Abstract
Simple Summary
Prostate cancer (PCa) is a hormone-dependent tumor characterized by a highly heterogeneous clinical outcome. This neoplastic process has become a leading cause of cancer worldwide, with over 1.4 million new cases and a total of 375,000 deaths in 2020. Despite the efforts to improve the diagnosis, risk stratification, and treatment of PCa patients, a number of challenges still need to be addressed. In this context, integration of different multi-omics datasets may represent a powerful approach for the development of novel metabolic signatures that could contribute to the clinical management of PCa patients. This review aims to provide the most relevant findings of recently published multi-omics studies with a particular focus on describing the metabolic alterations associated with PCa.
Abstract
Prostate cancer (PCa), one of the most frequently diagnosed cancers among men worldwide, is characterized by a diverse biological heterogeneity. It is well known that PCa cells rewire their cellular metabolism to meet the higher demands required for survival, proliferation, and invasion. In this context, a deeper understanding of metabolic reprogramming, an emerging hallmark of cancer, could provide novel opportunities for cancer diagnosis, prognosis, and treatment. In this setting, multi-omics data integration approaches, including genomics, epigenomics, transcriptomics, proteomics, lipidomics, and metabolomics, could offer unprecedented opportunities for uncovering the molecular changes underlying metabolic rewiring in complex diseases, such as PCa. Recent studies, focused on the integrated analysis of multi-omics data derived from PCa patients, have in fact revealed new insights into specific metabolic reprogramming events and vulnerabilities that have the potential to better guide therapy and improve outcomes for patients. This review aims to provide an up-to-date summary of multi-omics studies focused on the characterization of the metabolomic phenotype of PCa, as well as an in-depth analysis of the correlation between changes identified in the multi-omics studies and the metabolic profile of PCa tumors.
Keywords: prostate cancer, metabolism, multi-omics, metabolomics
1. Introduction
Prostate cancer (PCa) is the second most frequent cancer and represents the fifth leading cause of cancer-related death in men worldwide [1]. According to the Global Cancer Incidence, Mortality, and Prevalence (GLOBOCAN) database, new PCa cases were estimated to account for almost 1.4 million, with a total of 375,000 cancer-related deaths in 2020 [1]. Clinically, PCa is characterized by a heterogeneous behavior, ranging from indolent phenotypes to a rapid progression into an aggressive metastatic disease [2]. Early PCa diagnosis mainly relies on prostate-specific antigen (PSA) tests, although this screening method exhibits several limitations as it is prostate-specific but not cancer-specific [3], leading to overdiagnosis and overtreatment [4,5,6]. Thus, histopathological evaluation of biopsies, graded on the basis of the Gleason Score (GS) [7], is required to confirm the presence of PCa [8] and to determine the treatment strategy to follow [9]. However, prostate biopsy is an invasive procedure that might cause health complications (e.g., hematospermia, hematuria, fever, bleeding, urinary retention) [10,11]. In addition, although the grading system has been modified several times, there remains no classification scheme that allows accurately discriminating indolent from aggressive PCa stages [12]. Thus, there is a need for more precise and robust PCa biomarkers to improve diagnosis and risk stratification of patients.
In recent years, metabolic phenotyping has become a powerful approach for the identification of new molecular biomarkers and metabolic vulnerabilities that could represent novel therapeutic opportunities in oncological diseases [13,14,15,16,17,18]. Hence, several metabolomics analyses have been carried out on PCa samples (e.g., tissue, urine, serum, plasma, and seminal fluid) to characterize the specific metabolic profile associated with PCa progression and identify metabolic alterations that may potentially be used as clinical biomarkers (reviewed in [19,20,21,22]). Together, these studies have revealed a specific metabolic phenotype that could distinguish between healthy and PCa samples [23]. Healthy prostate cells accumulate high concentrations of zinc, which results in the inhibition of mitochondrial aconitase (ACO2) and consequently decreases citrate oxidation, thus disrupting the tricarboxylic acid (TCA) cycle metabolism [24]. In contrast, decreased zinc levels in PCa tumors enable the activation of ACO2 for citrate oxidation and subsequent re-establishment of the TCA cycle [23,25]. In line with this, metabolic studies have reported decreased citrate levels and increased concentrations of several TCA cycle intermediates (e.g., fumarate, malate, and succinate) in PCa tumor samples when compared with healthy prostate tissues, suggesting an increased TCA cycle metabolism [26,27,28,29]. In addition, other studies have reported lower levels of polyamines and sarcosine metabolism (e.g., spermine, spermidine, sarcosine) [29,30,31,32,33], as well as dysregulations of several amino acids (e.g., alanine, glutamate, arginine, tyrosine, phenylalanine) [26,27,28,34,35,36,37,38,39] and other metabolites involved in cellular membrane metabolism (e.g., choline, phospholipids) [26,27,40,41,42,43,44].
These metabolic alterations have been observed at different omics levels [45,46,47]. For instance, transcriptomics analyses facilitated the identification of three distinct metabolism-associated PCa clusters and the development of a six-gene metabolic signature associated with disease-free survival [47]. In addition, following a loss-of-function genetic screen, the glycolytic 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4) enzyme was identified as an essential gene for PCa cell survival and evaluated as a potential therapeutic target for PCa treatment [48]. On the other hand, proteomics analyses carried out on PCa cell lines and tissue samples revealed that enzymes involved in the ketogenic metabolism pathway were overexpressed in high-grade PCa [49]. Furthermore, the characterization of the proteomics landscape of exosomes, isolated from primary prostate epithelial and PCa cell lines, identified four exosomal proteins (PDCD6IP, FASN, XPO1, and ENO1) as potential new candidate biomarkers for PCa [50]. Moreover, lipidomics, an emerging omics approach [51], has also demonstrated its potential as an alternative diagnostic tool in PCa, revealing specific associations between alterations in glycerophospholipid metabolism and fatty-acid synthesis and oxidation with PCa progression [52,53].
In summary, the information derived from different omics studies offers new avenues for better understanding the biological and molecular processes underlying metabolic changes occurring during cancer progression, as well as for developing novel molecular biomarkers to improve the clinical management of cancer patients. Moreover, a number of studies have demonstrated that the combination of multi-omics data can provide deeper insight into the metabolic changes associated with the progression of different oncological diseases than any of these omics on their own [54,55,56,57]. Thus, the integration of different omics platforms has emerged as a powerful and promising strategy for the elucidation of potential genetic and epigenetic alterations, changes in gene expression levels and signaling pathways, and other biological dysregulations that could be driving metabolic rewiring during cancer progression. Hence, this review aims to provide the most relevant findings reported in recently published multi-omics-based studies focused on the analysis of metabolic alterations associated with PCa initiation and progression (Figure 1). To that end, a literature search was conducted on PubMed, using different combinations of the following terms: “(omics OR multi-omics OR omics integration OR (metabolomics OR lipidomics) AND (metabolomics OR lipidomics OR transcriptomics OR genomics OR epigenomics OR proteomics) AND (metabol * OR metabolic profiling OR metabolic phenotype)) AND prostate cancer”. Then, titles and abstracts of the selected publications were examined to evaluate their eligibility according to their relevance on the issue of interest and to determine their inclusion in the review.
Figure 1.
Graphical representation of different omics-based approaches and multi-omics analyses applied to the characterization of PCa-related metabolic alterations.
2. PCa Multi-Omics Studies
Between 2013 and 2021, 21 studies, focused on characterizing the specific metabolic profile associated with PCa, reported the integration of data from at least two different omics platforms. Tissue was by far the most frequently analyzed sample (15 studies), cell lines were harvested in four studies, biofluids (urine, serum) from Pca patients were collected in three of the studies, and only one of them relied on using murine models. Integration of transcriptomics and metabolomics was conducted in most of the studies, while transcriptomics, metabolomics, and lipidomics were only combined in two of them. Lastly, mass spectroscopy (MS) was preferentially chosen over nuclear magnetic resonance (NMR) as analytical platform for proteomics, metabolomics, or lipidomics analyses.
2.1. Benign Tissue vs. PCa Tumor
Nine of the studies discussed in this review relied on the analysis of benign prostate and PCa samples for identifying specific metabolic alterations associated with the metabolic phenotype of PCa patients (Table 1). Integration of transcriptomics and metabolomics data was the primary approach followed in these studies, and tissue samples were the biological specimens preferentially collected.
Table 1.
Most relevant metabolic alterations reported in recent multi-omics studies focused on the characterization of the specific metabolic phenotype of PCa patients.
| Study | Sample | Omics Data | Major Findings * |
|---|---|---|---|
| Meller et al. [58] | Tissue | M + T | ↑ ACC, ACLY, FASN, SCD, 2-hydroxybehenic acid, cerebronic acid, glycerol phosphate, palmitic acid, GSH/GSSG, and spermidine ↓ putrescine and spermine |
| Li et al. [59] | Tissue | L + T | ↑ PLA2s, free MUFA and PUFA, and LPLATs ↓ free SFA, PUFA-acyl, and ether-linked chains in PLs |
| Torrano et al. [60] | Cell lines | M + T | ↓ PGC1A, FAO, and TCA cycle |
| Lima et al. [61] | Tissue | L + M | ↑ amino-acid metabolism, nicotinate and nicotinamide metabolism, purine metabolism, and glycerophospholipid metabolism |
| Shao et al. [62] | Tissue | M + T | ↑ fumarate, malate, succinate, 2-hydroxyglutaric acid, 2-ketoglutarate, glutamine, glutamate, PDH, GLS, GLUD1, GLUD2, and BCAA degradation enzymes |
| Tessem et al. [63] | Tissue | M + T | ↑ ACLY, ACACA, FASN, SAT1, SMOX, SRM, and succinate, ↓ ACO1, SDHD, SUCLA2, putrescine, and citrate |
| Kaushik et al. [64] | Tissue | M + T | ↑ HBP, GNPNAT1, UAP1, and UDP-GlcNAc |
| Ren et al. [65] | Tissue | M + T | ↑ HBP, UDP-GlcNAc, and sphingosine |
| Lee et al. [66] | Urine | M + T | ↑ GOT1 and glutamate |
ACACA: acetyl-CoA carboxylase alpha, ACC: acetyl-CoA carboxylase, ACLY: ATP citrate lyase, ACO1: aconitase, BCAA: branched-chain amino acids, FAO: fatty-acid oxidation, FASN: fatty-acid synthase, GLS: glutaminase, GLUD1: glutamate dehydrogenase 1, GLUD2: glutamate dehydrogenase 2, GNPNAT1: glucosamine-phosphate N-acetyltransferase 1, GOT1: glutamate oxaloacetate transaminase 1, GSH: reduced glutathione, GSSG: oxidized glutathione, HBP: hexosamine biosynthesis pathway, L: lipidomics, LPLATs: lysophospholipid acyltransferase, M: metabolomics, MUFA: mono-unsaturated fatty acids, PDH: pyruvate dehydrogenase, PGC1A: PPARG coactivator 1 alpha, PLs: phospholipids, PLA2s: phospholipase A2, PUFA: polyunsaturated fatty acids, SAT1: spermidine/spermine N1-acetyltransferase 1, SCD: acyl-CoA desaturase, SDHD: succinate dehydrogenase complex subunit D, SFA: saturated fatty acids, SMOX: spermine oxidase, SRM: spermidine synthase, SUCLA2: succinate-CoA ligase ADP-forming beta subunit, T: transcriptomics, TCA: tricarboxylic acid, UAP1: UDP N-acetyl glucosamine pyrophosphate 1. * Direction of variation, considering the benign group as reference. Up and down arrows indicate direction of the variation observed in PCa samples.
Several of these studies reported alterations in enzymes and/or metabolites involved in fatty-acid metabolism. Among them, Meller et al. observed a highly deregulated metabolism of fatty acids, sphingolipids, and polyamines in malignant tissue [58]. Altered fatty-acid and sphingolipid metabolism was associated with increased expression of acetyl-CoA carboxylase (ACC), ATP citrate lyase (ACLY), fatty-acid synthase (FASN), and acyl-CoA desaturase (SCD) as well as with elevated concentrations of several fatty acids, such as 2-hydroxybehenic acid, cerebronic acid, glycerol phosphate, and palmitic acid. Furthermore, a higher ratio of reduced (GSH) to oxidized (GSSG) glutathione and alterations in the levels of several metabolites involved in polyamine metabolism, including putrescine, spermine, and spermidine, were detected in PCa tumors. These observations were based on the metabolomics, transcriptomics, and immunohistochemistry analysis of matched malignant and nonmalignant prostatectomy samples from 106 PCa patients. These results are in agreement with previous studies reporting FASN to be upregulated in PCa tumors [67,68,69], and SCD to promote PCa proliferation [70], as its inhibition resulted in a reduction in tumor growth [71]. Other studies reported higher glutathione reductase activity in PCa, leading to higher GSH levels, which could confer higher oxidative stress resistance to these tumors [72]. A recent study also showed that mTORC1 regulated polyamine synthesis as part of an essential oncogenic metabolic reprograming in PCa [73].
