affected but also often fall short of estimates from twin and family studies.38 Moreover, genome-wide methods produce narrow-sense heritability estimates due to additive effects from common SNPs,8 whereas twin estimates include both additive and non-additive genetic influences (e.g. dominance and epistasis),18 or broad-sense heritability, and with contributions from all variants, common and rare. However, recent work suggests that heritability is ‘recovered’ for complex traits such as human height and body mass index (BMI) when using sequencing data such that SNP-based heritabilities are in line with twin and family-based estimates.39,40 Thus, disagreement between biometric and SNP-based heritabilities is not universal. That substantial variation may be attributed to rare variants has also been observed for other complex disease traits such as prostate cancer41 and for phenotypes in other species such as yeast.42,43 The missing heritability is likely not due to simple additivity across common variants but also to contributions from rare variants as well as to non-additive effects including dominance and epistasis.42,44 Studies of rare variants and Alzheimer’s disease risk have observed effects for rare coding variants in genes such as ABCA7, BIN, NOTCH3, PLCG2, SORL1, TREM and ABI3 among others45–47 not captured by PRSs. Apart from a rare variant in TREM2 (p.Arg47His), little replication work has been reported.8 However, an Icelandic study observed a protective mutation in the APP gene (A673T), that codes for APP, with replication analyses suggesting that it predicted higher cognitive status scores among nursing home residents.48
Moreover, gene–environment interplay may increase estimates of genetic influences.49 For example, a correlation may be induced between genes and environments (rGE) whereby individuals at higher genetic risk may construct contexts that buffer expression of Alzheimer’s disease, such as engagement in physical or cognitive activities. Empirical examples of rGE for Alzheimer’s disease are rare. On the contrary, studies testing for gene–environment interaction (G × E) are more common for Alzheimer’s disease and related traits, typically evaluating APOE,49–51 e.g. risk for Alzheimer’s disease is magnified for those with APOE risk alleles who are also obese or have high blood pressure in midlife. Moreover, reports from the IGEMS consortium using a within-pair MZ twin design report small-to-moderate G × E effects across country and gender for cross-sectional measures of BMI, depressive symptoms, cognitive performance52 as well as grip strength.53 Furthermore, APOE may partly account for G × E effects for depressive symptoms and spatial reasoning whereby ɛ4 individuals may show less sensitivity to the environment.52
In conclusion, in the context of a Swedish twin study, the APOE region explains much of the measured genetic contribution to Alzheimer’s disease, with smaller contributions from other measured polygenic influences, yet much of the background genetic liability to risk is unexplained. Sensitive designs that capture all the measured genetic influences, such as the sequencing of rare variants, as well as models that evaluate direct and indirect contributions and gene–environment interplay may reconcile the high background heritability observed in twin and family studies with the extant estimates of measured polygenic risk from genome-wide approaches.
Supplementary Material
Abbreviations
- Aβ =
β-amyloid
- APP =
amyloid precursor protein
- GWAS =
genome-wide association study
- PRS =
polygenic risk score
Contributor Information
Ida K. Karlsson, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden Aging Research Network—Jönköping (ARN-J), School of Health and 6 Welfare, Jönköping University, Jönköping, Sweden.
Valentina Escott-Price, UK Dementia Research Institute at Cardiff, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK.
Margaret Gatz, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden; Center for Economic and Social Research, University of Southern California, Los Angeles, CA, USA.
John Hardy, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, Queen Square, London WC1N 3BG, UK; UK Dementia Research Institute at UCL and Department of Neurodegenerative Disease, UCL Institute of Neurology, University College London, London, UK; Reta Lila Weston Institute, UCL Queen Square Institute of Neurology, 1 Wakefield Street, London WC1N 1PJ, UK; UCL Movement Disorders Centre, University College London, London, UK; Institute for Advanced Study, The Hong Kong University of Science and Technology, Hong Kong SAR, China.
Nancy L. Pedersen, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden Department of Psychology, University of Southern California, Los Angeles, CA, USA.
Maryam Shoai, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, Queen Square, London WC1N 3BG, UK; UK Dementia Research Institute at UCL and Department of Neurodegenerative Disease, UCL Institute of Neurology, University College London, London, UK.
