Abstract
Amplification of the partial Cpn60 (or GroEL) gene segment has been used for identification of many bacteria, including Enterococcus species. To obtain more sequence data from groESL genes of Enterococcus faecalis, the full-length sequence of the E. faecalis groESL genes containing groES (285 bp), spacer (57 bp), and groEL (1,626 bp) was determined. A database search of GenBank revealed that the deduced E. faecalis GroES and GroEL proteins show significant homology to the GroES and GroEL proteins of other bacteria. The GroEL (groEL) of E. faecalis had the highest identity with Streptococcus pneumoniae (81.8% amino acid sequence identity and 73.0% nucleotide sequence identity), followed by Lactococcus zeae, while GroES (groES) had 60.2% (64.6%) identity with Lactobacillus zeae and 58.5% (66.2%) identity with Lactococcus lactis, followed by 57.0% (65.5%) identity with Bacillus subtilis. Based on the groES sequence, an E. faecalis-specific PCR assay was developed, and this PCR assay was positive for all the E. faecalis strains tested. Dot blot hybridization using either groES or groEL as the probe distinguished E. faecalis clearly from other species, indicating that both genes can be used as suitable targets for E. faecalis identification. Moreover, broad-range PCR-restriction fragment length polymorphism of groESL was designed to differentiate eight commonly encountered Enterococcus species. The Enterococcus species of reference strains could be easily differentiated on the basis of restriction patterns produced by HaeIII and RsaI. The DNA-based assays developed in this study provide an alternative to currently used methods of identification for clinically important enterococcal species.
Once considered harmless commensals of the intestinal tract, enterococci now rank among the leading causes of nosocomial infections (19). There are two major pathogenic species in humans, Enterococcus faecalis and E. faecium, with occasional infections being caused by other species (19). The increasing occurrence of high-level gentamicin-resistant (HLGR) and vancomycin-resistant enterococci (VRE) has become a major concern worldwide (13, 27).
Rapid identification of bacteria is important for effective patient management and reducing the spread of antibiotic resistance (2). Conventional identification methods, which are based on phenotypic and culture characteristics, require 2 to 3 days to provide results (7, 32). Currently, species identification of enterococci in many laboratories relies on automation or rapid kits. However, errors in automated identification systems are not easily detected, as species identification is based on an inbuilt database (8, 29). In addition, atypical phenotypic characteristics can lead to misidentification. Another approach to species identification may be the use of molecular methods. Several DNA-based techniques for identifying clinical isolates have been developed (5, 6, 15, 18). A variety of conserved genes, including 16S rRNA genes, the tRNA intergenic spacer, the d-alanine:d-alanine ligase gene (ddl gene), the sodA gene encoding superoxide dismutase, penicillin-binding protein 5, and the elongation factor tuf gene have been used for identification of enterococci (15, 20, 21, 23, 30).
The groESL genes (also known as cpn10/60 or hsp10/60), which encode 10-kDa (GroES) and 60-kDa (GroEL) heat shock proteins, are ubiquitous and evolutionarily highly conserved among bacteria (12). Recently, Goh et al. developed reverse checkerboard hybridization to identify Staphylococcus and Enterococcus species on the basis of amplification of partial chaperonin 60 gene sequences (10, 11). The goals of this study were to obtain the full-length sequences of groESL genes of E. faecalis and provide another approach for species identification.
MATERIALS AND METHODS
Bacterial strains.
E. faecalis ATCC 29212, E. faecium ATCC 19434, E. avium ATCC 14025, E. casseliflavus ATCC 25788, E. gallinarum ATCC 49573, E. raffinosus ATCC 49427, E. hirae ATCC 8043, and E. durans ATCC 19432 were obtained from the American Type Culture Collection (ATCC), Rockville, Md. Clinical isolates, including eight E. faecalis isolates, four E. faecium isolates, two E. avium isolates, four E. casseliflavus isolates, four E. gallinarum isolates, and two E. raffinosus isolates, were obtained from the Bacteriology Laboratory, National Taiwan University Hospital, a 2,000-bed teaching hospital in northern Taiwan.
DNA amplification and sequencing of a partial fragment by PCR.