Dysregulated lipid metabolism in PCa was also reported by Li et al., in a study focused on understanding the regulatory networks involved in adaptative transformation of lipid metabolism in PCa tissues [59]. Following a network-wide integrated mapping of lipid metabolism, including changes in the lipidome, transcript alterations, and post-transcriptional regulations, the authors observed a significant upregulation of de novo lipogenesis and a strengthened biosynthesis of phospholipids (PLs) via a de novo pathway in PCa lipogenesis, together with a reprogrammed composition in membrane PLs. Overall, percentages of free mono- and polyunsaturated fatty acids (MUFAs and PUFAs, respectively) were elevated, while free saturated fatty acids (SFA) were reduced. Moreover, activated PL remodeling was characterized by enhanced activities of phospholipase A2 (PLA2s) and reduced lysophospholipid acyltransferase (LPLATs), which contributed to increased MUFA-acyl residues and reduced PUFA-acyl and ether-linked chains in PCa PLs. In fact, lipogenesis upregulation has been described as a hallmark of invasive cancers and termed the “lipogenic phenotype” [74]. Furthermore, several studies have associated changes in the PL content of the cellular membrane with PCa aggressiveness [27,75,76].
The characterization of relevant master regulators contributing to the metabolic switch in PCa was also evaluated in a multi-omics study conducted by Torrano et al. [60]. In this study, the analysis of the expression levels of several metabolic coregulators in five different PCa datasets revealed that only alterations in the transcriptional coactivator PPARG coactivator 1 alpha (PPARGC1A or PGC1A), PPARG coactivator 1 beta (PPARGC1B or PGC1B), and histone deacetylase 1 (HDAC1) expression were present in the majority or all datasets. Among them, PGC1A was the only coregulator negatively associated with GS. Additional integrative metabolomics analysis demonstrated that the tumor suppressive activity of PGC1A was associated with a global metabolic rewiring, leading to an enhanced fatty-acid β-oxidation and TCA cycle activity. TCA cycle downregulation has also been associated with PCa progression in other multi-omics studies [77], while upregulation of TCA cycle activity has been observed when comparing PCa tumor vs. adjacent prostate tissue [62]. Notably, the results from these studies are in agreement with a previously undescribed two-step metabolic shift in the TCA cycle during PCa development and progression, which was recently identified by Latonen et al. [77]. Further in vitro and in vivo analyses performed in this study demonstrated the role of PGC1A in tumor progression and metastatic dissemination, with these results also being in agreement with recent findings [78]. Moreover, a recent study showed that downregulation of PGC1A could promote PCa aggressiveness through activation of the polyamine pathway [79].
The comparison of benign and PCa tissue samples has also revealed additional changes in energy-related metabolic pathways. Thus, in a study conducted by Lima et al., an analysis of the metabolomics and lipidomics profiles of benign and PCa tissues by NMR and MS revealed metabolic dysregulations associated with PCa development [61]. The multivariate statistical analyses revealed that the levels of 26 metabolites, including different amino acids, organic acids, and nucleotide derivatives, and 21 phospholipid species were significantly altered between both groups. Furthermore, a metabolic pathway analysis revealed 11 dysregulated metabolic pathways associated with PCa development. Dysregulations in these pathways were confirmed by strong correlations among metabolites participating in the same pathway. The main metabolic pathways associated with PCa were amino-acid metabolism, nicotinate and nicotinamide metabolism, purine metabolism, and glycerophospholipid metabolism. Notably, metabolites involved in these pathways were upregulated in PCa tissues, being in accordance with other results published in previous studies [21,22,23]. Many of these pathways provide metabolic intermediates for the TCA cycle, nucleotide synthesis, and lipid synthesis, thus contributing to the production of high levels of cellular building blocks required for rapid proliferation of cancer cells [13,80].
Shao et al. also reported accumulation and upregulation of metabolites and genes related to the TCA cycle in another multi-omics-based study [62]. Metabolomics and transcriptomics analysis of PCa tumors and matched adjacent normal tissues revealed significant accumulations of key TCA metabolic intermediates (malate, fumarate, succinate, and 2-hydroxyglutaric acid) and enrichment in genes from different anaplerotic routes, including those involved in pyruvate, glutamine catabolism, and branched-chain amino-acid (BCAA) degradation. Associations between TCA cycle and the potential anaplerotic routes were supported by increased expression of pyruvate dehydrogenase (PDH) complex, higher expression levels of different BCAA degradation genes, glutaminase (GLS) and glutamate dehydrogenase (GLUD1 and GLUD2), and higher α-ketoglutarate, glutamine, and glutamate levels. Dysregulations in the TCA cycle were also identified in PCa tissues by Tessem et al. after accounting for the confounding effect of stroma [63]. In this study, integration of metabolomics and transcriptomics data revealed associations between increased succinate levels, also observed in other studies [29,81], and downregulation of succinate-CoA ligase ADP-forming subunit beta (SUCLA2) and succinate dehydrogenase complex subunit D (SDHD). Additional observations included lower citrate levels and decreased expression of ACO1, together with overexpression of fatty-acid synthesis genes ACLY, acetyl-CoA carboxylase alpha (ACACA), and FASN, suggesting an enhanced fatty-acid synthesis in these tissues. Reduced citrate concentrations and increased lipid synthesis are considered relevant metabolic features of PCa [23,82]. Furthermore, the authors observed relevant associations between reduced putrescine levels and upregulation of spermidine synthase (SRM), as well as lower spermine and increased spermidine/spermine N1-acetyltransferase 1 (SAT1) and spermine oxidase (SMOX) expression. In agreement with these results, other authors also reported a reduction in spermine and putrescine levels [34,58,74,75], as well as an overexpression of enzymes involved in the polyamine pathway [83,84,85].
In another study conducted by Kaushik et al., transcriptomics and metabolomics analyses were integrated, using a pathway-centric analytical framework that enabled the combination of the rankings of biochemical pathways enriched independently by gene expression and metabolic profiles in a single significance score [64]. Following this analysis, the hexosamine biosynthesis pathway (HBP) was found to be the most enriched pathway in treatment-naïve localized PCa, when compared to benign adjacent prostate tissues. Moreover, in silico analysis showed that the expression of glucosamine-phosphate N-acetyltransferase 1 (GNPNAT1) and UDP N-acetyl glucosamine pyrophosphate 1 (UAP1) were significantly elevated in PCa tumors. In contrast, HBP genes were significantly downregulated in castrate-resistant prostate cancer (CRPC) in comparison with localized PCa. The opposite effect of the HBP on the growth of androgen-dependent PCa and CRPC cells suggests the existence of metabolic rewiring during PCa progression. Moreover, on the basis of different in vitro and in vivo approaches, the authors concluded that downregulation of HBP in CRPC cells modulates progression via either PI3K/Akt or specific protein 1 (SP1)-regulated expression of carbohydrate response element-binding protein (ChREBP), depending on the androgen receptor variant. Previous studies have also reported several metabolic rewiring mechanisms associated with different androgen receptor variants [86]. Lastly, in this study, the authors evaluated the therapeutic efficacy of UDP-GlcNAc treatment, alone and in combination with anti-androgen therapy, for the treatment of CRPC-like tumors bearing different androgen receptor variants. Notably, in vivo UDP-GlcNAc treatment significantly reduced the proliferation in all assayed CRPC-like tumors. These findings are particularly relevant as CRPC cells containing the AR-V7 variant are essentially resistant to anti-androgen therapy. Interestingly, Ren et al. also reported increased activity of the HBP in PCa compared to adjacent prostate tissues [65]. In both studies, UDP-GlcNAc, the end product of the HBP and a key substrate for the O-linked N-acetyl-glucosamine transferase (OGT), which plays a vital role in O-GlcNAcylated modification of proteins, was found to be increased in PCa tissues [64,65]. Interestingly, posttranslational O-GlcNAcylation of chromatin is a significant feature of enhancers in the PCa genome [46,87]. In addition to the HBP, Ren et al. reported metabolic perturbations in other metabolic pathways, including the metabolism of cysteine and methionine and nucleotide sugars, glycerophospholipids, lysine, and sphingolipids. Moreover, nine metabolites showed potential utility as metabolic PCa biomarkers. Among them, sphingosine demonstrated high specificity and sensitivity for distinguishing PCa from benign prostatic hyperplasia (BPH), particularly in patients with low PSA levels. Other metabolomics studies have also reported alterations in the HBP and sphingolipid metabolism when analyzing the metabolic profile of PCa patients [27,28].
More recently, Lee et al. carried out a transcriptomics and metabolomics analysis of urine liquid biopsies from BPH, prostatitis, and PCa patients with a focus on the identification of PCa-specific biomarkers and the discovery of novel therapeutic targets for PCa treatment [66]. Significantly enriched pathways in PCa patients included the TCA cycle and alanine, aspartate, and glutamate metabolism. Other metabolomics studies have also reported alterations in urine levels of metabolites involved in these pathways in PCa patients [36,88,89,90]. By examining the top 25 altered metabolites and corresponding genes, the authors identified a regulatory metabolic node that influenced both pathways and was mediated by changes in glutamate oxaloacetate transaminase 1 (GOT1)- and GOT2-related metabolism. Notably, GOT1 expression was higher in PCa patients, and glutamate, the product of GOT1, also exhibited elevated levels in these patients. Moreover, knock-down of GOT1 in LNCaP and PC3 cells resulted in a significant decrease in cell viability, consistent with previous studies where GOT1 repression suppressed tumor growth in different tumors [91,92]. Overall, these results suggest that the metabolic alterations observed in urine liquid biopsies obtained from PCa patients could reflect the specific changes already observed in PCa cells and tumors.
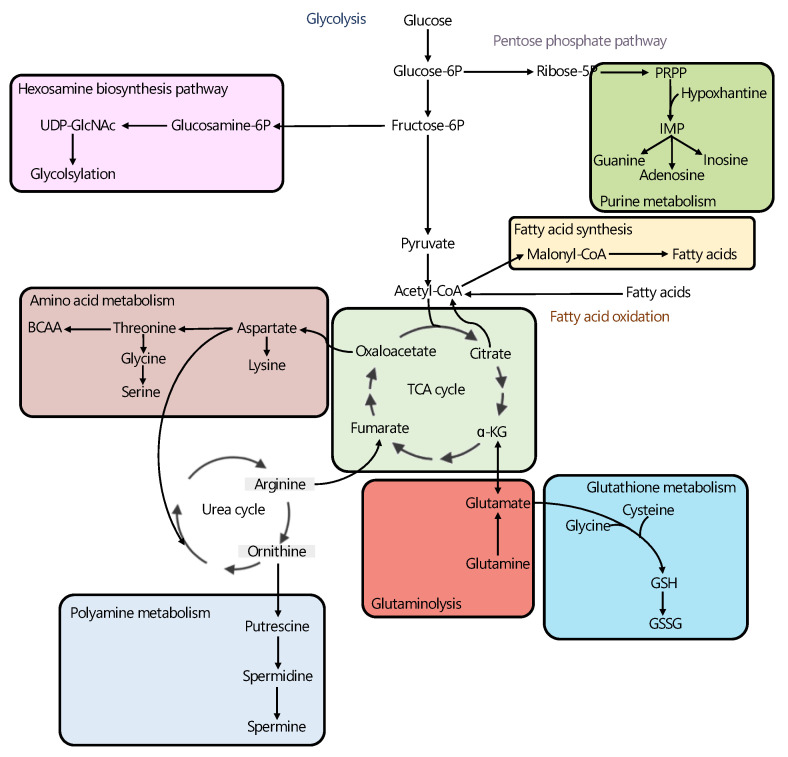
Altogether, in agreement with other studies where metabolomics was the only analytical platform used for analyzing the metabolic profile of PCa patients [39,40,41,93,94,95,96,97], the results from the multi-omics-based studies reviewed in this article suggest that the PCa-specific metabolic phenotype is characterized by alterations in the TCA cycle, polyamine synthesis, HBP, and nucleotide and lipid metabolism (Figure 2).
Figure 2.
Overview of metabolic pathways most consistently reported to be altered in PCa in the multi-omics studies reviewed in this article, including: hexosamine biosynthesis pathway [64,65], purine metabolism [61], fatty acid synthesis [58,59,63], amino acid metabolism [61], TCA cycle [60,62], glutathione metabolism [58], glutaminolysis [62,66] and polyamine metabolism [58,63]. Thick lines highlight the metabolic pathways found to be upregulated in PCa tumors when compared with benign prostate tissue. References corresponding to the multi-omic studies describing alterations in each metabolic pathway are included. α-KG: alpha-ketoglutarate, Fructose-6P: fructose-6-phosphate, Glucosamine-6P: glucosamine-6-phosphate, Glucose-6P: glucose-6-phosphate, IMP: inosine monophosphate, PRPP: phosphoribosyl diphosphate, Ribose-5P: ribose-5-phosphate.
