Abstract
Evidence of infection with Helicobacter species in pig stomach was investigated by the use of a PCR with Helicobacter genus-specific primers. Forty pig stomachs, each of four different ulcer lesion grades, 0, 1, 2, and 3 in the pars esophagea area, were collected from a slaughterhouse in Minnesota. Of 160 stomach samples examined, 102 (63.8%) were positive by the PCR assay. The 40 samples each of lesion grades 0, 1, 2, and 3 showed 22.5, 52.5, 85.0, and 95.0% PCR-positive results, respectively. There was a significant trend (P ≤ 0.01) in the proportions of PCR-positive cases relative to severity of the lesion. About 80% of the 16S rRNA gene was amplified, and PCR-restriction fragment length polymorphism (RFLP) patterns were analyzed. Of 102 PCR-positive samples, the PCR-RFLP patterns resulted in four different types, 32 samples being classified into type MN 1, 16 samples into type MN 2, 43 samples into type MN 3, and 11 samples into type MN 4. When the sequences of each RFLP type were compared to those reported in databases by using BLAST software, types MN 1, MN 2, MN 3, and MN 4 showed homologies of 97.3, 95.4, 96.7, and 99.5% with the 16S ribosomal DNA of Helicobacter flexispira taxon 3, Helicobacter sp. strains MIT 94-022 and MZ 640285, and Helicobacter suis, respectively. None of the 102 samples positive for the Helicobacter genus were positive with a primer set specific for Helicobacter pylori. Attempts to culture the organisms from selected stomachs in vitro were unsuccessful.
Helicobacter pylori was first described in 1984 (19), and it was proposed to be a cause of gastritis and peptic ulcers in humans. Subsequently, Helicobacter species have been isolated from various animal species in association with or without gastric ulcers (1, 4, 9, 10, 15, 17, 24, 25, 26, 29, 31). Most of the isolations have been made from different regions of the gastrointestinal system. H. pylori and other Helicobacter species were also isolated from livers, suggesting that Helicobacter species could cause hepatitis in humans and animal species (9, 10, 21, 31).
In swine, hyperkeratosis and ulceration of the pars esophagea and the cardiac mucosa in the stomach are a common problem throughout the world (11). There have been great herd-to-herd variations in the prevalence and severity of gastric ulcers. Abattoir surveys of the prevalence showed a range of 5 to 100%, with an average of 63.2% for the 13 studies in different countries (11). The ulcer lesions in pigs are characterized by various degrees of damage in the stratified squamous epithelium extending from the esophagus to the stomach. The damage ranges from mild hyperkeratosis to severe epithelial desquamation, ulceration, and acute hemorrhage. Multiple etiologies with different risk factors have been suggested as causes of gastric ulcers in swine. Feed processing, housing, management, and environmental factors are known to be linked to ulcer development. Affected pigs show clinical signs of anorexia, chronic anemia, decreased weight gain, acute gastric hemorrhage, and sudden death (8, 11). Gastric ulceration was reported as a common cause for sow mortality (2). Because these signs associated with gastric ulcers are commonly observed in grow-finish and breeding pigs, the economic losses are often significant on swine farms (11).
Spiral bacteria or helicobacter-like organisms in pig stomachs were first identified by histopathologic examination (22). Subsequently, Helicobacter species in pig stomachs have been detected by different diagnostic methods, including histology, urease test, immunohistochemistry, and PCR (1, 4, 5, 6, 7, 12, 18, 20, 23). The organisms present in the gastric lesions were initially referred to as Gastrospirillum suis (20). Recently, G. suis was reclassified into the genus Helicobacter by a phylogenetic analysis based on 16S ribosomal DNA (rDNA) sequence data, and then it was renamed Candidatus Helicobacter suis (5). In addition, there are several reports on the presence of H. pylori or H. pylori-like organisms in pig stomachs (7, 13, 22, 23, 29; Mendes et al., Am. J. Gastroenterol. 89:1296, 1994 [abstract]). However, H. pylori has not been detected in pig stomachs by some other researchers (1, 13, 22; Mendes et al., abstr.). Presently, the role of Helicobacter species in the pathogenesis of gastric ulcers in pigs is not known.
While H. pylori is the most common Helicobacter species in humans, H. heilmannii has also been observed in human gastric pathology (3, 16, 32). Recently, a 99.5% 16S rDNA sequence homology has been reported between H. heilmannii type 1 and Candidatus H. suis (4), while H. heilmannii type 1 and type 2 have been identified in pig stomachs (1, 4, 5, 32). If these results are true, both H. pylori and H. heilmannii could be recognized as zoonotic pathogens, and pigs can be a potential source for human Helicobacter infection. Public health concerns of animal Helicobacter infection have been expressed by some researchers (15; Mendes et al., abstr.).