Initially, degenerate PCR primers, 590F and 590R (Table 1), complementary to highly conserved regions of the groEL gene among eubacteria, were designed and used to amplify a 590-bp internal fragment of the groEL gene from enterococcal species. Genomic DNA was isolated and purified from Enterococcus species with a DNA isolation kit, Puregene (Gentra Systems, Inc., Minneapolis, Minn.), according to the manufacturer's instructions. The thermal cycling conditions were 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C, followed by a final extension of 7 min at 72°C. An amplified product of the expected size was subsequently sequenced.
TABLE 1.
PCR primers used in this study
| Primer name | Sequencea (5′ to 3′) | Gene and nucleotide positions |
|---|---|---|
| Universal amplification | ||
| 590F | GGNGACGGNACNACNACNGCAACNGT | groEL, 255–280 |
| 590R | TCNCCRAANCCNGGYGCNTTNACNGC | groEL, 844–819 |
| LA-PCR | ||
| C1 | GTACATATTGTCGTTAGAACGCGTAATACGACTCA | |
| C2 | CGTTAGAACGCGTAATACGACTCACTATAGGGAGA | |
| EL-1F | CAAGTCGCTGCTGTTTCATC | groEL, 435–454 |
| EL-2F | AACCAAATCGGCGAAACAAC | groEL, 1047–1066 |
| EL-3F | GGTGAATGGGTAAACATGGTTGAA | groEL, 1440–1463 |
| EL-1R | CATCCGCAATAATCAATAGT | groEL, 753–734 |
| EL-2R | CCTGATGAAACAGCAGCGAC | groEL, 457–438 |
| E. faecalis specific | ||
| EfGroES-F | GGAATTGTTCTTGCATCCGT | groES, 67–86 |
| EfGroES-R | ACAATTAAGTATTCTACGCC | groES, 251–232 |
| Broad-range PCR-RFLP | ||
| EntGroES-F | TTAAAACCATTAGGCGATCG | groES, 4–23 |
| EntGroEL-R | CCCATNCCCATNGANGGRTCCAT | groEL, 1613–1591 |
N, any nucleotide; R, purine; Y, pyrimidine.
Southern blot hybridization.
A 590-bp DNA product internal to the groEL gene of E. faecalis was used as the probe. Probes were produced by the PCR method described above and simultaneously labeled by incorporation of digoxigenin-11-dUTP (Boehringer Mannheim, Mannheim, Germany). E. faecalis DNA was digested with EcoRI or BamHI and then separated by agarose gel electrophoresis and transferred to nylon membranes (Hybond-N; Amersham Phamacia Biotech Inc., Piscataway, N.J.) using standard techniques. After prehybridization, membranes were hybridized with digoxigenin-labeled DNA fragments in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS)–50% formamide at 42°C for 16 h. After high-stringency washing (68oC with 0.1× SSC–0.1% SDS), the detection of hybridization was performed by using an antidigoxigenin antibody conjugated to alkaline phosphatase as a substrate (Boehringer Mannheim) according to the manufacturer's instructions.
Cloning and sequencing of groESL genes of E. faecalis by LA-PCR.
In order to obtain the entire groESL gene sequences, an LA-PCR in vitro cloning kit (Takara Shuzo Co., Tokyo, Japan) was used. The amplification was performed with one cassette primer (C1 or C2) supplied by the manufacturer and a target gene-specific primer (Table 1). Amplification fragments were subsequently sequenced on an Applied Biosystem model 377 sequencing system (Applied Biosystems, Foster City, Calif.) using the Taq BigDye-Deoxy Terminator cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions. The complete E. faecalis groESL sequence was collected by aligning and combining amplification fragments obtained by LA-PCR.
E. faecalis-specific PCR.
The sequence of E. faecalis groESL genes was compared with the published sequences of other bacteria. A pair of PCR primers derived from groES sequence was used to amplify a target region of 185 bp from E. faecalis. These primers were named EfGroES-F and EfGroES-R (Table 1). The PCR was carried out in a DNA thermal cycler (MJ Research, Inc., Watertown, Mass.) with 35 cycles of denaturation (94°C, 30 s), annealing (52°C, 1 min), and extension (72°C, 1 min), followed by a final extension step (72°C, 7 min). The PCR amplification products were analyzed by agarose gel electrophoresis in 1.5% agarose (FMC BioProducts, Rockland, Maine) and stained with ethidium bromide. A visible band of the appropriate size (185 bp) was considered to indicate a positive reaction.
Dot blot hybridization.