2.2. PCa Subtyping
Twelve of the multi-omics studies included in this review focused on the identification of metabolic alterations associated with specific subtypes of PCa (Table 2). Half of the studies combined transcriptomics and metabolomics analyses to characterize metabolic dysregulations in different subgroups of PCa patients, and the other half relied on the analysis of proteomics and lipidomics profiles. Tissue was the biological sample most often analyzed in these studies, whereas cell lines and biofluids were collected in only three and two studies, respectively. Different subgroups of PCa patients showed alterations in the TCA cycle and amino-acid, nucleotide, and lipid metabolism. Overall, these results correlate with metabolic changes, observed in previous studies, where metabolomics was the only analytical approach used to performed the analyses [26,27,28,75,98,99].
Table 2.
Most relevant metabolic alterations reported in recent multi-omics studies focused on the characterization the metabolic phenotypes of different PCa subtypes.
| Study | Sample | Omics Data | Group Comparison | Major Findings |
|---|---|---|---|---|
| Gómez-Cebrián et al. [100] | Urine, serum | M + T | Low- vs. high- grade PCa | High-grade: ↑ glucose, glycine, and 1-methylnicotinamide |
| Kiebish et al. [101] | Serum | L + M + P | non-BCR vs. BCR | BCR: ↑ TNC, APOA-IV, and 1-methyladenosine and ↓ phosphatidic acid |
| Liu et al. [102] | Tissue | G + M | PCa vs. metastatic | Metastatic Pca: ↑ CYP1A1, PNP, SMS, proline, cholesterol, sarcosine, spermidine, and spermine |
| Li et al. [103] | Tissue | M + T | PCa vs.metastatic | Metastatic PCa: ↓ histamine |
| Latonen et al. [77] | Tissue | E + G + P + T | PCa vs. CRPC | CRPC: ↓ ACO2, OGDH, SUCLG1, and IDH3A; ↑ MDH2 |
| Gao et al. [104] | Cell lines | L + M + T | LNCaP vs. SCNC | LNCaP: ↑ PHGDH, PSAT1, PSPH, TDH, GCAT, citrate, isocitrate, and succinate; ↓ fumarate, glutamate, glutamine, IDH1, GLUD1, GLUD2, carnitine, and short-chain acylcarnitines SCNC: ↑ lactate and LDH; ↓ G6P |
| Joshi et al. [105] | Cell lines | M + T | CPT1A KD vs. CPT1A OE | CPT1A OE: ↑ PHGDH, PSAT1, SHMT2, CTH, GSTO2, dimethylglycine, cystathionine, cystathionine, and cysteine; ↓ glycolysis |
| Chen et al. [106] | Cell lines | M + T | ARCaPE vs. ARCaPM | ARCaPM: ↑malate, ACO2, SDHA, aspartate, ASS1, and SRR; ↓ glycolysis, succinate, and citrate |
| Hansen et al. [107] | Tissue | L + M | ERGlow vs. ERGhigh | ERGhigh: ↑ ethanolamine, glycine, phosphocholine, phosphoethanolamine, ACACA, FASN, and SAT1; ↓ ACO2, citrate, spermine, putrescine, and glucose |
| Yan et al. [108] | Tissue | L + M + T | SPOP wt vs. SPOP-mutant | SPOP-mutant:↑ ACADL, ELOVL2, FH, fatty acids, fumarate, and malate |
| Andersen et al. [109] | Tissue | M + T | Low vs. high reactive stroma | High reactive stroma: ↑ taurine and leucine; ↓ citrate, spermine, and scyllo-inositol |
| Oberhuber et al. [110] | Tissue | M + P + T | STAT3low vs. STAT3high | STAT3low: ↑ OXPHOS, TCA cycle, ribosomal activity, pyruvate, fumarate, and malate; ↓ PDK4 |
ACACA: acetyl-CoA carboxylase alpha, ACADL: acyl-CoA dehydrogenase, long chain, ACO2: aconitase, APO-AIV: apolipoprotein A1V, ARCaP: androgen-repressed prostate cancer cell, ASS1: arginosuccinate synthase 1, BCR: biochemical recurrence, CPT1A: carnitine palmitoyl transferase I, CRPC: castrate-resistant prostate cancer, CTH: cystathionine gamma-lyase, CYP1A1: cytochrome P450 family 1 subfamily A member 1, E: epigenomics, ELOVL2: ELOVL fatty acid elongase 2, ERG: ETS transcription factor ERG, FASN: fatty-acid synthase, FH: fumarate hydratase, G: genomics, GSTO2: glutathione S-transferase omega 2, GCAT: glycine C-acetyltransferase, GLUD1: glutamate dehydrogenase 1, GLUD2: glutamate dehydrogenase 2, G6P: glucose-6-phosphate, IDH1: isocitrate dehydrogenase (NADP(+)) 1, IDH3A: isocitrate dehydrogenase (NAD(+)) 3 catalytic subunit alpha, KD: knockdown, L: lipidomics, LDH: lactate dehydrogenase, LNCaP: lymph node carcinoma of the prostate, M: metabolomics, OE: overexpressed, MDH2: malate dehydrogenase 2, OGDH: oxoglutarate dehydrogenase, OXPHOS: oxidative phosphorylation, P: proteomics, PCa: prostate cancer, PDK4: pyruvate dehydrogenase kinase 4, PHGDH: d-3-phosphoglycerate dehydrogenase, PNP: purine nucleoside phosphorylase, PSAT1: phosphohydroxythreonine aminotransferase, PSPH: phosphoserine phosphatase, SAT1: spermidine N(1)-acetyltransferase, SCNC: small-cell neuroendocrine carcinoma, SDHA: succinate dehydrogenase complex flavoprotein subunit A, SHMT2: serine hydroxymethyltransferase, SMS: spermine synthase, SPOP: Speckle-type POZ protein, SRR: serine racemase, STAT3: signal transducer and activator of transcription 3, SUCLG1: succinate-CoA ligase alpha subunit, T: transcriptomics, TCA: tricarboxylic acid, TDH: threonine dehydrogenase, TNC: tenascin C.
Several studies have revealed specific metabolic alterations in PCa patients by comparing prostate tumors of different grade. Furthermore, different systemic and local metabolic alterations have consistently been associated with PCa risk and progression [111,112,113,114,115,116]. In line with this, in a recent study by Gómez-Cebrián et al., the specific metabolomics profile of high-grade PCa patients was characterized on the basis of the alterations in metabolite levels identified in the serum and urine of PCa patients with different tumor grades [100]. A gene set enrichment analysis (GSEA) of three publicly available Pca transcriptomics datasets facilitated a targeted analysis of the metabolomics profiles, with a focus on metabolites involved in potentially altered metabolic pathways in high-grade Pca patients. Statistically significant alterations in the levels of glucose, glycine, and 1-methylnicotinamide were found in high-grade PCa patients. Interestingly, dysregulations in the levels of these metabolites could be associated with different metabolic changes previously observed in PCa patients [35,99,117,118,119]. Particularly, in other multi-omics studies based on the analysis of tissue samples, glycine levels were found to be higher in PCa tumors enriched in the TMPRSS2–ERG gene fusion set [107], and nicotinamide metabolism was elevated in PCa tissues when compared with benign tissues [61]. In addition, Kiebish et al. recently investigated the metabolic profile of presurgical serum samples of PCa patients with a focus on selecting serum metabolic biomarkers that could be valuable for predicting biochemical recurrence (BCR) [101]. In this study, the integration of proteomics, metabolomics, and lipidomics data from PCa patients facilitated the identification of four analytes (tenascin C (TNC), apolipoprotein A-IV (APOA-IV), 1-methyladenosine, and phosphatidic acid 18:0–22:0) as potential biomarkers to discriminate BCR from non-BCR patients. Of note, TNC expression levels in PCa tumor tissues and stroma have previously been reported to predict poor prognosis in PCa patients [120,121,122], and different serum studies have described apolipoproteins as a potential biomarker for PCa [123,124]. The authors evaluated the association between the levels of each individual biomarker and survival, and they found that higher levels of serum TNC, APO-AIV and 1-methyladenosine and lower concentration of phosphatidic acid increased the probability of disease progression. The predictive potential of these markers was further validated in a testing cohort of patients. Overall, the combination of the four biomolecules resulted in a model with a predictive performance for differentiating PCa patients with and without BCR characterized by an AUC of 0.78, a value that increased to 0.89 after adding the pathological T stage and the GS to the model.
Furthermore, other multi-omics studies have focused on the analysis of local metabolic changes, as reflected in the metabolic profile of PCa tissues and cell lines. In a multi-omics study conducted by Liu et al., the authors developed an approach to improve the accuracy of PCa classification and risk evaluation [102]. According to the combined analysis of genomics and metabolomics data from benign prostate samples, as well as localized and metastatic PCa samples, the authors generated classifier models that proved to be informative for Pca prognosis in additional datasets. Following this approach, they found that arginine and proline metabolism, purine metabolism, and steroid hormone biosynthesis were relevant metabolic pathways for the discrimination between localized and metastatic PCa. Next, topologically important genes and metabolites involved in these pathways were selected as promising markers for PCa prognosis. Selected genes and metabolites included cytochrome P450 family 1 subfamily A member 1 (CYP1A1), purine nucleoside phosphorylase (PNP), spermine synthase (SMS), proline, cholesterol, sarcosine, spermidine, and spermine. Interestingly, elevated PNP expression has been observed in aggressive PCa cells [125], whereas alterations in the levels of some of the topologically relevant metabolites have been associated with PCa progression and aggressiveness, including sarcosine [126,127], proline [99], and spermine [41,128]. Moreover, the classification method achieved a more accurate overall performance compared to other existing classification methods across additional datasets.
Efforts to discover dysregulated metabolic pathways in metastatic stages were also made in another multi-omics study conducted by Li et al. In this study, the authors proposed an analytical method, referred to as Subpathway-GM, aiming to identify biologically meaningful metabolic subpathways based on the combined analysis of metabolomics and transcriptomics data [103]. This method allowed the identification of disease-relevant subpathways that could go undetected on the basis of classical entire pathway identification methods. After applying this method to the analysis of a PCa dataset including data obtained from localized and metastatic tumors, 16 subpathways were identified as relevant in metastatic PCa. Among these metabolic routes, nine of them were involved in amino-acid metabolism, including glycine, serine, and threonine metabolism, tryptophan metabolism, cysteine, and methionine metabolism, and histidine metabolism. Interestingly, both tryptophan and histidine metabolism were not previously reported to be associated with metastatic PCa. Specifically, in the histidine metabolism pathway, the histamine region was accurately identified as a disease-relevant subpathway. On the basis of this information, the authors explored the effect of different histamine concentrations on PCa cell proliferation and migration. The results showed that high histamine concentrations inhibited cell migration in a dose-dependent manner, confirming that this metabolite could be associated with metastatic PCa. This finding is supplementary to other results included in previous studies where histamine altered the response to radiation in PCa tumors and significantly reduced proliferation of tumor cells compared with irradiation alone [129,130].
Other multi-omics studies have focused on the analysis of the metabolic profile associated with CRPC. Among them, the study by Latonen et al. was aimed at characterizing the distinct protein profiles of BPH, PCa, and CRPC patients [77]. To that end, the authors performed an integrated analysis of four different omics data. Following this experimental approach, it was found that gene copy number, DNA methylation, and RNA expression levels did not reliably predict proteomics changes in CRPC. These results suggested that proteomics data could be associated with alterations not detectable at the transcriptomic level. In fact, proteomics analyses revealed specific pathway alterations that were not previously reported in CRPC. Interestingly, no significant alterations were observed in the regulation of androgen receptor signaling at the mRNA or protein levels. The combined analysis by transcriptomics and proteomics identified alterations in different cell-cycle regulatory pathways, whereas changes in DNA repair pathways were only detected by proteomics. The combined analysis of the omics data also revealed a previously undescribed two-step modulation of the TCA cycle associated with metabolic changes occurring during PCa development and progression. This pathway exhibited two different metabolic shifts: a first one defined by the upregulation of most of TCA enzymes during initial PCa stages, and a second metabolic shift during PCa progression, involving the downregulation of ACO2, oxoglutarate dehydrogenase (OGDH), and succinate-CoA ligase alpha subunit (SUCLG1), together with elevated expression of malate dehydrogenase 2 (MDH2). Previous studies have already reported that PCa patients with MDH2 overexpression have a significantly shorter period of relapse-free survival, and that stable knockdown of MDH2 PCa cell lines decreased cell proliferation and increased docetaxel sensitivity, all suggesting that MDH2 inhibition could be a viable strategy to target CRPC [131].