To date, evidence of infection with Helicobacter species in U.S. pigs has not been demonstrated, and information on its prevalence on swine farms is not available. The purpose of the present study was to investigate evidence of infection with Helicobacter species, including H. pylori, in slaughter pigs in Minnesota by PCR assays. The PCR-positive samples were further classified into different types by PCR-restriction fragment length polymorphism (RFLP), partial 16S rDNA sequencing, and phylogenetic analysis.
MATERIALS AND METHODS
Stomach sample collection.
Over 400 pig stomachs were examined grossly for gastric ulcer lesions during four different visits to a slaughterhouse in Minnesota. The stomachs were graded according to the severity of the gastric lesion. A standard method was used to grade the gross stomach lesions; 0 for no lesion, 1 for evidence of parakeratosis, 2 for erosions of the epithelium, and 3 for active ulcers and cicatrization (11). Then, 40 samples from each grade were randomly selected in this study. The stomachs were placed in separate plastic bags and transported to the laboratory within 3 h under chilled conditions. A tissue segment (1 cm2) from each stomach was collected from the border area of grossly normal and pars esophagea lesion, placed in a bottle containing sterile phosphate-buffered saline (PBS; pH 7.2), and stored at −80°C until PCR analysis. To avoid cross-contamination, different sets of sterile scissors and pinchers were used for each stomach during sampling.
Culture.
Twelve stomach samples with severe gastric ulcers (grade 3) were randomly collected and cultured on Trypticase soy agar plates that contained 5% sheep blood (25). Additionally, the samples were inoculated onto brucella agar plates containing 7% fetal bovine serum and selected antibiotics in an anaerobic gas jar with 5% O2, 10% CO2, 85% N2, and H2 in an atmosphere with 95% humidity at 37°C for up to 7 days (24).
DNA extraction.
The superficial cell layers and mucus were scraped from each stomach sample with a surgical blade. The DNA from the scrapings was extracted using the DNeasy tissue kit (Qiagen Inc, Valencia, Calif.) according to the manufacturer's instructions.
Cloning and sequencing of the PCR products.
A pGEM-T Easy Vector system I (Promega Corp. Madison, Wis.) was used for cloning PCR products. The PCR products were purified from low-melting-point agarose gels with a QIAEXII kit (Qiagen Inc, Valencia, Calif.) according to the manufacturer's instructions. Purified PCR product was ligated with 50 ng of pGEM-T Easy Vector at 15°C overnight and then transferred into pBlueScript Escherichia coli cells. The Luria-Bertani agar (Bio 101 Inc, Carlsbad, Calif.) plates containing ampicillin (50 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-d-galactopyranoside) (40 μg/ml) were used to select clones. Plasmid DNA was isolated from E. coli by the QIAprep spin miniprep kit (Qiagen Inc.).
Preparation of primers.
Oligonucleotide primers were synthesized at the DNA Core Facility, University of Minnesota, St. Paul, Minn. Several primers were used for PCR, as shown in Table 1. The selection of primers for PCR amplification was based on published GenBank data. Primers Hcom 1, Hcom 2, and Hcom 3 specific to the Helicobacter genus were prepared on the basis of the 16S rRNA gene sequence of H. suis. Primers HP 1 and HP 2 specific to H. pylori were also prepared.
TABLE 1.
Primers used for PCR and sequencing
| Primera | Nucleotide sequence (5′ → 3′) | Positions | Type of analysis | Cycle
|
|
|---|---|---|---|---|---|
| Temp (°C) | Time (min) | ||||
| Hcom 1 | GTA AAG GCT CAC CAA GGC TAT | 187–207 | PCR and sequencing | 94 | 1 |
| Hcom 2 | CCA CCT ACC TCT CCC ACA CTC | 576–556 | 63 | 1 | |
| Hcom 3 | TTA TCA CCG CAA CAT GGC TGA TTT G | 1286–1262 | 72 | 1 | |
| Hcom 4 | GAG TGT GGG AGA GGT AGG TGG | 556–576 | Sequencing | ||
| HP 1 | CCT AAC CAA TTG AGC CAA GAA G | 1176–1197 | PCR | 94 | 1 |
| 56 | 1 | ||||
| HP 2 | CTT TCT AAC ACT AAC GCG CTC A | 1579–1558 | 72 | 1 | |
Enzymatic digestion of amplified DNA.