Probes were produced with the PCR method and simultaneously labeled by incorporation of digoxigenin-11-dUTP (Boehringer Mannheim). For each strain tested, 300 ng of chromosomal DNA was denatured by heating at 96°C for 10 min and spotted onto Hybond-N nylon membranes (Amersham Pharmacia Biotech Inc.). DNA was then fixed onto the filter by UV treatment at an intensity of 120 mJ/cm2 for 3 min on a UV cross-linker. The prehybridization and hybridization temperatures were both 42°C. All filters were prehybridized for 1 h in 5× SSC. Hybridization was carried out overnight with the heat-denatured probe. Detection was performed by using an antidigoxigenin antibody conjugated to alkaline phosphatase as a substrate (Boehringer Mannheim) according to the manufacturer's instructions.
Intraspecies polymorphism.
The intraspecies polymorphism of groES or groEL was investigated by sequencing groES and groEL partial fragments among E. faecalis clinical isolates including two vancomycin-susceptible Enterococcus (VSE) and two VRE isolates.
Broad-range PCR-RFLP.
Based on the sequence obtained, primers EntGroES-F and EntGroEL-R were derived to amplify the entire groESL region of DNA from Enterococcus species by PCR-restriction fragment length polymorphism (RFLP). The amplification product was subsequently digested with the restriction enzymes HaeIII and RsaI (Gibco-BRL, Gaithersburg, Md.). After incubation, the DNA fragments were subjected to gel electrophoresis (FMC BioProducts), stained with ethidium bromide, and photographed under UV light.
Nucleotide sequence accession number.
The complete sequence of the E. faecalis groESL has been submitted to the GenBank database under accession number AF335185.
RESULTS
Nucleotide sequence of the groESL genes.
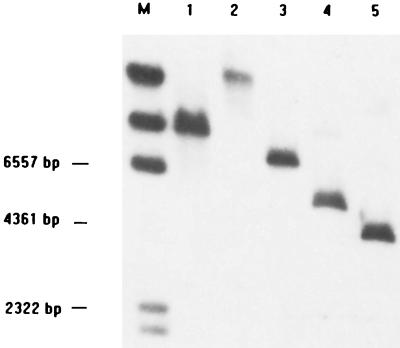
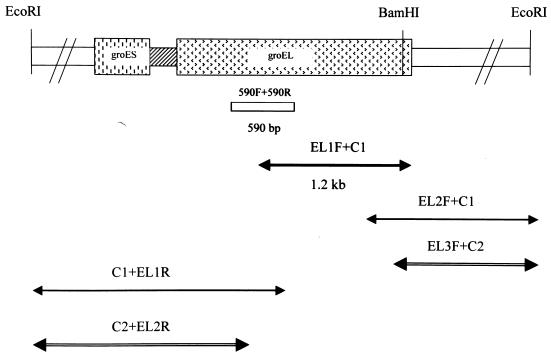
Initially, PCR primers complementary to highly conserved regions of the groEL gene among eubacteria were derived and used to amplify a 590-bp portion of the groEL gene from E. faecalis. Genomic E. faecalis DNA was used as a template in PCR. After sequencing and homology searches with databases from gene banks, this fragment yielded the highest mating scores for bacterial groEL genes. Southern hybridization of the 590-bp groEL partial fragment to E. faecalis genomic DNA digested with BamHI and EcoRI showed the specific hybridization of a 3-kb BamHI fragment and a 5-kb EcoRI fragment (Fig. 1). Then, the LA-PCR was performed. A 1.2-kb DNA fragment was amplified from BamHI-digested genomic DNA by the primers EL-1F and C1 (Fig. 2). Subsequently, two other PCR fragments were obtained by nested LA-PCR from EcoRI-digested genomic DNA, and the sequences of these fragments (EL3F+C2 and C2+EL2R) were determined (Fig. 2). Therefore, the full-length sequence of E. faecalis groESL operon was obtained.
FIG. 1.
Southern hybridization of the 590-bp groEL internal fragment to E. faecalis genomic DNA digested with restriction enzymes shows the probe hybridized to a 5-kb EcoRI fragment and 3-kb BamHI fragment. Lane M, markers; lanes 1 to 5, digested with XbaI, PstI, HindIII, EcoRI, and BamHI, respectively.
FIG. 2.
Schematic illustration of amplification fragments and restriction sites of E. faecalis groESL genes. The box between groES and groEL is spacer. The 590-bp fragment was used as the probe for Southern blot. The arrows below the restriction map indicate the amplified and sequenced fragments. The primers used in LA-PCR are indicated above the arrows.