Additionally, other multi-omics studies have focused on characterizing metabolic dysregulations associated with specific PCa subtypes. In this context, Gao et al. integrated transcriptomics and metabolomics data to characterize the metabolic profile of two main types of PCa, adenocarcinoma (LNCaP), and small-cell neuroendocrine carcinoma (SCNC) [104]. By conducting an individual GSEA on SCNC and adenocarcinoma cell lines, a total of 62 and 112 genes, respectively, were found to be upregulated in each subgroup. Metabolomics and lipidomics analyses also revealed significant differences in 25 metabolite clusters. In particular, the LNCaP phenotype was characterized by an increased serine biosynthesis, a finding supported by elevated levels of serine, glycine, and threonine concentrations and higher expression of phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1), phosphoserine phosphatase (PSPH), threonine dehydrogenase (TDH), and glycine C-acetyltransferase (GCAT). This cell line also exhibited increased levels of citrate, isocitrate, and succinate, together with higher expression of many enzymes involved in the TCA, as well as decreased levels of fumarate, glutamate, and glutamine and lower expression of isocitrate dehydrogenase (NADP(+)) 1 (IDH1), GLUD1, and GLUD2, an indication of a citrate accumulation phenotype. Furthermore, an enhanced alpha-linoleic acid, arachidonic acid, linoleic acid, fatty-acid, and sphingolipid metabolism was also observed in the LNCaP group, along with a reduced fatty-acid oxidation activity, suggested by the lower levels of carnitine and some short-chain acylcarnitines and the overexpression of genes involved in biosynthesis, as well as the use of acylcarnitines and members of the acyl-coenzyme A synthetase family. On the other hand, SCNC was characterized by an enhanced glycerolipid, glycerophospholipid, and ether lipid metabolism, as well as by an elevated pyruvate metabolism, which was supported by lower levels of glucose-6-phosphate and higher lactate concentrations together with increased expression of lactate dehydrogenase isoforms (LDHA and LDHB). Although a limited number of samples were included in this pilot study, the results highlight the potential of multi-omics approaches for the identification of novel therapeutic targets in specific subgroups of PCa. Furthermore, the integrated analysis of transcriptomics and metabolomics data carried out by Joshi et al. revealed an enhanced lipid catabolism in the carnitine palmitoyl transferase I (CPT1A) overexpressed (OE) phenotype, which was also associated with the elevated concentration of acyl-carnitine and higher lipase activity [105]. In this study, the analysis of molecular differences between CPT1A gain- and loss-of-function cellular models revealed genetic and metabolomics vulnerabilities associated with the progression to neuroendocrine differentiation in PCa. Cellular models overexpressing CPT1A were characterized by enhanced lipid metabolism, glycine and serine metabolism, and glutathione homeostasis. In addition, the OE phenotype exhibited lower glycolysis as glucose was preferentially shunted toward de novo serine biosynthesis. This finding was correlated with the increased expression of key serine/glycine pathway genes, including PHGDH, PSAT1, and serine hydroxymethyltransferase (SHMT2), together with elevated levels of some metabolites involved in the folate cycle (e.g., dimethylglycine and cystathionine). Furthermore, although cells overexpressing CPT1A showed increased levels of mitochondrial reactive oxygen species (ROS), elevated concentrations of metabolites involved in glutathione homeostasis, including overexpression of cystathionine gamma-lyase (CTH) and glutathione S-transferase omega 2 (GSTO2), were also found, indicating a key role of CPT1A in supporting adaptation to stress and antioxidant defense production. Lastly, the analysis of data derived from patients, available from public databases, provided evidence that lipid catabolism driven by CPT1A was associated with more aggressive disease, suggesting that CPT1A activity could rewire metabolism to promote growth and transformation in these patients.
Other multi-omics studies have focused on characterizing the metabolic features of PCa cells undergoing epithelial–mesenchymal transition (EMT). In the study carried out by Chen et al., two subclones derived from the androgen-repressed prostate cancer cell (ARCaP) line that exhibited epithelial and mesenchymal phenotypes, ARCaPE and ARCaPM, respectively, were used as EMT PCa models [106]. Integration of transcriptomics and metabolomics data revealed lower levels of glycolysis intermediates and decreased expression of several glucose metabolism-related genes in ARCAPM, indicating a downregulation of glucose metabolism. In addition, this phenotype was characterized by exhibiting higher malate levels, as well as by overexpressing ACO2 and succinate dehydrogenase complex flavoprotein subunit A (SDHA) enzymes. At the same time, authors found lower succinate and citrate levels, suggesting that TCA might be fueled by glutamine and aspartate in addition to glucose in these cells. Notably, upregulation of ACO2 has been identified as an important event in prostate carcinogenesis [23], whereas lower citrate levels have been observed in PCa when compared to non-cancer epithelium [29,132]. Furthermore, malate has been associated with Gleason progression [99] and found to be altered between different PCa stages [133]. Additionally, increased aspartate and aspartate-derived metabolite levels and upregulation of important enzymes involved in aspartate metabolism, including arginosuccinate synthase 1 (ASS1) and serine racemase (SRR), were observed in ARCaPM cells, suggesting an enhanced aspartate metabolism.
A combination of metabolomics and transcriptomics data was also used by Hansen et al. to identify changes in PCa metabolism related to the TMPRSS2-ERG gene fusion [107]. In this study, PCa patients were classified in two cohorts, ERGlow or ERGhigh, as a function of specific enrichment of the ERG fusion gene set [134,135]. Multivariate analysis of metabolomic data revealed decreased concentrations of citrate, spermine, putrescine, and glucose, and higher levels of ethanolamine, glycine, phosphocholine, and phosphoethanolamine in ERGhigh PCa patients included in two independent patient cohorts. Furthermore, a targeted analysis of genes involved in the metabolic pathways associated with these metabolic changes revealed an upregulation of genes involved in the polyamine pathway, together with a decrease in relevant genes in the TCA cycle and increased lipogenic phenotype. In particular, N(1)-acetyltransferase (SAT1), involved in spermine depletion, was highly expressed in ERGhigh tumors. In addition, this group of patients also exhibited decreased expression of ACO2 and elevated activity of the lipogenic enzymes ACACA and FASN, indicating that citrate might be preferentially derived from de novo lipid synthesis in these tumors. Several studies have also reported increased expression of FASN [67,68,136] and enhanced de novo fatty-acid synthesis in PCa [137] and PCa invasiveness [138]. Interestingly, in a different multi-omics study performed by Yan et al., the integration of data from three different omics platforms was used to analyze correlations between Speckle-type POZ protein (SPOP) mutations and changes in PCa metabolism [108]. SPOP, a cullin-based E3 ubiquitin ligase, has been identified as one of the most frequently mutated genes in PCa [139]. Several studies have shown that SPOP could directly bind to androgen receptor and contribute to its ubiquitination and degradation [140]. Interestingly, the authors found a strong upregulation of acyl-CoA dehydrogenase, long chain (ACADL), and ELOVL fatty-acid elongase 2 (ELOVL2) together with an increase in the levels of most fatty acids in SPOP-mutated patients. Relevant upregulations were also observed in the levels of two key intermediates of the TCA cycle (malate and fumarate) and fumarate hydratase (FH). Although FH, ELOVL2, and ACADL were identified as key genes in SPOP-mutated PCa patients in this study, their oncogenic role in PCa still needs to be proven.
Multi-omics studies have also focused on exploring specific metabolic alterations associated with PCa progression. Andersen et al. focused on identifying correlations between changes in genes and metabolites and high reactive stroma content in tumors [109], as it has been linked to worse clinical outcome and earlier BCR in Pca [141,142,143,144,145]. High reactive stroma samples were characterized by elevated levels of taurine and leucine, as well as by decreased levels of citrate, spermine, and scyllo-inositol. Interestingly, metastatic CRPC has previously been defined as leucine-dependent [146,147], and leucine deprivation has been shown to inhibit PCa growth [148]. The metabolic changes observed in high reactive stroma samples, together with the results from a gene enrichment analysis, indicated that immune processes and extracellular matrix remodeling were particularly important in these tumors. In a more recent study, Oberhuber et al. evaluated the correlation between the PCa transcriptomics and proteomics profiles with signal transducer and activator of transcription 3 (STAT3) expression looking for biomarkers associated with earlier BCR [110]. The integrative multi-omics analysis revealed enhanced oxidative phosphorylation (OXPHOS), TCA cycle, and ribosomal activity in the STAT3low group of tumors. These findings were also observed in a PCa murine model, which showed enrichment of ribosomal gene sets and elevated TCA cycle and OXPHOS, as well as elevated pyruvate, fumarate, and malate levels in xenografts with loss of STAT3. The authors also observed that pyruvate dehydrogenase kinase 4 (PDK4) was significantly downregulated in STAT3low samples. Expression of PDK4 has already been reported to be significantly altered when comparing PCa patients with healthy individuals [149]. The analysis of the correlation between PDK4 expression and BCR in primary and metastatic tumors demonstrated its ability to predict disease recurrence independently of diagnostic risk factors, such as grading, staging, and PSA levels, thus suggesting its potential as a promising independent prognostic biomarker for distinguishing between a good and bad prognostic PCa.
3. Future Perspectives and Conclusions
Altered cell metabolism is a well-established hallmark of cancer [150]. Metabolism is dysregulated to support the metabolic requirements of uncontrolled proliferation in cancer cells [151,152]. This rewiring of cellular metabolism leads to characteristic metabolic phenotypes that can be used for the development of effective screening methods for early cancer detection, patient selection strategies, or evaluation of treatment responses [153,154]. Altered metabolism also results in unique metabolic vulnerabilities that can be exploited to develop novel therapeutic strategies in cancer, some of which are being evaluated in preclinical models or clinical trials [17,155,156,157]. Recently, the availability and advances in the development of different analytical platforms have prompted the application of new omics approaches for the characterization of specific cancer-associated metabolic phenotypes. Particularly, metabolomics approaches have greatly contributed to metabolically characterize the profile of PCa patients and to discover specific alterations associated with this disease [22,158,159]. However, compared with other omics (e.g., genomics, transcriptomics), the metabolome coverage is limited, thus adding difficulty to the final interpretation of the results [160]. In this context, the integration of different omics datasets could represent a powerful strategy to develop more robust and consistent metabolic signatures with a clinical impact on the management of cancer patients [161].
In this review, the most relevant findings reported in multi-omics studies focused on the characterization of the metabolic phenotype associated with PCa were summarized. Overall, the most frequently reported metabolic alterations associated with PCa onset and progression include differences in the TCA cycle, polyamine synthesis, HBP, and nucleotide and lipid metabolism, and the most widely applied multi-omics approach was the combination of transcriptomics and metabolomics data. In most of the reviewed studies, the different omics datasets were separately analyzed and only combined for the final interpretation of the metabolic changes. In this scenario, the development and implementation of novel computational tools, focused on the integrated analysis of different omics datasets that enable the assessment of the interplay between the different components of a biological system, would be greatly valuable [162,163]. Furthermore, although some studies included a vast number of samples [61,63,101,107,109], a major limitation in the majority of these studies was the lack of an external cohort of PCa patients/samples for confirming the reproducibility and robustness of the results. Thus, future studies including larger sample sizes and external datasets to increase the statistical power of the analyses and validate the findings of selected metabolites, together with confirmatory experiments to evaluate the clinical significance of these metabolic findings, are required. Lastly, access to publicly accessible databases integrating all metabolic alterations reported in the literature, associated with each tumor subtype, would greatly contribute to our understanding of the metabolic heterogeneity in PCa [164,165].
Author Contributions
Conceptualization, N.G.-C., L.P.-C. and A.P.-L.; writing—original draft preparation, N.G.-C. with input from L.P.-C.; writing—review and editing, J.L.P., L.P.-C. and A.P.-L. All authors have read and agreed to the published version of the manuscript.