Restriction enzymes HhaI and MboI were used because these enzymes have been widely used to differentiate Helicobacter species and their usefulness has been demonstrated (24). A 10-μl sample of each PCR product was digested with 10 U of each enzyme for 3 h at 37°C in buffers recommended by the manufacturer. The digested samples were analyzed by electrophoresis using 2% agarose containing ethidium bromide. The restriction fragments were separated at 70 V in 1× Tris-borate-EDTA buffer for 60 min and examined by transillumination before being photographed.
Nucleotide sequencing.
The nucleotide sequencing of the amplified products were carried out by the Advanced Genetic Analysis Center of the University of Minnesota with a DNA sequencer (model 377; Applied BioSystems, Inc., Foster City, Calif.) and a Taq Dye Deoxy terminator cycle sequencing kit (Applied BioSystems, Inc.) by using additional (Hcom 4) Helicobacter primers (Table 1). The sequence analyses were resolved with the ABI Prism collection program (Perkin-Elmer, Foster City, Calif.).
Statistical analysis.
The chi-square test was used to examine the relationship between gastric ulcer lesion grades and PCR-positive rates. A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
The GenBank database accession numbers are AF327023 for MN-1, AF327024 for MN-2, AF327025 for MN-3, and AF327026 for MN-4.
RESULTS
PCR of DNA from stomach tissue samples.
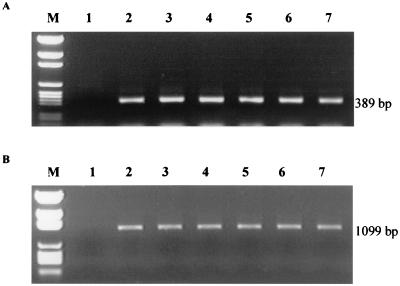
Of 160 stomach samples examined, 102 (63.8%) showed positive results by PCR assay using Helicobacter genus-specific primer sets Hcom 1 and Hcom 2. Utilization of the Hcom 1-Hcom 2 primers and Hcom 1-Hcom 3 primer sets allowed the amplification of a 389-bp fragment and a 1,099-bp fragment of the expected size, respectively (Fig. 1). Results of PCR for the stomach samples with different ulcer lesion grades are shown in Table 2. Of the 40 samples analyzed for each lesion grade, 22.5, 52.5, 85.0, and 95.0% were PCR positive for grades 0, 1, 2, and 3, respectively. There was a significant trend (P ≤ 0.01) in the proportion of PCR-positive cases relative to the severity of the gross lesion. As the lesion score increased, the number of PCR-positive cases rose. Primers specific to H. pylori (HP 1 and HP 2) were also used, but all 102 samples that were positive with Helicobacter genus-specific primers were negative.
FIG. 1.
Electrophoresis of DNA amplified by PCR using Helicobacter genus-specific primers Hcom 1 and Hcom 2 (A) and Hcom 1 and Hcom 3 (B) for pig stomach samples with different ulcer lesion grades. Lane M, pGEM DNA markers; lane 1, grade 0; lanes 2 and 3, grade 1; lanes 4 and 5, grade 2; lanes 6 and 7, grade 3.
TABLE 2.
Identification of Helicobacter species by PCR in pig stomachs with different lesion grades
| Lesion grade | No. of samples examined | No. (%) PCR positive with:
|
|
|---|---|---|---|
| Primers Hcom 1 and Hcom 2 | Primers Hcom 1 and Hcom 3 | ||
| 0 | 40 | 9 (22.5) | 9 (22.5) |
| 1 | 40 | 21 (52.5) | 21 (52.5) |
| 2 | 40 | 34 (85.0) | 34 (85.0) |
| 3 | 40 | 38 (95.0) | 38 (95.0) |
| Total | 160 | 102 (63.8) | 102 (63.8) |
RFLP types.
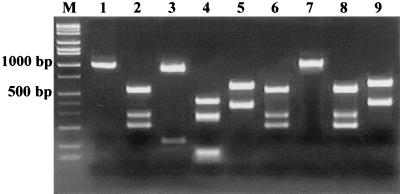
A fragment of 1,099 bp was amplified with primers Hcom 1 and Hcom 3 from all 102 samples that were positive by the PCR assays. The PCR products showing a single band of the expected size (1,099 bp) were subjected to RFLP analysis with HhaI and MboI. The PCR-RFLP results indicated that the Helicobacter species identified from the pig stomachs could be classified into four different types, MN 1, MN 2, MN 3, and MN 4, on the basis of the presence of zero, one, two, or three recognition sites for the enzymes, respectively (Fig. 2).
FIG. 2.