The sequence obtained revealed the presence of two open reading frames (ORF) of 285 nucleotides and 1,626 nucleotides separated by 57 nucleotides. Comparative analysis of these nucleotide sequences with those in the genetic databases showed that the deduced E. faecalis ORF 1 and ORF 2 proteins showed significant homology with the GroES and GroEL proteins of other bacteria (Table 2). The GroES (groES) homolog of E. faecalis had 60.2% amino acid sequence identity (64.6% nucleotide sequence identity) with Lactobacillus zeae and 58.5% (66.2%) identity with Lactococcus lactis, followed by 57.0% (65.5%) identity with Bacillus subtilis, while the GroEL (groEL) homolog of E. faecalis had highest homology (81.8% amino acid sequence identity and 73.0% nucleotide sequence identity) with Streptococcus pneumoniae, followed by 79.7% (71.7%) identity with Lactococcus zeae. The groES homolog (ORF 1) is 285 bp long, and 94 amino acids were deduced. A putative ribosome-binding site (GGAGG) was located 8 nucleotides upstream of the start codon (GTG) of ORF 1. A potential translation initiation codon (AUG) of the second ORF (ORF 2) was located 57 bp downstream of the stop codon (TAA) of ORF 1. ORF 2 (groEL homolog) is 1,626 bp long, and 541 amino acids were deduced. A repeat sequence (positions 766 to 792 in GenBank accession number AF335185) was observed upstream of ORF 1 and was identical with CIRCE-like element, a well-conserved inverted repeat involved in groE regulation in gram-positive bacteria. A large inverted repeat which may function as a rho-independent transcriptional terminator was located downstream of OFR 2.
TABLE 2.
Sequence identity among inferred GroES and GroEL genes from E. faecalis and related bacteria
| Speciesa | Amino acid (nucleotide) sequence identity with E. faecalis gene product (gene)
|
|
|---|---|---|
| GroES (groES) | GroEL (groEL) | |
| Lactobacillus zeae | 60.2 (64.6) | 79.7 (71.7) |
| Lactococcus lactis | 58.5 (66.2) | 79.5 (74.6) |
| Bacillus subtilis | 57.0 (65.5) | 77.5 (70.2) |
| Bacillus stearothermophilus | 57.0 (64.6) | 77.7 (73.4) |
| Streptococcus pneumoniae | 46.8 (58.5) | 81.8 (73.0) |
| Staphylococcus aureus | 47.8 (58.6) | 71.2 (71.3) |
Development of an E. faecalis-specific PCR.
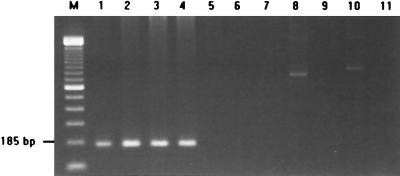
Since the groES sequences vary more than groEL among organisms, E. faecalis-specific identification of DNA was tested with the primers based on E. faecalis groES sequences. The specificity of the primers was tested with eight ATCC strains and 24 clinical isolates of various Enterococcus species. PCRs with the primer pair EfGroES-F/EfGroES-R identified all E. faecalis isolates. All E. faecalis isolates tested produced the expected 185-bp amplicon (Fig. 3). Moreover, the sequences of the 185-bp amplicon generated from four clinical isolates perfectly matched the reference sequence from the ATCC strain. There was no amplification product of 185 bp from 16 clinical isolates of other species.
FIG. 3.
E. faecalis-specific PCR amplification. Specific amplification of the 185-bp DNA fragment was detected only in E. faecalis isolates. M, 100-bp DNA ladder (Gibco-BRL). Lanes 1 to 4, E. faecalis clinical isolates; lane 5, E. faecium; lane 6, E. durans; lane 7, E. hirae; lane 8, E. avium; lane 9, E. gallinarum; lane 10, E. casseliflavus; lane 11, negative control.
Dot blot hybridization.
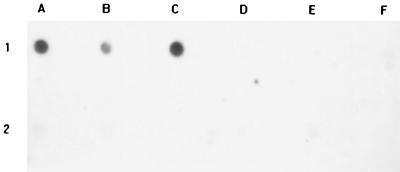
Dot blot hybridization was performed on eight reference strains and 24 clinical isolates. The amplification products of groES or groEL from E. faecalis were used as probes. Examples of results using E. faecalis groES as probe are shown in Fig. 4. Only DNAs from E. faecalis showed a strong hybridization signal. No hybridization signal was detected by dot blot analysis of DNAs from non-E. faecalis strains.