Funding
N.G.-C. and L.P.-C. are supported by the European Regional Development Fund (FEDER) and Conselleria de Innovación, Universidades, Ciencia y Sociedad Digital (GV/2021/154). N.G.-C., L.P.-C., and A.P.-L. are supported by MCIN (PID2020-115875RB-I00/AEI/10.13039/501100011033).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Sathianathen N.J., Konety B.R., Crook J., Saad F., Lawrentschuk N. Landmarks in Prostate Cancer. Nat. Rev. Urol. 2018;15:627–642. doi: 10.1038/s41585-018-0060-7. [DOI] [PubMed] [Google Scholar]
- 3.Schalken J., Dijkstra S., Baskin-Bey E., van Oort I. Potential Utility of Cancer-Specific Biomarkers for Assessing Response to Hormonal Treatments in Metastatic Prostate Cancer. Ther. Adv. Urol. 2014;6:245–252. doi: 10.1177/1756287214545328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etzioni R., Penson D.F., Legler J.M., di Tommaso D., Boer R., Gann P.H., Feuer E.J. Overdiagnosis Due to Prostate-Specific Antigen Screening: Lessons from U.S. Prostate Cancer Incidence Trends. J. Natl. Cancer Inst. 2002;94:981–990. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- 5.Postma R., Schröder F.H. Screening for Prostate Cancer. Eur. J. Cancer. 2005;41:825–833. doi: 10.1016/j.ejca.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 6.Ziglioli F., Granelli G., Cavalieri D., Bocchialini T., Maestroni U. What Chance Do We Have to Decrease Prostate Cancer Overdiagnosis and Overtreatment? A Narrative Review. Acta Biomed. 2019;90:423–426. doi: 10.23750/abm.v90i4.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleason D.F., Mellinger G.T. Prediction of Prognosis for Prostatic Adenocarcinoma by Combined Histological Grading and Clinical Staging. J. Urol. 1974;111:58–64. doi: 10.1016/S0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 8.Hübner N., Shariat S., Remzi M. Prostate Biopsy: Guidelines and Evidence. Curr. Opin. Urol. 2018;28:354–359. doi: 10.1097/MOU.0000000000000510. [DOI] [PubMed] [Google Scholar]
- 9.Mottet N., van den Bergh R.C.N., Briers E., Van den Broeck T., Cumberbatch M.G., De Santis M., Fanti S., Fossati N., Gandaglia G., Gillessen S., et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021;79:243–262. doi: 10.1016/j.eururo.2020.09.042. [DOI] [PubMed] [Google Scholar]
- 10.Kawachi M.H., Bahnson R.R., Barry M., Busby J.E., Carroll P.R., Carter H.B., Catalona W.J., Cookson M.S., Epstein J.I., Etzioni R.B., et al. NCCN Clinical Practice Guidelines in Oncology: Prostate Cancer Early Detection. J. Natl. Compr. Cancer Netw. 2010;8:240–262. doi: 10.6004/jnccn.2010.0016. [DOI] [PubMed] [Google Scholar]
- 11.Das C.J., Razik A., Sharma S., Verma S. Prostate Biopsy: When and How to Perform. Clin. Radiol. 2019;74:853–864. doi: 10.1016/j.crad.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Miles B., Ittmann M., Wheeler T., Sayeeduddin M., Cubilla A., Rowley D., Bu P., Ding Y., Gao Y., Lee M., et al. Moving Beyond Gleason Scoring. Arch. Pathol. Lab. Med. 2019;143:565–570. doi: 10.5858/arpa.2018-0242-RA. [DOI] [PubMed] [Google Scholar]
- 13.Zadra G., Loda M. Metabolic Vulnerabilities of Prostate Cancer: Diagnostic and Therapeutic Opportunities. Cold Spring Harb. Perspect. Med. 2018;8:a030569. doi: 10.1101/cshperspect.a030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turanli B., Zhang C., Kim W., Benfeitas R., Uhlen M., Arga K.Y., Mardinoglu A. Discovery of Therapeutic Agents for Prostate Cancer Using Genome-Scale Metabolic Modeling and Drug Repositioning. EBioMedicine. 2019;42:386–396. doi: 10.1016/j.ebiom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Márquez J., Matés J.M. Tumor Metabolome: Therapeutic Opportunities Targeting Cancer Metabolic Reprogramming. Cancers. 2021;13:314. doi: 10.3390/cancers13020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pemovska T., Bigenzahn J.W., Srndic I., Lercher A., Bergthaler A., César-Razquin A., Kartnig F., Kornauth C., Valent P., Staber P.B., et al. Metabolic Drug Survey Highlights Cancer Cell Dependencies and Vulnerabilities. Nat. Commun. 2021;12:7190. doi: 10.1038/s41467-021-27329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stine Z.E., Schug Z.T., Salvino J.M., Dang C.V. Targeting Cancer Metabolism in the Era of Precision Oncology. Nat. Rev. Drug Discov. 2021;3:2–22. doi: 10.1038/s41573-021-00339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Reyes I., Chandel N.S. Cancer Metabolism: Looking Forward. Nat. Rev. Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 19.Lima A.R., de Lourdes Bastos M., Carvalho M., de Pinho P.G. Biomarker Discovery in Human Prostate Cancer: An Update in Metabolomics Studies. Transl. Oncol. 2016;9:357–370. doi: 10.1016/j.tranon.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Cebrián N., Rojas-Benedicto A., Albors-Vaquer A., López-Guerrero J.A., Pineda-Lucena A., Puchades-Carrasco L. Metabolomics Contributions to the Discovery of Prostate Cancer Biomarkers. Metabolites. 2019;9:48. doi: 10.3390/metabo9030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kdadra M., Höckner S., Leung H., Kremer W., Schiffer E. Metabolomics Biomarkers of Prostate Cancer: A Systematic Review. Diagnostics. 2019;9:21. doi: 10.3390/diagnostics9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima A.R., Pinto J., Amaro F., Bastos M.D., Carvalho M., Guedes de Pinho P. Advances and Perspectives in Prostate Cancer Biomarker Discovery in the Last 5 Years through Tissue and Urine Metabolomics. Metabolites. 2021;11:181. doi: 10.3390/metabo11030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eidelman E., Twum-Ampofo J., Ansari J., Siddiqui M.M. The Metabolic Phenotype of Prostate Cancer. Front. Oncol. 2017;7:131. doi: 10.3389/fonc.2017.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad F., Cherukuri M.K., Choyke P.L. Metabolic Reprogramming in Prostate Cancer. Br. J. Cancer. 2021;125:1185–1196. doi: 10.1038/s41416-021-01435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lima A.R., Araújo A.M., Pinto J., Jerónimo C., Henrique R., de Lourdes Bastos M., Carvalho M., de Pinho P.G. Discrimination between the Human Prostate Normal and Cancer Cell Exometabolome by GC-MS. Sci. Rep. 2018;8:5539. doi: 10.1038/s41598-018-23847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng H., Dong B., Ning J., Shao X., Zhao L., Jiang Q., Ji H., Cai A., Xue W., Gao H. NMR-Based Metabolomics Analysis Identifies Discriminatory Metabolic Disturbances in Tissue and Biofluid Samples for Progressive Prostate Cancer. Clin. Chim. Acta. 2020;501:241–251. doi: 10.1016/j.cca.2019.10.046. [DOI] [PubMed] [Google Scholar]
- 27.Dudka I., Thysell E., Lundquist K., Antti H., Iglesias-Gato D., Flores-Morales A., Bergh A., Wikström P., Gröbner G. Comprehensive Metabolomics Analysis of Prostate Cancer Tissue in Relation to Tumor Aggressiveness and TMPRSS2-ERG Fusion Status. BMC Cancer. 2020;20:437. doi: 10.1186/s12885-020-06908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franko A., Shao Y., Heni M., Hennenlotter J., Hoene M., Hu C., Liu X., Zhao X., Wang Q., Birkenfeld A.L., et al. Human Prostate Cancer Is Characterized by an Increase in Urea Cycle Metabolites. Cancers. 2020;12:1814. doi: 10.3390/cancers12071814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giskeødegård G.F., Bertilsson H., Selnæs K.M., Wright A.J., Bathen T.F., Viset T., Halgunset J., Angelsen A., Gribbestad I.S., Tessem M.-B. Spermine and Citrate as Metabolic Biomarkers for Assessing Prostate Cancer Aggressiveness. PLoS ONE. 2013;8:e62375. doi: 10.1371/journal.pone.0062375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serkova N.J., Gamito E.J., Jones R.H., O’Donnell C., Brown J.L., Green S., Sullivan H., Hedlund T., Crawford E.D. The Metabolites Citrate, Myo-Inositol, and Spermine Are Potential Age-Independent Markers of Prostate Cancer in Human Expressed Prostatic Secretions. Prostate. 2008;68:620–628. doi: 10.1002/pros.20727. [DOI] [PubMed] [Google Scholar]
- 31.Lynch M.J., Nicholson J.K. Proton MRS of Human Prostatic Fluid: Correlations between Citrate, Spermine, and Myo-Inositol Levels and Changes with Disease. Prostate. 1997;30:248–255. doi: 10.1002/(SICI)1097-0045(19970301)30:4<248::AID-PROS4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 32.Cheng L.L., Wu C., Smith M.R., Gonzalez R.G. Non-Destructive Quantitation of Spermine in Human Prostate Tissue Samples Using HRMAS 1H NMR Spectroscopy at 9.4 T. FEBS Lett. 2001;494:112–116. doi: 10.1016/S0014-5793(01)02329-8. [DOI] [PubMed] [Google Scholar]
- 33.Tsoi T.-H., Chan C.-F., Chan W.-L., Chiu K.-F., Wong W.-T., Ng C.-F., Wong K.-L. Urinary Polyamines: A Pilot Study on Their Roles as Prostate Cancer Detection Biomarkers. PLoS ONE. 2016;11:e0162217. doi: 10.1371/journal.pone.0162217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar D., Gupta A., Mandhani A., Sankhwar S.N. Metabolomics-Derived Prostate Cancer Biomarkers: Fact or Fiction? J. Proteome Res. 2015;14:1455–1464. doi: 10.1021/pr5011108. [DOI] [PubMed] [Google Scholar]
- 35.Kumar D., Gupta A., Mandhani A., Sankhwar S.N. NMR Spectroscopy of Filtered Serum of Prostate Cancer: A New Frontier in Metabolomics. Prostate. 2016;76:1106–1119. doi: 10.1002/pros.23198. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Rambla C., Puchades-Carrasco L., García-Flores M., Rubio-Briones J., López-Guerrero J.A., Pineda-Lucena A. Non-Invasive Urinary Metabolomic Profiling Discriminates Prostate Cancer from Benign Prostatic Hyperplasia. Metabolomics. 2017;13:52. doi: 10.1007/s11306-017-1194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giskeødegård G.F., Hansen A.F., Bertilsson H., Gonzalez S.V., Kristiansen K.A., Bruheim P., Mjøs S.A., Angelsen A., Bathen T.F., Tessem M.-B. Metabolic Markers in Blood Can Separate Prostate Cancer from Benign Prostatic Hyperplasia. Br. J. Cancer. 2015;113:1712–1719. doi: 10.1038/bjc.2015.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn P., Smith I.C., Leboldus L., Littman C., Somorjai R.L., Bezabeh T. The Classification of Benign and Malignant Human Prostate Tissue by Multivariate Analysis of 1H Magnetic Resonance Spectra. Cancer Res. 1997;57:3398–3401. [PubMed] [Google Scholar]
- 39.Dereziński P., Klupczynska A., Sawicki W., Pałka J.A., Kokot Z.J. Amino Acid Profiles of Serum and Urine in Search for Prostate Cancer Biomarkers: A Pilot Study. Int. J. Med. Sci. 2017;14:1–12. doi: 10.7150/ijms.15783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madhu B., Shaw G.L., Warren A.Y., Neal D.E., Griffiths J.R. Response of Degarelix Treatment in Human Prostate Cancer Monitored by HR-MAS 1H NMR Spectroscopy. Metabolomics. 2016;12:120. doi: 10.1007/s11306-016-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braadland P.R., Giskeødegård G., Sandsmark E., Bertilsson H., Euceda L.R., Hansen A.F., Guldvik I.J., Selnæs K.M., Grytli H.H., Katz B., et al. Ex Vivo Metabolic Fingerprinting Identifies Biomarkers Predictive of Prostate Cancer Recurrence Following Radical Prostatectomy. Br. J. Cancer. 2017;117:1656–1664. doi: 10.1038/bjc.2017.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kühn T., Floegel A., Sookthai D., Johnson T., Rolle-Kampczyk U., Otto W., von Bergen M., Boeing H., Kaaks R. Higher Plasma Levels of Lysophosphatidylcholine 18:0 Are Related to a Lower Risk of Common Cancers in a Prospective Metabolomics Study. BMC Med. 2016;14:13. doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts M.J., Richards R.S., Chow C.W.K., Buck M., Yaxley J., Lavin M.F., Schirra H.J., Gardiner R.A. Seminal Plasma Enables Selection and Monitoring of Active Surveillance Candidates Using Nuclear Magnetic Resonance-Based Metabolomics: A Preliminary Investigation. Prostate Int. 2017;5:149–157. doi: 10.1016/j.prnil.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin H.-M., Mahon K.L., Weir J.M., Mundra P.A., Spielman C., Briscoe K., Gurney H., Mallesara G., Marx G., Stockler M.R., et al. A Distinct Plasma Lipid Signature Associated with Poor Prognosis in Castration-Resistant Prostate Cancer. Int. J. Cancer. 2017;141:2112–2120. doi: 10.1002/ijc.30903. [DOI] [PubMed] [Google Scholar]