Chandra A. Reynolds, Department of Psychology, University of California—Riverside, Riverside, CA, USA
Funding
This work was supported by the National Institutes of Health, National Institute on Aging R01 AG08724, R01 AG17561, R01 AG028555, RF1 AG058068 and R01 AG060470. The manuscript content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests
I.K.K. receives funding from Swedish Research Council for Health, Working Life and Welfare (2018-01201) and the National Institutes of Health R01AG060470. V.E.-P. receives funding from the Dementia Research Institute (UKDRI-3003), MRC Centre for Neuropsychiatric Genetics and Genomics Centre (MR/L010305/1), EU Joint Programme—Neurodegenerative Disease Research (JPND) (MR/T04604X/1). M.G. receives funding from the National Institute of Health Grant Nos RF1AG058068, R01AG060470, R01AG059329, P01AG055367, R01ES025888, RF1AG054068, R01AG056163, RF1AG054442, 1U2C AG060408 and 5U01 AG054580. The work of J.H. on this project was supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK. J.H. receives funding from Medical Research Council (award number MR/N026004/1), Wellcome Trust Hardy (award number 202903/Z/16/Z), Dolby Family Fund, National Institute for Health Research University College London Hospitals Biomedical Research Centre, BRCNIHR Biomedical Research Centre at University College London Hospitals NHS Foundation Trust and University College London. N.L.P. receives funding from the National Institutes of Health Grants Nos R01AG059329 and R01AG060470. M.S. received funding from the Fidelity Charitable Foundation. C.A.R. receives funding from the National Institutes of Health (NIH) including RF1AG058068, R01AG060470, R01AG059329, R01AG050595 and R01AG046938. The authors declare no other competing interests.
Supplementary material
Supplementary material is available at Brain Communications online.
References
- 1. Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. [DOI] [PubMed] [Google Scholar]
- 2. Sims R, Hill M, Williams J. The multiplex model of the genetics of Alzheimer’s disease. Nat Neurosci. 2020;23(3):311–322. [DOI] [PubMed] [Google Scholar]
- 3. Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Escott-Price V, Sims R, Bannister C, et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138(Pt 12):3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Escott-Price V, Myers A, Huentelman M, Shoai M, Hardy J. Polygenic risk score analysis of Alzheimer’s disease in cases without APOE4 or APOE2 alleles. J Prev Alzheimers Dis. 2019;6(1):16–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann Neurol. 2017;82(2):311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging. 2017;49:214.e7–214.e11. [DOI] [PubMed] [Google Scholar]
- 8. Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer’s disease: Evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15(8):857–868. [DOI] [PubMed] [Google Scholar]
- 9. Ridge PG, Hoyt KB, Boehme K, et al. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol Aging. 2016;41:200.e13–200.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kunkle BW, Grenier-Boley B, Sims R, et al. Author Correction: Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51(9):1423–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Q, Sidorenko J, Couvy-Duchesne B, et al. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat Commun. 2020;11(1):4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leonenko G, Baker E, Stevenson-Hoare J, et al. Identifying individuals with high risk of Alzheimer’s disease using polygenic risk scores. Nat Commun. 2021;12(1):4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abondio P, Sazzini M, Garagnani P, et al. The genetic variability of APOE in different human populations and its implications for longevity. Genes (Basel). 2019;10(3):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soerensen M, Dato S, Tan Q, et al. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age (Dordr). 2013;35(2):487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heffernan AL, Chidgey C, Peng P, Masters CL, Roberts BR. The neurobiology and age-related prevalence of the epsilon4 allele of apolipoprotein E in Alzheimer’s disease cohorts. J Mol Neurosci. 2016;60(3):316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKay GJ, Silvestri G, Chakravarthy U, et al. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. Am J Epidemiol. 2011;173(12):1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coventry WL, Keller MC. Estimating the extent of parameter bias in the classical twin design: A comparison of parameter estimates from extended twin-family and classical twin designs. Twin Res Hum Genet. 2005;8(3):214–223. [DOI] [PubMed] [Google Scholar]
- 19. Magnusson PK, Almqvist C, Rahman I, et al. The Swedish Twin Registry: Establishment of a biobank and other recent developments. Twin Res Hum Genet. 2013;16(1):317–329. [DOI] [PubMed] [Google Scholar]
- 20. Gatz M, Fratiglioni L, Johansson B, et al. Complete ascertainment of dementia in the Swedish Twin Registry: The HARMONY study. Neurobiol Aging. 2005;26(4):439–447. [DOI] [PubMed] [Google Scholar]
- 21. Finkel D, Pedersen N. Processing speed and longitudinal trajectories of change for cognitive abilities: The Swedish adoption/twin study of aging. Neuropsychol, Dev Cogn Sec B Aging Neuropsychol Cogn. 2004;11(2-3):325–345. [Google Scholar]
- 22. Gold CH, Malmberg B, McClearn GE, Pedersen NL, Berg S. Gender and health: A study of older unlike-sex twins. J Gerontol: Soc Sci. 2002;57(3):S168–S176. [DOI] [PubMed] [Google Scholar]
- 23. McClearn GE, Johansson B, Berg S, et al. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science. 1997;276(5318):1560- 156–3.. [DOI] [PubMed] [Google Scholar]
- 24. Beam CR, Kaneshiro C, Jang JY, Reynolds CA, Pedersen NL, Gatz M. Differences between women and men in incidence rates of Dementia and Alzheimer’s disease. J Alzheimers Dis. 2018;64(4):1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beam CR, Kaneshiro C, Jang JY, Reynolds CA, Pedersen NL, Gatz M. A twin study of sex differences in genetic risk for all Dementia, Alzheimer’s disease (AD), and non-AD Dementia. J Alzheimers Dis. 2020;76(2):539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rizzuto D, Feldman AL, Karlsson IK, Dahl Aslan AK, Gatz M, Pedersen NL. Detection of dementia cases in two Swedish health registers: A validation study. J Alzheimers Dis. 2018;61(4):1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Folstein Marshal F, Folstein Susan E, et al. Mini-mental state. J Psychiatr Res. 1975;12:189–198. ( 10.1016/0022-3956(75)90026-6) [DOI] [PubMed] [Google Scholar]
- 28. WHO Collaborating Centre for Drug Statistics Methodology . ATC/DDD classification. WHO Drug Information. 2017;31(4):629–634. [Google Scholar]
- 29. Reynolds CA, Zavala C, Gatz M, et al. Sortilin receptor 1 predicts longitudinal cognitive change. Research Support, N.I.H., Extramural. Research Support, Non-U.S. Gov't. Neurobiol Aging. 2013;34(6):1710.e11–1710.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. 1000 Genomes Project Consortium . An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Archer KJ, Hedeker D, Nordgren R, Gibbons RD. mixor: An R package for longitudinal and clustered ordinal response modeling 2015.