FIG. 4.
Dot blot hybridization using E. faecalis groES as the probe, showing the species specificity. All positive hybridization (1A, 1B, and 1C) was obtained from E. faecalis isolates. 1A, E. faecalis ATCC 29212; 1B and 1C, E. faecalis clinical isolates. All negative reactions were from E. faecium (1D), E. cecorum (1E), E. durans (1F), E. casseliflavus (2A), E. gallinarum (2B and 2C), E. hirae (2D), and E. avium (2E and 2F).
Differentiation of Enterococcus species by groESL broad-range PCR-RFLP.
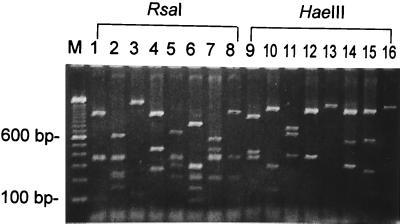
A PCR amplification was performed with a pair of primers, EntGroES-F and EntGroEL-R, based on the E. faecalis groESL genes sequence obtained in this study. PCR-RFLP analysis was carried out with eight Enterococcus ATCC reference strains. A major PCR-amplified product of approximately 1.9 kb in length was detected. These fragments correspond to nearly the entire length of groESL genes. The amplified PCR products were subsequently subjected to two sets of restriction enzyme digestion, with RsaI and HaeIII being used individually. The analysis showed that the RFLP profiles of PCR products from each species of Enterococcus were quite distinguishable, except for HaeIII digests between E. avium and E. hirae (Fig. 5).
FIG. 5.
Broad-range PCR-RFLP of groESL genes among eight enterococcal species. Lane M, DNA size markers. Lanes 1 to 8, RsaI digestion; lanes 10 to 16, HaeIII. Lanes 1 and 9, E. faecalis; lanes 2 and 10, E. faecium; lanes 3 and 11, E. casseliflavus; lanes 4 and 12, E. gallinarum; lanes 5 and 13, E. avium; lanes 6 and 14, E. raffinosus; lanes 7 and 15, E. durans; and lanes 8 and 16, E. hirae.
A total of eight E. faecalis clinical isolates were tested by this analysis. All isolates tested, including vancomycin-susceptible and vancomycin-resistant E. faecalis isolates, showed RFLP patterns identical to that of the reference strain, indicating intraspecies uniformity.
DISCUSSION
Accurate species identification of enterococci has become important with the wide prevalence of acquired vancomycin resistance. Conventional methods of identification of Enterococcus species are time-consuming (7, 32). Errors can happen and are not easily detected in automated identification systems, and supplemental testing is sometimes required for identification (14). Tsakris et al. found that 14 E. faecalis isolates with HLGR were misidentified as E. durans when using a semiautomated system (29). The development of rapid and sensitive DNA-based assays may improve the speed and accuracy of diagnosis of enterococcal infections. In this study, we describe the full-length sequencing of groESL genes from E. faecalis and the application of species identification.
Using the LA-PCR method, the complete sequence of the E. faecalis groESL genes containing the putative promoter region, ORF 1 (groES homolog, 285 bp), spacer (57 bp), and ORF 2 (groEL homolog, 1,626 bp) was determined. The sequence data of groESL genes from E. faecalis showed that the gene structure was similar to those of most bacterial species studied (3, 24, 26, 28). The deduced amino acid sequences of ORF 1 (94 amino acids) and ORF 2 (541 amino acids) proteins exhibited a high degree of overall identity with homologous bacterial GroESs and GroELs, respectively. The homology search with published gene sequences in the database revealed that the GroES (groES) sequence of E. faecalis had highest identity with Lactobacillus zeae (60.2% amino acid identity and 64.6% nucleotide identity) and Lactococcus lactis (58.5% amino acid identity and 66.2% nucleotide identity), followed by Bacillus subtilis, while the GroEL (groEL) sequence of E. faecalis had highest identity with S. pneumoniae (81.8% amino acid identity and 73.0% nucleotide identity), followed by Lactococcus zeae (79.7% amino acid identity and 71.7% nucleotide identity). The data presented here suggest a close relationship between these organisms. This result is generally in agreement with the data obtained from 5S rRNA, 16s RNA and grpE reported by Ahmad et al. (1). Studies of the phylogenetic relationship by Ahmad et al. also found that Streptococcus species, L. lactis, and E. faecalis formed groups within the low-G+C gram-positive bacteria.