- 45.Marzec J., Ross-Adams H., Pirrò S., Wang J., Zhu Y., Mao X., Gadaleta E., Ahmad A.S., North B.V., Kammerer-Jacquet S.-F., et al. The Transcriptomic Landscape of Prostate Cancer Development and Progression: An Integrative Analysis. Cancers. 2021;13:345. doi: 10.3390/cancers13020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh R., Mills I.G. The Interplay Between Prostate Cancer Genomics, Metabolism, and the Epigenome: Perspectives and Future Prospects. Front. Oncol. 2021;11:704353. doi: 10.3389/fonc.2021.704353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Zhang R., Liang F., Zhang L., Liang X. Identification of Metabolism-Associated Prostate Cancer Subtypes and Construction of a Prognostic Risk Model. Front. Oncol. 2020;10:598801. doi: 10.3389/fonc.2020.598801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ros S., Santos C.R., Moco S., Baenke F., Kelly G., Howell M., Zamboni N., Schulze A. Functional Metabolic Screen Identifies 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 4 as an Important Regulator of Prostate Cancer Cell Survival. Cancer Discov. 2012;2:328–343. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- 49.Saraon P., Cretu D., Musrap N., Karagiannis G.S., Batruch I., Drabovich A.P., van der Kwast T., Mizokami A., Morrissey C., Jarvi K., et al. Quantitative Proteomics Reveals That Enzymes of the Ketogenic Pathway Are Associated with Prostate Cancer Progression. Mol. Cell. Proteom. 2013;12:1589–1601. doi: 10.1074/mcp.M112.023887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duijvesz D., Burnum-Johnson K.E., Gritsenko M.A., Hoogland A.M., Vredenbregt-van den Berg M.S., Willemsen R., Luider T., Paša-Tolić L., Jenster G. Proteomic Profiling of Exosomes Leads to the Identification of Novel Biomarkers for Prostate Cancer. PLoS ONE. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang K., Han X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016;41:954–969. doi: 10.1016/j.tibs.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Zhu Y., Jijiwa M., Nasu M., Ai J., Dai S., Jiang B., Zhang J., Huang G., Deng Y. Identification of Plasma Lipid Species as Promising Diagnostic Markers for Prostate Cancer. BMC Med. Inform. Decis. Mak. 2020;20:223. doi: 10.1186/s12911-020-01242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buszewska-Forajta M., Pomastowski P., Monedeiro F., Walczak-Skierska J., Markuszewski M., Matuszewski M., Markuszewski M.J., Buszewski B. Lipidomics as a Diagnostic Tool for Prostate Cancer. Cancers. 2021;13:2000. doi: 10.3390/cancers13092000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasin Y., Seldin M., Lusis A. Multi-Omics Approaches to Disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hakimi A.A., Reznik E., Lee C.-H., Creighton C.J., Brannon A.R., Luna A., Aksoy B.A., Liu E.M., Shen R., Lee W., et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo X., Yu H., Song Y., Sun T. Integration of Metabolomic and Transcriptomic Data Reveals Metabolic Pathway Alteration in Breast Cancer and Impact of Related Signature on Survival. J. Cell. Physiol. 2019;234:13021–13031. doi: 10.1002/jcp.27973. [DOI] [PubMed] [Google Scholar]
- 57.Hoang L.T., Domingo-Sabugo C., Starren E.S., Willis-Owen S.A.G., Morris-Rosendahl D.J., Nicholson A.G., Cookson W.O.C.M., Moffatt M.F. Metabolomic, Transcriptomic and Genetic Integrative Analysis Reveals Important Roles of Adenosine Diphosphate in Haemostasis and Platelet Activation in Non-Small-Cell Lung Cancer. Mol. Oncol. 2019;13:2406–2421. doi: 10.1002/1878-0261.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meller S., Meyer H.-A., Bethan B., Dietrich D., Maldonado S.G., Lein M., Montani M., Reszka R., Schatz P., Peter E., et al. Integration of Tissue Metabolomics, Transcriptomics and Immunohistochemistry Reveals ERG- and Gleason Score-Specific Metabolomic Alterations in Prostate Cancer. Oncotarget. 2016;7:1421–1438. doi: 10.18632/oncotarget.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J., Ren S., Piao H., Wang F., Yin P., Xu C., Lu X., Ye G., Shao Y., Yan M., et al. Integration of Lipidomics and Transcriptomics Unravels Aberrant Lipid Metabolism and Defines Cholesteryl Oleate as Potential Biomarker of Prostate Cancer. Sci. Rep. 2016;6:20984. doi: 10.1038/srep20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torrano V., Valcarcel-Jimenez L., Cortazar A.R., Liu X., Urosevic J., Castillo-Martin M., Fernández-Ruiz S., Morciano G., Caro-Maldonado A., Guiu M., et al. The Metabolic Co-Regulator PGC1α Suppresses Prostate Cancer Metastasis. Nat. Cell Biol. 2016;18:645–656. doi: 10.1038/ncb3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lima A.R., Carvalho M., Aveiro S.S., Melo T., Domingues M.R., Macedo-Silva C., Coimbra N., Jerónimo C., Henrique R., de Bastos M.L., et al. Comprehensive Metabolomics and Lipidomics Profiling of Prostate Cancer Tissue Reveals Metabolic Dysregulations Associated with Disease Development. J. Proteome Res. 2021 doi: 10.1021/acs.jproteome.1c00754. [DOI] [PubMed] [Google Scholar]
- 62.Shao Y., Ye G., Ren S., Piao H.-L., Zhao X., Lu X., Wang F., Ma W., Li J., Yin P., et al. Metabolomics and Transcriptomics Profiles Reveal the Dysregulation of the Tricarboxylic Acid Cycle and Related Mechanisms in Prostate Cancer. Int. J. Cancer. 2018;143:396–407. doi: 10.1002/ijc.31313. [DOI] [PubMed] [Google Scholar]
- 63.Tessem M.-B., Bertilsson H., Angelsen A., Bathen T.F., Drabløs F., Rye M.B. A Balanced Tissue Composition Reveals New Metabolic and Gene Expression Markers in Prostate Cancer. PLoS ONE. 2016;11:e0153727. doi: 10.1371/journal.pone.0153727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaushik A.K., Shojaie A., Panzitt K., Sonavane R., Venghatakrishnan H., Manikkam M., Zaslavsky A., Putluri V., Vasu V.T., Zhang Y., et al. Inhibition of the Hexosamine Biosynthetic Pathway Promotes Castration-Resistant Prostate Cancer. Nat. Commun. 2016;7:11612. doi: 10.1038/ncomms11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren S., Shao Y., Zhao X., Hong C.S., Wang F., Lu X., Li J., Ye G., Yan M., Zhuang Z., et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol. Cell. Proteom. 2016;15:154–163. doi: 10.1074/mcp.M115.052381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee B., Mahmud I., Marchica J., Dereziński P., Qi F., Wang F., Joshi P., Valerio F., Rivera I., Patel V., et al. Integrated RNA and Metabolite Profiling of Urine Liquid Biopsies for Prostate Cancer Biomarker Discovery. Sci. Rep. 2020;10:3716. doi: 10.1038/s41598-020-60616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swinnen J.V., Roskams T., Joniau S., Van Poppel H., Oyen R., Baert L., Heyns W., Verhoeven G. Overexpression of Fatty Acid Synthase Is an Early and Common Event in the Development of Prostate Cancer. Int. J. Cancer. 2002;98:19–22. doi: 10.1002/ijc.10127. [DOI] [PubMed] [Google Scholar]
- 68.Van de Sande T., Roskams T., Lerut E., Joniau S., Van Poppel H., Verhoeven G., Swinnen J.V. High-Level Expression of Fatty Acid Synthase in Human Prostate Cancer Tissues Is Linked to Activation and Nuclear Localization of Akt/PKB. J. Pathol. 2005;206:214–219. doi: 10.1002/path.1760. [DOI] [PubMed] [Google Scholar]
- 69.Myers J.S., von Lersner A.K., Sang Q.-X.A. Proteomic Upregulation of Fatty Acid Synthase and Fatty Acid Binding Protein 5 and Identification of Cancer- and Race-Specific Pathway Associations in Human Prostate Cancer Tissues. J. Cancer. 2016;7:1452–1464. doi: 10.7150/jca.15860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim S.-J., Choi H., Park S.-S., Chang C., Kim E. Stearoyl CoA Desaturase (SCD) Facilitates Proliferation of Prostate Cancer Cells through Enhancement of Androgen Receptor Transactivation. Mol. Cells. 2011;31:371–377. doi: 10.1007/s10059-011-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peck B., Schug Z.T., Zhang Q., Dankworth B., Jones D.T., Smethurst E., Patel R., Mason S., Jiang M., Saunders R., et al. Inhibition of Fatty Acid Desaturation Is Detrimental to Cancer Cell Survival in Metabolically Compromised Environments. Cancer Metab. 2016;4:6. doi: 10.1186/s40170-016-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freitas M., Baldeiras I., Proença T., Alves V., Mota-Pinto A., Sarmento-Ribeiro A. Oxidative Stress Adaptation in Aggressive Prostate Cancer May Be Counteracted by the Reduction of Glutathione Reductase. FEBS OpenBio. 2012;2:119–128. doi: 10.1016/j.fob.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zabala-Letona A., Arruabarrena-Aristorena A., Martín-Martín N., Fernandez-Ruiz S., Sutherland J.D., Clasquin M., Tomas-Cortazar J., Jimenez J., Torres I., Quang P., et al. MTORC1-Dependent AMD1 Regulation Sustains Polyamine Metabolism in Prostate Cancer. Nature. 2017;547:109–113. doi: 10.1038/nature22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Menendez J.A., Lupu R. Fatty Acid Synthase (FASN) as a Therapeutic Target in Breast Cancer. Expert Opin. Ther. Targets. 2017;21:1001–1016. doi: 10.1080/14728222.2017.1381087. [DOI] [PubMed] [Google Scholar]
- 75.Vandergrift L.A., Decelle E.A., Kurth J., Wu S., Fuss T.L., DeFeo E.M., Halpern E.F., Taupitz M., McDougal W.S., Olumi A.F., et al. Metabolomic Prediction of Human Prostate Cancer Aggressiveness: Magnetic Resonance Spectroscopy of Histologically Benign Tissue. Sci. Rep. 2018;8:4997. doi: 10.1038/s41598-018-23177-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X., Mei H., Agee J., Brown T., Mao J. Racial Differences in Distribution of Fatty Acids in Prostate Cancer and Benign Prostatic Tissues. Lipids Health Dis. 2019;18:189. doi: 10.1186/s12944-019-1130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Latonen L., Afyounian E., Jylhä A., Nättinen J., Aapola U., Annala M., Kivinummi K.K., Tammela T.T.L., Beuerman R.W., Uusitalo H., et al. Integrative Proteomics in Prostate Cancer Uncovers Robustness against Genomic and Transcriptomic Aberrations during Disease Progression. Nat. Commun. 2018;9:1176. doi: 10.1038/s41467-018-03573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valcarcel-Jimenez L., Macchia A., Crosas-Molist E., Schaub-Clerigué A., Camacho L., Martín-Martín N., Cicogna P., Viera-Bardón C., Fernández-Ruiz S., Rodriguez-Hernandez I., et al. PGC1α Suppresses Prostate Cancer Cell Invasion through ERRα Transcriptional Control. Cancer Res. 2019;79:6153–6165. doi: 10.1158/0008-5472.CAN-19-1231. [DOI] [PubMed] [Google Scholar]
- 79.Kaminski L., Torrino S., Dufies M., Djabari Z., Haider R., Roustan F.-R., Jaune E., Laurent K., Nottet N., Michiels J.-F., et al. PGC1α Inhibits Polyamine Synthesis to Suppress Prostate Cancer Aggressiveness. Cancer Res. 2019;79:3268–3280. doi: 10.1158/0008-5472.CAN-18-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin J., Ren W., Huang X., Deng J., Li T., Yin Y. Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 2018;9:1697. doi: 10.3389/fimmu.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kami K., Fujimori T., Sato H., Sato M., Yamamoto H., Ohashi Y., Sugiyama N., Ishihama Y., Onozuka H., Ochiai A., et al. Metabolomic Profiling of Lung and Prostate Tumor Tissues by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Metabolomics. 2013;9:444–453. doi: 10.1007/s11306-012-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu X., Daniels G., Lee P., Monaco M.E. Lipid Metabolism in Prostate Cancer. Am. J. Clin. Exp. Urol. 2014;2:111–120. [PMC free article] [PubMed] [Google Scholar]