PCR-RFLP patterns of Hcom 1 and Hcom 3 products with MboI and HhaI restriction enzymes. Lane M, Hi-Lo DNA size markers; lane 1, without enzyme digestion; lanes 3, 5, 7, and 9, digested with MboI; lanes 2, 4, 6, and 8, digested with HhaI. Type MN 1, lanes 2 and 3; type MN 2, lanes 4 and 5; type MN 3, lanes 6 and 7; type MN 4, lanes 8 and 9.
As shown in Fig. 2, the results of RFLP analysis resolved by agarose gel electrophoresis were almost identical to the predicted fragments based on the nucleotide sequence data. Of 102 PCR-positive samples, PCR-RFLP pattern analysis results showed that 32 samples (31.4%) were classified as type MN 1, 16 samples (15.7%) were type MN 2, 43 samples (42.2%) were type MN 3, and 11 samples (10.8%) were type MN 4.
16S rDNA sequencing.
In order to determine the Helicobacter species in pigs, sequencing of the Hcom 1 and Hcom 3 PCR products was performed on both strands with or without preliminary cloning using a commercially available sequencing kit (Taq Dye Deoxy Terminator Cycle sequencing kit; Applied Biosystems, Inc.,). When these sequences were compared to those present in databases by using BLAST software, each type showed a high homology with the 16S rDNA of known Helicobacter species. Type MN 1 showed 97.3% homology with H. flexispira taxon 3 (GenBank accession no. AF225547); type MN 2 showed 95.4% homology with a rodent isolate, MIT-94-022 (GenBank accession no. AF225550); type MN 3 showed 96.7% homology with the MZ 640285 strain, a Helicobacter species isolated from an abdominal abscess from a human patient (GenBank accession no. AJ011431); and type MN 4 showed 99.5% homology with H. suis (GenBank accession no. AF127028) (Table 3).
TABLE 3.
Nucleotide sequence similarity among 16S rRNA genes of Helicobacter speciesa
| Organism or strain | % Similarity or difference
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pig- MN 1 | pig- MN 2 | pig- MN 3 | pig- MN 4 | MZ- 640285 | H. bilis | H. canadensis | H. canis | H. cinaedi | H. felis | H. fennenniae | H. flexispira taxon 3 | H. heilmannii | H. hepaticus | MIT 94-022 | H. pullorum | H. pylori | H. rodentium | H. salomonis | H. suis | |
| pig-MN 1 | 93.9 | 96.1 | 90.7 | 96.5 | 93.9 | 95.4 | 93.9 | 94.9 | 89.9 | 92.9 | 97.3 | 89.9 | 95.3 | 95.4 | 94.6 | 89.6 | 93.1 | 89.4 | 90.4 | |
| pig-MN 2 | 5.0 | 92.8 | 89.2 | 92.9 | 92.0 | 94.4 | 92.3 | 92.0 | 89.7 | 91.7 | 93.9 | 89.6 | 92.6 | 95.4 | 93.4 | 89.5 | 91.3 | 88.9 | 89.3 | |
| pig-MN 3 | 3.5 | 6.2 | 89.6 | 96.7 | 93.8 | 95.1 | 93.4 | 94.6 | 89.5 | 92.1 | 96.2 | 89.8 | 94.8 | 93.8 | 94.0 | 89.6 | 93.3 | 88.9 | 89.4 | |