- 33. Neale MC, Hunter MD, Pritikin JN, et al. OpenMx 2.0: Extended structural equation and statistical modeling. Psychometrika. 2016;81(2):535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dolan CV, Huijskens RCA, Minică CC, Neale MC, Boomsma DI. Incorporating polygenic risk scores in the ACE twin model to estimate A–C covariance. Behav Genet. 2021; 51(3):237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verhulst B, Prom-Wormley E, Keller M, Medland S, Neale MC. Type I error rates and parameter bias in multivariate behavioral genetic models. Behav Genet. 2019;49(1):99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Medland SE, Neale MC, Eaves LJ, Neale BM. A note on the parameterization of Purcell’s G x E model for ordinal and binary data. Behav Genet. 2009;39(2):220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verhulst B. A power calculator for the classical twin design. Behav Genet. 2017;47(2):255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14(7):507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wainschtein P, Jain DP, Yengo L, et al. Recovery of trait heritability from whole genome sequence data. bioRxiv. 2019;588020. [Google Scholar]
- 40. Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47(10):1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mancuso N, Rohland N, Rand KA, et al. The contribution of rare variation to prostate cancer heritability. Nat Genet. 2016;48(1):30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fournier T, Abou Saada O, Hou J, Peter J, Caudal E, Schacherer J. Extensive impact of low-frequency variants on the phenotypic landscape at population-scale. Elife. 2019;8:e49258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bloom JS, Boocock J, Treusch S, et al. Rare variants contribute disproportionately to quantitative trait variation in yeast. Elife. 2019;8:e49212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang H, Bennett DA, De Jager PL, Zhang QY, Zhang HY. Genome-wide epistasis analysis for Alzheimer’s disease and implications for genetic risk prediction. Alzheimers Res Ther. 2021;13(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sims R, van der Lee SJ, Naj AC, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat Genet. 2017;49(9):1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patel D, Mez J, Vardarajan BN, et al. Association of rare coding mutations with Alzheimer disease and other dementias among adults of European ancestry. JAMA Netw Open. 2019;2(3):e191350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vardarajan BN, Ghani M, Kahn A, et al. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol. 2015;78(3):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jonsson T, Atwal JK, Steinberg S, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488(7409):96–99. [DOI] [PubMed] [Google Scholar]
- 49. Dunn AR, O’Connell KMS, Kaczorowski CC. Gene-by-environment interactions in Alzheimer’s disease and Parkinson’s disease. Neurosci Biobehav Rev. 2019;103:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Folley S, Zhou A, Llewellyn DJ, Hyppönen E. Physical activity, APOE genotype, and cognitive decline: Exploring gene-environment interactions in the UK biobank. J Alzheimer’s Dis. 2019;71(3):741–750. [DOI] [PubMed] [Google Scholar]
- 51. Head D, Bugg JM, Goate AM, et al. Exercise engagement as a moderator of the effects of APOE genotype on amyloid deposition. Arch Neurol. 2012;69(5):636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reynolds CA, Gatz M, Christensen K, et al. Gene-environment interplay in physical, psychological, and cognitive domains in mid to late adulthood: Is APOE a variability gene? Behav Genet. 2016;46(1):4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petersen I, Pedersen NL, Rantanen T, et al. GxE interaction influences trajectories of hand grip strength. Behav Genet. 2016;46(1):20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data were generated at the Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden. The derived data supporting the findings of this study are available from the corresponding author on request.