E. faecalis groES and groEL each have a ribosome-binding site. The putative ribosome-binding site sequence (GGAGG) of E. faecalis GroES was identical to that of S. pneumoniae. The putative ribosome-binding site sequence of E. faecalis GroEL was GGTGA. The sequence data obtained in this study suggest that the E. faecalis groES gene may utilize an uncommon start codon, GTG. To confirm that this uncommon start codon sequence is correct, we performed another PCR, amplifying a fragment covering this region from E. faecalis ATCC reference strain and two clinical isolates. All revealed the same results. The importance of the GTG start codon in Enterococcus is unknown. Non-AUG initiation codons usually act to limit the expression of a gene product at the translational level (24). Others have reported that L. helveticus utilizes UUG as the start codon of GroES genes (3). Upstream of the groES, a putative CIRCE sequence was also identified. The sequence of CIRCE was conserved in most gram-positive bacteria and some gram-negative bacteria (28, 31, 33). In the groESL and dnaK operons of Bacillus subtilis and many other gram-positive bacteria, CIRCE is located between the transcription start point and the start codon of the first ORF (31, 33).
The C terminus of E. faecalis GroEL was PSMGMGGMM. This sequence is also conserved in many gram-positive bacteria, such as PSMGMGGMI in L. lactis and PSMMGGMM in S. pneumoniae. Comparison of these sequences shows that the GroEL of gram-positive bacteria usually consists of only one GGM at the C terminus. In contrast, the C terminus of most gram-negative bacteria consists of three tandem repeats of the GGM. A large inverted repeat which may function as a rho-independent transcriptional terminator was located downstream of the groEL gene. The termination sequence of the inverted repeat was similar to others (24).
The spacer length between the GroES translation termination codon and the putative translation start codon for GroEL was 57 nucleotides. The spacer length usually varies among different species. The spacer length is 15 bp in S. pneumoniae, 75 bp in Staphylococcus aureus, 87 bp in L. lactis, 36 bp in L. zeae, 46 bp in B. subtilis, and 45 bp in E. coli. Whether the spacer length or sequence is specific for each enterococcal species needs to be determined.
HSP60 genes are ubiquitous in both prokaryotes and eukaryotes and encode highly conserved housekeeping proteins which are essential for the survival of cells. They are more variable than the 16S rRNA gene sequence and are therefore potentially useful for the identification of genetically related species. HSP60 gene has been used as a target for species identification of enterococci and many other bacteria. For example, analysis of the hsp60 gene based on PCR, PCR-RFLP, or direct sequencing has previously been used for the identification of Mycobacterium species, Staphylococcus species, Streptococcus iniae, Ehrlichia species, and other species (4, 9, 11, 22, 25, 26). Since groES seems to be more variable than groEL among different organisms and the specific primers based on groES may be more easily found to differentiate E. faecalis from other organisms, we tried to use groES as an alternative target for species identification. Based on the sequence determined, an E. faecalis-specific PCR was developed. No false positives or false negatives were observed. This assay may facilitate the accurate identification of E. faecalis. Other approaches using HSP10 as a target for identification have been reported. For example, LaVerda and Byrne used monoclonal antibodies against HSP10 to identify Chlamydia trachomatis (16).
The dot blot hybridization results from testing eight ATCC strains and 24 clinical isolates in this study and the DNA sequencing of the PCR fragments showing complete identity from four clinical isolates are highly suggestive that not only the groEL but also groES gene can be a useful target for species identification.
The sequence obtained in this study was from E. faecalis ATCC 29212. It is different from the strain (ATCC 19434) used by Goh et al. for (10) Cpn60 partial sequencing. The deduced amino acid residues of groEL agreed with the published Cpn60 partial fragment (184 amino acids, 552 nucleotides) but differed in one amino acid residue and two nucleotides. The observed difference in this region may represent strain-to-strain variations. Intraspecies variation in this region was further tested for four unrelated clinical isolates, two VRE and two VSE. The results showed that the similarity of nucleotide sequences were very high (greater than 99% identity) in this 590-bp region, and the deduced amino acid sequences from three isolates were identical to that of ATCC 29212 and one is identical to that of ATCC 19434. Intraspecies variation of groEL among Bartonella and Ehrlichia species has been studied by Marston et al. and Sumner et al., respectively, and their reports revealed that some sequence divergence may be evident between strains from different countries (17, 26).