| pig-MN 4 | 6.7 | 8.1 | 7.7 | 91.1 | 90.2 | 90.8 | 91.2 | 90.2 | 96.0 | 90.4 | 90.6 | 95.8 | 90.4 | 89.4 | 90.8 | 94.2 | 90.0 | 95.0 | 99.5 | |
| MZ-640285 | 2.9 | 6.2 | 2.4 | 6.3 | 96.6 | 96.6 | 95.9 | 97.5 | 91.2 | 94.4 | 97.9 | 91.6 | 95.3 | 95.1 | 97.0 | 91.9 | 96.0 | 90.4 | 91.1 | |
| H. bilis | 5.2 | 7.1 | 5.2 | 7.3 | 3.3 | 95.3 | 98.6 | 98.6 | 90.5 | 95.0 | 95.4 | 90.0 | 96.6 | 95.0 | 96.0 | 91.9 | 95.3 | 89.1 | 90.2 | |
| H. canadensis | 3.7 | 5.4 | 3.9 | 6.6 | 2.7 | 3.9 | 95.6 | 95.2 | 90.7 | 93.8 | 97.6 | 91.2 | 96.3 | 96.0 | 97.2 | 92.4 | 95.7 | 90.0 | 90.8 | |
| H. canis | 5.2 | 6.9 | 5.2 | 7.1 | 3.3 | 0.9 | 3.6 | 97.7 | 91.4 | 94.7 | 95.4 | 91.3 | 96.3 | 94.7 | 95.7 | 92.8 | 95.2 | 90.3 | 91.2 | |
| H. cinaedi | 4.4 | 7.0 | 4.6 | 7.2 | 2.4 | 1.2 | 4.1 | 1.8 | 90.2 | 94.5 | 96.4 | 90.0 | 96.3 | 94.7 | 95.3 | 91.0 | 95.4 | 88.7 | 90.2 | |
| H. felis | 7.2 | 7.9 | 7.6 | 3.5 | 6.1 | 6.8 | 6.4 | 6.4 | 7.2 | 89.9 | 90.2 | 98.4 | 90.7 | 89.6 | 91.2 | 94.6 | 90.2 | 97.4 | 96.0 | |
| H. fennenniae | 5.0 | 6.3 | 5.5 | 6.2 | 4.2 | 3.4 | 4.3 | 3.3 | 3.9 | 6.2 | 92.9 | 89.8 | 94.3 | 93.4 | 94.5 | 91.0 | 93.7 | 89.3 | 90.2 | |
| H. flexispira taxon | 2.1 | 5.7 | 3.0 | 6.7 | 1.6 | 3.9 | 2.2 | 3.9 | 3.1 | 7.0 | 5.1 | 90.3 | 95.7 | 96.1 | 95.4 | 90.8 | 94.6 | 89.4 | 90.6 | |
| H. heilmannii | 7.3 | 7.9 | 7.4 | 3.8 | 5.9 | 7.2 | 6.1 | 6.5 | 7.3 | 1.5 | 6.4 | 6.9 | 90.7 | 90.3 | 91.9 | 94.6 | 90.3 | 97.8 | 95.8 | |
| H. hepaticus | 4.5 | 6.3 | 4.8 | 6.9 | 3.8 | 2.6 | 3.0 | 3.0 | 3.1 | 6.5 | 3.4 | 3.7 | 6.4 | 95.4 | 94.6 | 91.2 | 93.9 | 89.9 | 90.4 | |
| MIT 94-022 | 3.6 | 3.9 | 5.0 | 7.1 | 3.9 | 4.1 | 3.3 | 4.3 | 4.4 | 6.8 | 4.5 | 3.7 | 6.3 | 3.5 | 94.6 | 90.9 | 92.7 | 89.0 | 89.2 | |
| H. pullorum | 4.1 | 5.4 | 4.5 | 6.3 | 2.3 | 3.3 | 1.8 | 3.2 | 3.9 | 5.6 | 3.6 | 3.4 | 5.2 | 3.9 | 3.6 | 92.8 | 95.9 | 90.6 | 90.8 | |
| H. pylori | 7.4 | 7.6 | 7.3 | 5.2 | 5.6 | 5.6 | 4.9 | 5.3 | 6.4 | 4.8 | 5.6 | 6.3 | 4.9 | 5.8 | 5.9 | 4.2 | 91.9 | 94.0 | 94.1 | |
| H. rodentium | 5.7 | 7.3 | 5.6 | 7.1 | 3.7 | 4.2 | 3.4 | 4.0 | 4.2 | 6.6 | 4.6 | 4.6 | 6.4 | 4.9 | 5.4 | 3.4 | 5.3 | 89.0 | 90.0 | |
| H. salomonis | 7.8 | 8.4 | 8.2 | 4.7 | 6.9 | 7.7 | 7.1 | 7.1 | 8.0 | 2.4 | 6.8 | 7.7 | 1.9 | 7.0 | 6.9 | 6.2 | 5.4 | 7.4 | 94.8 | |
| H. suis | 7.1 | 8.1 | 7.8 | 0.4 | 6.4 | 7.4 | 6.7 | 7.2 | 7.3 | 3.6 | 6.5 | 6.8 | 3.9 | 7.0 | 7.4 | 6.4 | 5.2 | 7.2 | 4.9 | |
The values above the diagonal are uncorrected percent similarity, and the values below the diagonal are percent difference corrected for multiple base changes by the Clustal method. MZ-640285 and MIT 94-022 are Helicobacter sp. strains.
Phylogenetic analysis.