- 83.Rhodes D.R., Barrette T.R., Rubin M.A., Ghosh D., Chinnaiyan A.M. Meta-Analysis of Microarrays: Interstudy Validation of Gene Expression Profiles Reveals Pathway Dysregulation in Prostate Cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 84.Goodwin A.C., Jadallah S., Toubaji A., Lecksell K., Hicks J.L., Kowalski J., Bova G.S., De Marzo A.M., Netto G.J., Casero R.A. Increased Spermine Oxidase Expression in Human Prostate Cancer and Prostatic Intraepithelial Neoplasia Tissues. Prostate. 2008;68:766–772. doi: 10.1002/pros.20735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng Q., Wong C.Y.-P., Cheuk I.W.-Y., Teoh J.Y.-C., Chiu P.K.-F., Ng C.-F. The Emerging Clinical Role of Spermine in Prostate Cancer. Int. J. Mol. Sci. 2021;22:4382. doi: 10.3390/ijms22094382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shafi A.A., Putluri V., Arnold J.M., Tsouko E., Maity S., Roberts J.M., Coarfa C., Frigo D.E., Putluri N., Sreekumar A., et al. Differential Regulation of Metabolic Pathways by Androgen Receptor (AR) and Its Constitutively Active Splice Variant, AR-V7, in Prostate Cancer Cells. Oncotarget. 2015;6:31997–32012. doi: 10.18632/oncotarget.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kinnaird A., Zhao S., Wellen K.E., Michelakis E.D. Metabolic Control of Epigenetics in Cancer. Nat. Rev. Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 88.Clos-Garcia M., Loizaga-Iriarte A., Zuñiga-Garcia P., Sánchez-Mosquera P., Rosa Cortazar A., González E., Torrano V., Alonso C., Pérez-Cormenzana M., Ugalde-Olano A., et al. Metabolic Alterations in Urine Extracellular Vesicles Are Associated to Prostate Cancer Pathogenesis and Progression. J. Extracell. Vesicles. 2018;7:1470442. doi: 10.1080/20013078.2018.1470442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernández-Peralbo M.A., Gómez-Gómez E., Calderón-Santiago M., Carrasco-Valiente J., Ruiz-García J., Requena-Tapia M.J., Luque de Castro M.D., Priego-Capote F. Prostate Cancer Patients-Negative Biopsy Controls Discrimination by Untargeted Metabolomics Analysis of Urine by LC-QTOF: Upstream Information on Other Omics. Sci. Rep. 2016;6:38243. doi: 10.1038/srep38243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yumba-Mpanga A., Struck-Lewicka W., Wawrzyniak R., Markuszewski M., Roslan M., Kaliszan R., Markuszewski M.J. Metabolomic Heterogeneity of Urogenital Tract Cancers Analyzed by Complementary Chromatographic Techniques Coupled with Mass Spectrometry. Curr. Med. Chem. 2019;26:216–231. doi: 10.2174/0929867324666171006150326. [DOI] [PubMed] [Google Scholar]
- 91.Kremer D.M., Nelson B.S., Lin L., Yarosz E.L., Halbrook C.J., Kerk S.A., Sajjakulnukit P., Myers A., Thurston G., Hou S.W., et al. GOT1 Inhibition Promotes Pancreatic Cancer Cell Death by Ferroptosis. Nat. Commun. 2021;12:4860. doi: 10.1038/s41467-021-24859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., et al. Glutamine Supports Pancreatic Cancer Growth through a KRAS-Regulated Metabolic Pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goto T., Terada N., Inoue T., Kobayashi T., Nakayama K., Okada Y., Yoshikawa T., Miyazaki Y., Uegaki M., Utsunomiya N., et al. Decreased Expression of Lysophosphatidylcholine (16:0/OH) in High Resolution Imaging Mass Spectrometry Independently Predicts Biochemical Recurrence after Surgical Treatment for Prostate Cancer. Prostate. 2015;75:1821–1830. doi: 10.1002/pros.23088. [DOI] [PubMed] [Google Scholar]
- 94.Struck-Lewicka W., Kordalewska M., Bujak R., Yumba Mpanga A., Markuszewski M., Jacyna J., Matuszewski M., Kaliszan R., Markuszewski M.J. Urine Metabolic Fingerprinting Using LC-MS and GC-MS Reveals Metabolite Changes in Prostate Cancer: A Pilot Study. J. Pharm. Biomed. Anal. 2015;111:351–361. doi: 10.1016/j.jpba.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 95.Mondul A.M., Moore S.C., Weinstein S.J., Karoly E.D., Sampson J.N., Albanes D. Metabolomic Analysis of Prostate Cancer Risk in a Prospective Cohort: The Alpha-Tocolpherol, Beta-Carotene Cancer Prevention (ATBC) Study. Int. J. Cancer. 2015;137:2124–2132. doi: 10.1002/ijc.29576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang J., Mondul A.M., Weinstein S.J., Karoly E.D., Sampson J.N., Albanes D. Prospective Serum Metabolomic Profile of Prostate Cancer by Size and Extent of Primary Tumor. Oncotarget. 2017;8:45190–45199. doi: 10.18632/oncotarget.16775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lima A.R., Pinto J., Azevedo A.I., Barros-Silva D., Jerónimo C., Henrique R., de Lourdes Bastos M., Guedes de Pinho P., Carvalho M. Identification of a Biomarker Panel for Improvement of Prostate Cancer Diagnosis by Volatile Metabolic Profiling of Urine. Br. J. Cancer. 2019;121:857–868. doi: 10.1038/s41416-019-0585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jung K., Reszka R., Kamlage B., Bethan B., Stephan C., Lein M., Kristiansen G. Tissue Metabolite Profiling Identifies Differentiating and Prognostic Biomarkers for Prostate Carcinoma. Int. J. Cancer. 2013;133:2914–2924. doi: 10.1002/ijc.28303. [DOI] [PubMed] [Google Scholar]
- 99.McDunn J.E., Li Z., Adam K.-P., Neri B.P., Wolfert R.L., Milburn M.V., Lotan Y., Wheeler T.M. Metabolomic Signatures of Aggressive Prostate Cancer. Prostate. 2013;73:1547–1560. doi: 10.1002/pros.22704. [DOI] [PubMed] [Google Scholar]
- 100.Gómez-Cebrián N., García-Flores M., Rubio-Briones J., López-Guerrero J.A., Pineda-Lucena A., Puchades-Carrasco L. Targeted Metabolomics Analyses Reveal Specific Metabolic Alterations in High-Grade Prostate Cancer Patients. J. Proteome Res. 2020;19:4082–4092. doi: 10.1021/acs.jproteome.0c00493. [DOI] [PubMed] [Google Scholar]
- 101.Kiebish M.A., Cullen J., Mishra P., Ali A., Milliman E., Rodrigues L.O., Chen E.Y., Tolstikov V., Zhang L., Panagopoulos K., et al. Multi-Omic Serum Biomarkers for Prognosis of Disease Progression in Prostate Cancer. J. Transl. Med. 2020;18:10. doi: 10.1186/s12967-019-02185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu W., Bai X., Liu Y., Wang W., Han J., Wang Q., Xu Y., Zhang C., Zhang S., Li X., et al. Topologically Inferring Pathway Activity toward Precise Cancer Classification via Integrating Genomic and Metabolomic Data: Prostate Cancer as a Case. Sci. Rep. 2015;5:13192. doi: 10.1038/srep13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li C., Han J., Yao Q., Zou C., Xu Y., Zhang C., Shang D., Zhou L., Zou C., Sun Z., et al. Subpathway-GM: Identification of Metabolic Subpathways via Joint Power of Interesting Genes and Metabolites and Their Topologies within Pathways. Nucleic Acids Res. 2013;41:e101. doi: 10.1093/nar/gkt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao B., Lue H.-W., Podolak J., Fan S., Zhang Y., Serawat A., Alumkal J.J., Fiehn O., Thomas G.V. Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma. Metabolites. 2019;9:82. doi: 10.3390/metabo9050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Joshi M., Kim J., D’Alessandro A., Monk E., Bruce K., Elajaili H., Nozik-Grayck E., Goodspeed A., Costello J.C., Schlaepfer I.R. CPT1A Over-Expression Increases Reactive Oxygen Species in the Mitochondria and Promotes Antioxidant Defenses in Prostate Cancer. Cancers. 2020;12:3431. doi: 10.3390/cancers12113431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Y., Wang K., Liu T., Chen J., Lv W., Yang W., Xu S., Wang X., Li L. Decreased Glucose Bioavailability and Elevated Aspartate Metabolism in Prostate Cancer Cells Undergoing Epithelial-Mesenchymal Transition. J. Cell. Physiol. 2020;235:5602–5612. doi: 10.1002/jcp.29490. [DOI] [PubMed] [Google Scholar]
- 107.Hansen A.F., Sandsmark E., Rye M.B., Wright A.J., Bertilsson H., Richardsen E., Viset T., Bofin A.M., Angelsen A., Selnæs K.M., et al. Presence of TMPRSS2-ERG Is Associated with Alterations of the Metabolic Profile in Human Prostate Cancer. Oncotarget. 2016;7:42071–42085. doi: 10.18632/oncotarget.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan M., Qi H., Li J., Ye G., Shao Y., Li T., Liu J., Piao H.-L., Xu G. Identification of SPOP Related Metabolic Pathways in Prostate Cancer. Oncotarget. 2017;8:103032–103046. doi: 10.18632/oncotarget.21460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Andersen M.K., Rise K., Giskeødegård G.F., Richardsen E., Bertilsson H., Størkersen Ø., Bathen T.F., Rye M., Tessem M.-B. Integrative Metabolic and Transcriptomic Profiling of Prostate Cancer Tissue Containing Reactive Stroma. Sci. Rep. 2018;8:14269. doi: 10.1038/s41598-018-32549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oberhuber M., Pecoraro M., Rusz M., Oberhuber G., Wieselberg M., Haslinger P., Gurnhofer E., Schlederer M., Limberger T., Lagger S., et al. STAT3-Dependent Analysis Reveals PDK4 as Independent Predictor of Recurrence in Prostate Cancer. Mol. Syst. Biol. 2020;16:e9247. doi: 10.15252/msb.20199247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cacciatore S., Wium M., Licari C., Ajayi-Smith A., Masieri L., Anderson C., Salukazana A.S., Kaestner L., Carini M., Carbone G.M., et al. Inflammatory Metabolic Profile of South African Patients with Prostate Cancer. Cancer Metab. 2021;9:29. doi: 10.1186/s40170-021-00265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Labbé D.P., Zadra G., Yang M., Reyes J.M., Lin C.Y., Cacciatore S., Ebot E.M., Creech A.L., Giunchi F., Fiorentino M., et al. High-Fat Diet Fuels Prostate Cancer Progression by Rewiring the Metabolome and Amplifying the MYC Program. Nat. Commun. 2019;10:4358. doi: 10.1038/s41467-019-12298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Röhnisch H.E., Kyrø C., Olsen A., Thysell E., Hallmans G., Moazzami A.A. Identification of Metabolites Associated with Prostate Cancer Risk: A Nested Case-Control Study with Long Follow-up in the Northern Sweden Health and Disease Study. BMC Med. 2020;18:187. doi: 10.1186/s12916-020-01655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schmidt J.A., Fensom G.K., Rinaldi S., Scalbert A., Appleby P.N., Achaintre D., Gicquiau A., Gunter M.J., Ferrari P., Kaaks R., et al. Pre-Diagnostic Metabolite Concentrations and Prostate Cancer Risk in 1077 Cases and 1077 Matched Controls in the European Prospective Investigation into Cancer and Nutrition. BMC Med. 2017;15:122. doi: 10.1186/s12916-017-0885-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu H., Chen Y., Gu M., Liu C., Chen Q., Zhan M., Wang Z. Fatty Acid Metabolism Reprogramming in Advanced Prostate Cancer. Metabolites. 2021;11:765. doi: 10.3390/metabo11110765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moazzami A.A., Zhang J.-X., Kamal-Eldin A., Aman P., Hallmans G., Johansson J.-E., Andersson S.-O. Nuclear Magnetic Resonance-Based Metabolomics Enable Detection of the Effects of a Whole Grain Rye and Rye Bran Diet on the Metabolic Profile of Plasma in Prostate Cancer Patients. J. Nutr. 2011;141:2126–2132. doi: 10.3945/jn.111.148239. [DOI] [PubMed] [Google Scholar]
- 117.Wright J.L., Plymate S.R., Porter M.P., Gore J.L., Lin D.W., Hu E., Zeliadt S.B. Hyperglycemia and Prostate Cancer Recurrence in Men Treated for Localized Prostate Cancer. Prostate Cancer Prostatic Dis. 2013;16:204–208. doi: 10.1038/pcan.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wulaningsih W., Holmberg L., Garmo H., Zethelius B., Wigertz A., Carroll P., Lambe M., Hammar N., Walldius G., Jungner I., et al. Serum Glucose and Fructosamine in Relation to Risk of Cancer. PLoS ONE. 2013;8:e54944. doi: 10.1371/journal.pone.0054944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Varisli L. Identification of New Genes Downregulated in Prostate Cancer and Investigation of Their Effects on Prognosis. Genet. Test. Mol. Biomark. 2013;17:562–566. doi: 10.1089/gtmb.2012.0524. [DOI] [PubMed] [Google Scholar]
- 120.Ni W.-D., Yang Z.-T., Cui C.-A., Cui Y., Fang L.-Y., Xuan Y.-H. Tenascin-C Is a Potential Cancer-Associated Fibroblasts Marker and Predicts Poor Prognosis in Prostate Cancer. Biochem. Biophys. Res. Commun. 2017;486:607–612. doi: 10.1016/j.bbrc.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 121.San Martin R., Pathak R., Jain A., Jung S.Y., Hilsenbeck S.G., Piña-Barba M.C., Sikora A.G., Pienta K.J., Rowley D.R. Tenascin-C and Integrin A9 Mediate Interactions of Prostate Cancer with the Bone Microenvironment. Cancer Res. 2017;77:5977–5988. doi: 10.1158/0008-5472.CAN-17-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mishra P., Kiebish M.A., Cullen J., Srinivasan A., Patterson A., Sarangarajan R., Narain N.R., Dobi A. Genomic Alterations of Tenascin C in Highly Aggressive Prostate Cancer: A Meta-Analysis. Genes Cancer. 2019;10:150–159. doi: 10.18632/genesandcancer.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Malik G., Ward M.D., Gupta S.K., Trosset M.W., Grizzle W.E., Adam B.-L., Diaz J.I., Semmes O.J. Serum Levels of an Isoform of Apolipoprotein A-II as a Potential Marker for Prostate Cancer. Clin. Cancer Res. 2005;11:1073–1085. [PubMed] [Google Scholar]