Besides the identification of E. faecalis by species-specific PCR assay, identification of other species was also be performed by broad-range PCR-RFLP. The PCR-RFLP of groESL with RsaI distinguished clearly between the type strains of eight commonly encountered Enterococcus species. The HaeIII digestions distinguished most species but not E. avium and E. durans. Preliminary data from 24 clinical isolates of Enterococcus species showed that species-specific patterns were observed. However, whether more than one pattern existed in clinical isolates of same species is unknown, and more isolates are needed to test.
In conclusion, our results indicated that the E. faecalis species-specific PCR, dot blot hybridization, and broad-range PCR-RFLP of groESL genes used in this study are relatively simple and accurate in the identification of at least eight species of commonly encountered Enterococcus, especially for those with atypical phenotypes. The procedure described in this paper offers a rapid, convenient, and effective tool to identify E. faecalis and other species. The PCR-based assays developed in this study provide an alternative to currently used methods for identification of clinically important enterococcal species. Determining the sequences of groESL among other Enterococcus species has been undertaken. After obtaining these sequences, specific PCR of other species and phylogeny will be developed.
ACKNOWLEDGMENT
This work was supported by grant NSC 89–2314-B-002–269 from the National Science Council of Taiwan.
REFERENCES
- 1.Ahmad S, Selvapandiyan A, Bhatnagar R K. Phylogenetic analysis of Gram-positive bacteria based on grpE, encoded by the dnaK operon. Int J Syst Evol Microbiol. 2000;50:1761–1766. doi: 10.1099/00207713-50-5-1761. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron M G, Ouellette M. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J Clin Microbiol. 1998;36:2169–2172. doi: 10.1128/jcm.36.8.2169-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broadbent J R, Oberg C J, Wei L. Characterization of the Lactobacillus helveticus groESL operon. Res Microbiol. 1998;149:247–253. doi: 10.1016/s0923-2508(98)80300-8. [DOI] [PubMed] [Google Scholar]
- 4.Chae J, Foley J E, Stephen J, Madigan J E. Comparison of the nucleotide sequences of 16S rRNA, 444 Ep-ank, and groESL heat shock operon genes in naturally occurring Ehrlichia equi and human granulocytic ehrlichiosis agent isolates from northern California. J Clin Microbiol. 2000;38:1364–1369. doi: 10.1128/jcm.38.4.1364-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Descheemaeker P, Lammens C, Pot B, Vandamme P, Goossens H. Evaluation of arbitrarily primed PCR analysis and pulsed-field gel electrophoresis of large genomic DNA fragments for identification of enterocooi important in human medicine. Int J Syst Bacteriol. 1997;47:555–561. doi: 10.1099/00207713-47-2-555. [DOI] [PubMed] [Google Scholar]
- 6.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 2000;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Garrote F, Cercenado E, Bouza E. Evaluation of a new system, VITEK 2: for identification and antimicrobial susceptibility testing of enterococci. J Clin Microbiol. 2000;38:2108–2111. doi: 10.1128/jcm.38.6.2108-2111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh S H, Driedger D, Gillett S, Low D E, Hemmingsen S M, Amos M, Chan D, Lovgren M, Willey B M, Shaw C, Smith J A. Streptococcus iniae, a human and animal pathogen: specific identification by the chaperonin 60 gene identification method. J Clin Microbiol. 1998;36:2164–2166. doi: 10.1128/jcm.36.7.2164-2166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh S H, Facklam R R, Chang M, Hill J E, Tyrrell G J, Burns E C M, Chan D, He C, Rahim T, Shaw C, Hemmingsen S M. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J Clin Microbiol. 2000;38:3953–3959. doi: 10.1128/jcm.38.11.3953-3959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh S H, Potter S, Wood J O, Hemmingsen S M, Reynolds R P, Chow A W. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J Clin Microbiol. 1996;34:818–823. doi: 10.1128/jcm.34.4.818-823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemmingsen S M, Woolford C S, van der Vies M, Tilly K, Dennis D T, Georgopoulos C P, Hendrix R W, Ellis R J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature (London) 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- 13.Hsueh P R, Teng L J, Pan H J, Chen Y C, Wang L H, Chang S C, Ho S W, Luh K T. Emergence of vancomycin-resistant enterococci at a university hospital in Taiwan: persistence of multiple species and multiple clones. Infect Control Hosp Epidemiol. 1999;20:828–833. doi: 10.1086/501592. [DOI] [PubMed] [Google Scholar]