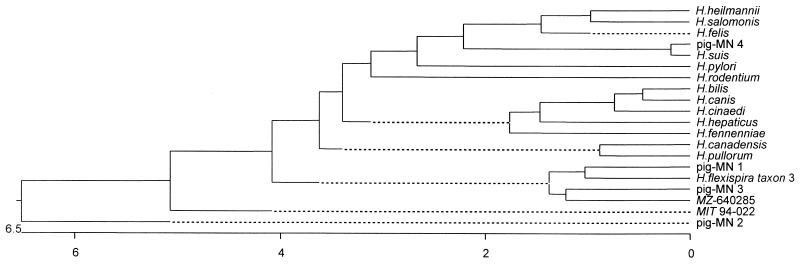
Helicobacter 16S rDNA gene sequences were compared to similar sequences reported for the genus Helicobacter. A phylogenetic tree using the Clustal method with weighted residues was constructed (Fig. 3). Phylogenetic results indicated that RFLP types MN 1 and MN 2 had a close relationship with H. flexispira taxon 3 and MIT 94-022 strain of rodent origin, respectively. The RFLP type MN 3 was the closest to MZ 640258, and type MN 4 had a relationship with previously reported swine isolates of H. suis.
FIG. 3.
Phylogenetic tree of Helicobacter species on the basis of 16S rDNA sequence distances. The scale bar indicates the percent difference in nucleotide sequences determined by measuring the lengths of the horizontal lines connecting any two species. MZ-640285 and MIT 94-022 are Helicobacter sp. strains.
DISCUSSION
The results of this study show a high prevalence (63.8%) of Helicobacter species infection in slaughter pigs by the PCR analysis. Similar high prevalences of infection were reported by Cantet et al. (1) and Thiberge et al. (27), in which 80 and 86.6%, respectively, of the pig samples were positive for Helicobacter species using PCR assays. In contrast to these reports, Queiroz et al. (22) and Grasso et al. (12) reported that 10.8 and 9.4% of the pigs had Helicobacter species infection by histological examination, respectively. This difference may be explained partly by the diagnostic tests employed, either histology or PCR assay.
An association between Helicobacter infection and stomach ulceration in pigs has been suggested (20, 23; Mendes et al., abstr.), but it has not been confirmed adequately. The present results have demonstrated a significant association: the higher the lesion grade of ulceration, the higher the detection rate of Helicobacter infection (Table 2). Queiroz et al. (23) reported similar results. They found Helicobacter species more frequently in the stomachs with ulcers (100%) and in those with preulcer lesions (90%) than in the stomachs with a grossly normal pars esophagea (35%). A large-scale study may be necessary to conclude whether there is a definitive association between the detection of Helicobacter species and the presence of gastric ulcers in swine. However, the present data suggest that infection with Helicobacter species can be one of the contributing factors for gastric ulcer development in pigs.
Helicobacter species reported from pigs were mostly Candidatus H. suis (4, 5), G. suis (20), H. heilmannii type 1 (1, 22), and type 2 (1). By sequence analysis, high homology was shown among them. Our results also demonstrated a high homology. In addition, Helicobacter species in pigs could be classified into at least four different types by PCR-RFLP analysis. Most interesting, some of the strains had high homology with Helicobacter species of rodent or human origin. H. flexispira taxon 3 and strain MIT-94-022 were isolated from rodents, and strain MZ 640285 is a Helicobacter species isolated from an abdominal abscess from a patient with X-linked hypogammaglobulinemia (14). These sequence similarities suggest that one or more Helicobacter species can colonize the pig stomach.
There have been several reports that H. pylori and H. heilmannii can colonize the stomach of pigs (7, 13, 20, 23; Mendes et al., abstr.). Eaton et al (7) described the possibility that commercial pigs may carry H. pylori naturally. Other researchers also supported this, detecting evidence of H. pylori infection in pigs (17, 29). However, the results by Grasso et al. (13), Mendes et al. (abstr.), Queiroz et al. (22), and Cantet et al. (1), as well as the results of this study, failed to detect H. pylori in pig stomachs. Further studies are required to determine if H. pylori can infect pigs under natural conditions.
While the role of Helicobacter species in the pathogenesis of gastric ulcers is not known, evidence of Helicobacter infection in pigs is now well demonstrated. The present results have confirmed the evidence of H. suis infection and identified novel Helicobacter species with high homology of human and rat origin in pigs. Although it is not known how Helicobacter species could be transmitted to pigs, wild rats may play a role in transmission under field conditions. Excretion of helicobacters in feces (28) and subsequent contamination of food or water has also been suggested (30). There are many questions to be answered concerning helicobacter infection in swine. An interesting question remains whether swine and pork products infected with Helicobacter species can be a source of human infection. This question is particularly important, since publicity has recently intensified regarding emerging food-borne pathogens.
ACKNOWLEDGMENTS
This work was supported by a Food Safety Initiative grant from the College of Veterinary Medicine, University of Minnesota.