- 124.Klee E.W., Bondar O.P., Goodmanson M.K., Dyer R.B., Erdogan S., Bergstralh E.J., Bergen H.R., Sebo T.J., Klee G.G. Candidate Serum Biomarkers for Prostate Adenocarcinoma Identified by MRNA Differences in Prostate Tissue and Verified with Protein Measurements in Tissue and Blood. Clin. Chem. 2012;58:599–609. doi: 10.1373/clinchem.2011.171637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kojima S., Chiyomaru T., Kawakami K., Yoshino H., Enokida H., Nohata N., Fuse M., Ichikawa T., Naya Y., Nakagawa M., et al. Tumour Suppressors MiR-1 and MiR-133a Target the Oncogenic Function of Purine Nucleoside Phosphorylase (PNP) in Prostate Cancer. Br. J. Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sreekumar A., Poisson L.M., Rajendiran T.M., Khan A.P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R.J., Li Y., et al. Metabolomic Profiles Delineate Potential Role for Sarcosine in Prostate Cancer Progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 127.Khan A.P., Rajendiran T.M., Ateeq B., Asangani I.A., Athanikar J.N., Yocum A.K., Mehra R., Siddiqui J., Palapattu G., Wei J.T., et al. The Role of Sarcosine Metabolism in Prostate Cancer Progression. Neoplasia. 2013;15:491–501. doi: 10.1593/neo.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maxeiner A., Adkins C.B., Zhang Y., Taupitz M., Halpern E.F., McDougal W.S., Wu C.-L., Cheng L.L. Retrospective Analysis of Prostate Cancer Recurrence Potential with Tissue Metabolomic Profiles. Prostate. 2010;70:710–717. doi: 10.1002/pros.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Johansson S., Landström M., Hellstrand K., Henriksson R. The Response of Dunning R3327 Prostatic Adenocarcinoma to IL-2, Histamine and Radiation. Br. J. Cancer. 1998;77:1213–1219. doi: 10.1038/bjc.1998.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Johansson S., Landström M., Henriksson R. Alterations of Tumour Cells, Stroma and Apoptosis in Rat Prostatic Adenocarcinoma Following Treatment with Histamine, Interleukin-2 and Irradiation. Anticancer Res. 1999;19:1961–1969. [PubMed] [Google Scholar]
- 131.Liu Q., Harvey C.T., Geng H., Xue C., Chen V., Beer T.M., Qian D.Z. Malate Dehydrogenase 2 Confers Docetaxel Resistance via Regulations of JNK Signaling and Oxidative Metabolism. Prostate. 2013;73:1028–1037. doi: 10.1002/pros.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Andersen M.K., Giskeødegård G.F., Tessem M.-B. Metabolic Alterations in Tissues and Biofluids of Patients with Prostate Cancer. Curr. Opin. Endocr. Metab. Res. 2020;10:23–28. doi: 10.1016/j.coemr.2020.02.003. [DOI] [Google Scholar]
- 133.Thysell E., Surowiec I., Hörnberg E., Crnalic S., Widmark A., Johansson A.I., Stattin P., Bergh A., Moritz T., Antti H., et al. Metabolomic Characterization of Human Prostate Cancer Bone Metastases Reveals Increased Levels of Cholesterol. PLoS ONE. 2010;5:e14175. doi: 10.1371/journal.pone.0014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Markert E.K., Mizuno H., Vazquez A., Levine A.J. Molecular Classification of Prostate Cancer Using Curated Expression Signatures. Proc. Natl. Acad. Sci. USA. 2011;108:21276–21281. doi: 10.1073/pnas.1117029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rye M.B., Bertilsson H., Drabløs F., Angelsen A., Bathen T.F., Tessem M.-B. Gene Signatures ESC, MYC and ERG-Fusion Are Early Markers of a Potentially Dangerous Subtype of Prostate Cancer. BMC Med. Genom. 2014;7:50. doi: 10.1186/1755-8794-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shah U.S., Dhir R., Gollin S.M., Chandran U.R., Lewis D., Acquafondata M., Pflug B.R. Fatty Acid Synthase Gene Overexpression and Copy Number Gain in Prostate Adenocarcinoma. Hum. Pathol. 2006;37:401–409. doi: 10.1016/j.humpath.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 137.Zadra G., Ribeiro C.F., Chetta P., Ho Y., Cacciatore S., Gao X., Syamala S., Bango C., Photopoulos C., Huang Y., et al. Inhibition of de Novo Lipogenesis Targets Androgen Receptor Signaling in Castration-Resistant Prostate Cancer. Proc. Natl. Acad. Sci. USA. 2019;116:631–640. doi: 10.1073/pnas.1808834116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bastos D.C., Ribeiro C.F., Ahearn T., Nascimento J., Pakula H., Clohessy J., Mucci L., Roberts T., Zanata S.M., Zadra G., et al. Genetic Ablation of FASN Attenuates the Invasive Potential of Prostate Cancer Driven by Pten Loss. J. Pathol. 2021;253:292–303. doi: 10.1002/path.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.-P., White T.A., Stojanov P., Van Allen E., Stransky N., et al. Exome Sequencing Identifies Recurrent SPOP, FOXA1 and MED12 Mutations in Prostate Cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.An J., Wang C., Deng Y., Yu L., Huang H. Destruction of Full-Length Androgen Receptor by Wild-Type SPOP, but Not Prostate-Cancer-Associated Mutants. Cell Rep. 2014;6:657–669. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ayala G., Tuxhorn J.A., Wheeler T.M., Frolov A., Scardino P.T., Ohori M., Wheeler M., Spitler J., Rowley D.R. Reactive Stroma as a Predictor of Biochemical-Free Recurrence in Prostate Cancer. Clin. Cancer Res. 2003;9:4792–4801. [PubMed] [Google Scholar]
- 142.Yanagisawa N., Li R., Rowley D., Liu H., Kadmon D., Miles B.J., Wheeler T.M., Ayala G.E. Stromogenic Prostatic Carcinoma Pattern (Carcinomas with Reactive Stromal Grade 3) in Needle Biopsies Predicts Biochemical Recurrence-Free Survival in Patients after Radical Prostatectomy. Hum. Pathol. 2007;38:1611–1620. doi: 10.1016/j.humpath.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 143.Tomas D., Spajić B., Milosević M., Demirović A., Marusić Z., Kruslin B. Intensity of Stromal Changes Predicts Biochemical Recurrence-Free Survival in Prostatic Carcinoma. Scand. J. Urol. Nephrol. 2010;44:284–290. doi: 10.3109/00365599.2010.485578. [DOI] [PubMed] [Google Scholar]
- 144.Billis A., Meirelles L., Freitas L.L.L., Polidoro A.S., Fernandes H.A., Padilha M.M., Magna L.A., Reis L.O., Ferreira U. Adenocarcinoma on Needle Prostatic Biopsies: Does Reactive Stroma Predicts Biochemical Recurrence in Patients Following Radical Prostatectomy? Int. Braz. J. Urol. 2013;39:320–327. doi: 10.1590/S1677-5538.IBJU.2013.03.04. [DOI] [PubMed] [Google Scholar]
- 145.McKenney J.K., Wei W., Hawley S., Auman H., Newcomb L.F., Boyer H.D., Fazli L., Simko J., Hurtado-Coll A., Troyer D.A., et al. Histologic Grading of Prostatic Adenocarcinoma Can Be Further Optimized: Analysis of the Relative Prognostic Strength of Individual Architectural Patterns in 1275 Patients From the Canary Retrospective Cohort. Am. J. Surg. Pathol. 2016;40:1439–1456. doi: 10.1097/PAS.0000000000000736. [DOI] [PubMed] [Google Scholar]
- 146.Tee A.R. Metastatic Castration-Resistant Prostate Cancer Hungers for Leucine. J. Natl. Cancer Inst. 2013;105:1427–1428. doi: 10.1093/jnci/djt252. [DOI] [PubMed] [Google Scholar]
- 147.Wang Q., Tiffen J., Bailey C.G., Lehman M.L., Ritchie W., Fazli L., Metierre C., Feng Y.J., Li E., Gleave M., et al. Targeting Amino Acid Transport in Metastatic Castration-Resistant Prostate Cancer: Effects on Cell Cycle, Cell Growth, and Tumor Development. J. Natl. Cancer Inst. 2013;105:1463–1473. doi: 10.1093/jnci/djt241. [DOI] [PubMed] [Google Scholar]
- 148.Otsuki H., Kimura T., Yamaga T., Kosaka T., Suehiro J.-I., Sakurai H. Prostate Cancer Cells in Different Androgen Receptor Status Employ Different Leucine Transporters. Prostate. 2017;77:222–233. doi: 10.1002/pros.23263. [DOI] [PubMed] [Google Scholar]
- 149.Mengual L., Ars E., Lozano J.J., Burset M., Izquierdo L., Ingelmo-Torres M., Gaya J.M., Algaba F., Villavicencio H., Ribal M.J., et al. Gene Expression Profiles in Prostate Cancer: Identification of Candidate Non-Invasive Diagnostic Markers. Actas Urol. Esp. 2014;38:143–149. doi: 10.1016/j.acuro.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 150.Pavlova N.N., Thompson C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hsu P.P., Sabatini D.M. Cancer Cell Metabolism: Warburg and Beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 152.Vander Heiden M.G., Lunt S.Y., Dayton T.L., Fiske B.P., Israelsen W.J., Mattaini K.R., Vokes N.I., Stephanopoulos G., Cantley L.C., Metallo C.M., et al. Metabolic Pathway Alterations That Support Cell Proliferation. Cold Spring Harb. Symp. Quant. Biol. 2011;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 153.Cheung P.K., Ma M.H., Tse H.F., Yeung K.F., Tsang H.F., Chu M.K.M., Kan C.M., Cho W.C.S., Ng L.B.W., Chan L.W.C., et al. The Applications of Metabolomics in the Molecular Diagnostics of Cancer. Expert Rev. Mol. Diagn. 2019;19:785–793. doi: 10.1080/14737159.2019.1656530. [DOI] [PubMed] [Google Scholar]
- 154.Burton C., Ma Y. Current Trends in Cancer Biomarker Discovery Using Urinary Metabolomics: Achievements and New Challenges. Curr. Med. Chem. 2019;26:5–28. doi: 10.2174/0929867324666170914102236. [DOI] [PubMed] [Google Scholar]
- 155.Lyssiotis C.A., Nagrath D. Metabolic Reprogramming and Vulnerabilities in Cancer. Cancers. 2019;12:90. doi: 10.3390/cancers12010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Harris A.L. Development of Cancer Metabolism as a Therapeutic Target: New Pathways, Patient Studies, Stratification and Combination Therapy. Br. J. Cancer. 2020;122:1–3. doi: 10.1038/s41416-019-0666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Luengo A., Gui D.Y., Vander Heiden M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017;24:1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gholizadeh N., Pundavela J., Nagarajan R., Dona A., Quadrelli S., Biswas T., Greer P.B., Ramadan S. Nuclear Magnetic Resonance Spectroscopy of Human Body Fluids and in Vivo Magnetic Resonance Spectroscopy: Potential Role in the Diagnosis and Management of Prostate Cancer. Urol. Oncol. 2020;38:150–173. doi: 10.1016/j.urolonc.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 159.Manzi M., Riquelme G., Zabalegui N., Monge M.E. Improving Diagnosis of Genitourinary Cancers: Biomarker Discovery Strategies through Mass Spectrometry-Based Metabolomics. J. Pharm. Biomed. Anal. 2020;178:112905. doi: 10.1016/j.jpba.2019.112905. [DOI] [PubMed] [Google Scholar]
- 160.Pinu F.R., Beale D.J., Paten A.M., Kouremenos K., Swarup S., Schirra H.J., Wishart D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites. 2019;9:76. doi: 10.3390/metabo9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Menyhárt O., Győrffy B. Multi-Omics Approaches in Cancer Research with Applications in Tumor Subtyping, Prognosis, and Diagnosis. Comput. Struct. Biotechnol. J. 2021;19:949–960. doi: 10.1016/j.csbj.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lewis J.E., Kemp M.L. Integration of Machine Learning and Genome-Scale Metabolic Modeling Identifies Multi-Omics Biomarkers for Radiation Resistance. Nat. Commun. 2021;12:2700. doi: 10.1038/s41467-021-22989-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Eicher T., Kinnebrew G., Patt A., Spencer K., Ying K., Ma Q., Machiraju R., Mathé A.E.A. Metabolomics and Multi-Omics Integration: A Survey of Computational Methods and Resources. Metabolites. 2020;10:202. doi: 10.3390/metabo10050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Das T., Andrieux G., Ahmed M., Chakraborty S. Integration of Online Omics-Data Resources for Cancer Research. Front. Genet. 2020;11:578345. doi: 10.3389/fgene.2020.578345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen J., Liu X., Shen L., Lin Y., Shen B. CMBD: A Manually Curated Cancer Metabolic Biomarker Knowledge Database. Database. 2021;2021:baaa094. doi: 10.1093/database/baaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]