We thank Dominiek Maes for statistical analysis and Connie Gebhart for critical reading of the manuscript.
REFERENCES
- 1.Cantet F, Magras C, Marais A, Federichi M, Megraud F. Helicobacter species colonizing pig stomach: molecular characterization and determination of prevalence. Appl Environ Microbiol. 1999;65:4672–4676. doi: 10.1128/aem.65.10.4672-4676.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chagnon M, D'Allaire S, Drolet R. A prospective study of sow mortality in breeding herds. Can J Vet Res. 1991;55:180–185. [PMC free article] [PubMed] [Google Scholar]
- 3.Debongnie J C, Donnay M, Mairesse J, Lamy V, Dekoninck X, Ramdani B. Gastric ulcers and Helicobacter heilmannii. Eur J Gastroenterol Hepatol. 1998;10:251–254. doi: 10.1097/00042737-199803000-00011. [DOI] [PubMed] [Google Scholar]
- 4.De Groote D, Van Doorn L J, Ducatelle R, Verschauuren A, Haesebrouck F, Quint W G V, Jalava K, Vandamme P. “Candidatus Helicobacter suis,” a gastric helicobacter from pigs and its phylogenetic relatedness to other gastrospirilla. Int J Syst Bacteriol. 1999;49:1769–1777. doi: 10.1099/00207713-49-4-1769. [DOI] [PubMed] [Google Scholar]
- 5.De Groote D, Ducatelle R, van Doorn L J, Tilmant K, Verschuuren A, Haesebrouck F. Detection of “Candidatus Helicobacter suis” in gastric samples of pigs by PCR: comparison with other invasive diagnostic techniques. J Clin Microbiol. 2000;38:1131–1135. doi: 10.1128/jcm.38.3.1131-1135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton K A, Morgan D R, Krakowka S. Persistence of Helicobacter pylori in conventionalized piglets. J Infect Dis. 1990;161:1299–1301. doi: 10.1093/infdis/161.6.1299. [DOI] [PubMed] [Google Scholar]
- 8.Elber A R W, Hessing M J C, Tielen M J M, Vos J H. Growth and oesophagogastric lesions in finishing pigs offered pelleted feed ad libitum. Vet Rec. 1995;136:588–590. doi: 10.1136/vr.136.23.588. [DOI] [PubMed] [Google Scholar]
- 9.Fox J G, Yan L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel helicobacter isolated from bile, livers, and intestines of aged, inbred mouse strain. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox J G, Drolet R, Higgins R, Messier S, Yan L, Coleman B E, Paster B J, Dewhirst F E. Helicobacter canis isolated from a dog liver with multifocal necrotizing hepatitis. J Clin Microbiol. 1996;34:2479–2482. doi: 10.1128/jcm.34.10.2479-2482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friendship R. Gastric ulcers. In: Straw B E, D'Allaire S, Mengeling W L, Taylor D J, editors. Diseases of swine. 8th ed. Ames, Iowa: Iowa State University Press; 1999. pp. 685–694. [Google Scholar]
- 12.Grasso G M, Sammarco M L, Ripabelli G, Ruberto G A, Iannitto G. In situ mapping of urease-positive areas in porcine gastric mucosa. Microbios. 1995;82:245–249. [PubMed] [Google Scholar]
- 13.Grasso G M, Ripabelli G, Sammarco M L, Ruberto A, Iannitto G. Prevalence of Helicobacter-like organisms in porcine gastric mucosa: a study of swine slaughtered in Italy. Comp Immun Microbiol Infect Dis. 1996;19:213–217. doi: 10.1016/0147-9571(96)00007-0. [DOI] [PubMed] [Google Scholar]
- 14.Han S-R, Schindel C, Genitsariotis R, Märker-Hermann E, Bhakdi S, Maeurer M J. Identification of a unique Helicobacter species by 16S rRNA gene analysis in an abdominal abscess from a patient with X-linked hypogammaglobulinemia. J Clin Microbiol. 2000;38:2740–2742. doi: 10.1128/jcm.38.7.2740-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handt L K, Fox J G, Dewhirst F E, Fraser G J, Paster B J, Yan L L, Rozmiarek H, Rufo R, Stalis I H. Helicobacter pylori isolated from the domestic cat: public health implications. Infect Immun. 1994;62:2367–2374. doi: 10.1128/iai.62.6.2367-2374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heilmann K L, Borchard F. Gastritis due to spiral shaped bacteria other than Helicobacter pylori: clinical, histological, and ultrastructural findings. Gut. 1991;32:137–140. doi: 10.1136/gut.32.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones D M, Elridge J. Gastric campylobacter-like organisms from man (C. pyloridis) compared with GCLO strains from the pig, baboon and ferret. In: Kaijser B, Falsen E, editors. Campylobacter IV. Goteborg, Sweden: University of Goteborg; 1988. p. 44. [Google Scholar]
- 18.Krakowka S, Eaton K A, Rings D M, Morgan D R. Gastritis induced by Helicobactor pylori in gnotobiotic piglets. Rev Infect Dis. 1991;13:S681–S685. doi: 10.1093/clinids/13.supplement_8.s681. [DOI] [PubMed] [Google Scholar]
- 19.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1314. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 20.Mendes E N, Queiroz D M M, Rocha G A, Moura S B, Leite V H R, Fonseca M E F. Ultrastructure of a spiral micro-organism from pig gastric mucosa (‘Gastrospirillum suis’) J Med Microbiol. 1990;33:61–66. doi: 10.1099/00222615-33-1-61. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson H O, Taneera J, Castedal M, Glatz E, Olsson R, Wadstrom T. Identification of Helicobacter pylori and other Helicobacter species by PCR, hybridization, partial DNA sequencing in human liver samples from patients with primary sclerosing cholangitis or primary biliary cirrhosis. J Clin Microbiol. 2000;38:1072–1076. doi: 10.1128/jcm.38.3.1072-1076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Queiroz D M M, Rocha G A, Mendes E N, Large A P, Carvalho A C T, Barbosa A J A. A spiral microorganism in the stomach of pigs. Vet Microbiol. 1990;24:199–204. doi: 10.1016/0378-1135(90)90067-6. [DOI] [PubMed] [Google Scholar]
- 23.Queiroz D M M, Rocha G A, Mendes E N, De Moura S B, De Oliveria A M, Miranda D. Association between Helicobacter and gastric ulcer disease of the pars esophagea in swine. Gastroenterology. 1996;111:19–27. doi: 10.1053/gast.1996.v111.pm8698198. [DOI] [PubMed] [Google Scholar]
- 24.Riley L K, Franklin C L, Hook R R, Jr, Besch-Williford C. Identification of murine helicobacters by PCR and restriction enzyme analyses. J Clin Microbiol. 1996;34:942–946. doi: 10.1128/jcm.34.4.942-946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons J H, Riley L K, Besch-Williford C, Franklin C L. Helicobacter mesocricetorum sp. nov., a novel helicobacter isolated from the feces of Syrian hamsters. J Clin Microbiol. 2000;38:1811–1817. doi: 10.1128/jcm.38.5.1811-1817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanley J, Linton D, Burens A P, Dewhirst F E, On S L, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov. genotype and phenotype of a new species isolated from poultry and from human with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 27.Thiberge J M, Bedel A, Huerre M, Pichard F, Labigne A. Comparison of several diagnostic tests for the detection of Helicobacter infection in swine. Gut. 1997;41(Suppl. 1):A125. [Google Scholar]
- 28.Thomas J E, Gibson G R, Darboe M K, Dale A, Weaver L T. Isolation of Helicobacter pylori from human faeces. Lancet. 1992;340:1194–1195. doi: 10.1016/0140-6736(92)92894-l. [DOI] [PubMed] [Google Scholar]
- 29.Vaira D, Ferron P, Negrini R, Cavazzini L, Holton J, Ainley C, Londei M, Vergura M, Dei R, Colecchia A, Tayler D, Pieracci F, Nenci I, Gandolfi L, Barbara L. Detection of Helicobacter pylori-like organisms in stomach of some food-source animals using a monoclonal antibody. Ital J Gastroenterol. 1992;24:181–184. [PubMed] [Google Scholar]
- 30.Velazquez M, Feirtag J M. Helicobacter pylori: characteristics, pathogenicity, detection methods and mode of transmission implicating foods and water. Int J Food Microbiol. 1999;53:95–104. doi: 10.1016/s0168-1605(99)00160-9. [DOI] [PubMed] [Google Scholar]
- 31.Ward J M, Anver M R, Haines D C, Benvensite R E. Chronic active hepatitis in mice caused by Helicobacter hepaticus. Am J Pathol. 1994;145:959–968. [PMC free article] [PubMed] [Google Scholar]
- 32.Yeomans N D. Helicobacter heilmannii (formerly Gastrospirillum): association with pig and human gastric pathology. Gastroenterology. 1996;111:244–259. doi: 10.1053/gast.1996.v111.agast961110244. [DOI] [PubMed] [Google Scholar]