- 14.Iwen P C, Rupp M E, Schreckenberger P C, Hinrichs S H. Evaluation of the revised MicroScan dried overnight gram-positive identification panel to identify Enterococcus species. J Clin Microbiol. 1999;37:3756–3758. doi: 10.1128/jcm.37.11.3756-3758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ke D, Picard F J, Martineau F, Ménard C, Roy P H, Ouellette M, Bergeron M G. Development of PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497–3503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaVerda D, Byrne G I. Use of monoclonal antibodies to facilitate identification, cloning, and purification of Chlamydia trachomatis hsp10. J Clin Microbiol. 1997;35:1209–1215. doi: 10.1128/jcm.35.5.1209-1215.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marston E L, Sumner J W, Regnery R L. Evaluation of intraspecies genetic variation within the 60 kDa heat-shock protein gene (groEL) of Bartonella species. Int J Syst Bacteriol. 1999;49:1015–1023. doi: 10.1099/00207713-49-3-1015. [DOI] [PubMed] [Google Scholar]
- 18.Monstein H J, Quednau M, Samuelsson A, Ahrné S, Lsaksson B, Jonasson J. Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology. 1998;144:1171–1179. doi: 10.1099/00221287-144-5-1171. [DOI] [PubMed] [Google Scholar]
- 19.Mundy L M, Sahm D F, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13:513–522. doi: 10.1128/cmr.13.4.513-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel R, Piper K E, Rouse M S, Steckelberg J M, Uhl J R, Kohner P, Hopkins M K, Cockerill III F R, Kline R C. Determination of 16S rRNA sequences of Enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J Clin Microbiol. 1998;36:3399–3407. doi: 10.1128/jcm.36.11.3399-3407.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poyart C, Quesnes G, Trieu-Cuot P. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J Clin Microbiol. 2000;38:415–418. doi: 10.1128/jcm.38.1.415-418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rastogi N, Goh K S, Berchel M. Species-specific identification of Mycobacterium leprae by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J Clin Microbiol. 1999;37:2016–2019. doi: 10.1128/jcm.37.6.2016-2019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbi C, Signoretto C, Boaretti M, Canepari P. The gene coding for penicillin-binding protein 5 of Enterococcus faecalis is useful for the development of a species-specific DNA probe. Microb Drug Resist. 1996;2:215–218. doi: 10.1089/mdr.1996.2.215. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt A, Schiesswohl M, Volker U, Hecker M, Schumann W. Cloning, sequencing, mapping, and transcriptional analysis of the groESL operon from Bacillus subtilis. J Bacteriol. 1992;174:3993–3999. doi: 10.1128/jb.174.12.3993-3999.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steingrube V A, Gibson J L, Brown B A, Zhang Y, Wilson R W, Rajagopalan M, Wallace R J., Jr PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J Clin Microbiol. 1995;33:149–153. doi: 10.1128/jcm.33.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumner J W, Storch G A, Buller R S, Liddell A M, Stockham S L, Rikihisa Y, Messenger S, Paddock C D. PCR amplification and phylogenetic analysis of groESL operon sequences from Ehrlichia ewingii and Ehrlichia muris. J Clin Microbiol. 2000;38:2746–2749. doi: 10.1128/jcm.38.7.2746-2749.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng L J, Liaw S J, Hsueh P R, Ho S W, Luh K T. Heterogeneity of resistance elements in clinical isolates of enterococci with high-level gentamicin resistance. J Formos Med Assoc. 1998;97:855–859. [PubMed] [Google Scholar]
- 28.Thies F L, Weishaupt A, Karch H, Hartung H P, Giegerich G. Cloning, sequencing and molecular analysis of the Campylobacter jejuni groESL bicistronic operon. Microbiology. 1999;145:89–98. doi: 10.1099/13500872-145-1-89. [DOI] [PubMed] [Google Scholar]
- 29.Tsakris A, Woodford N S, Pournaras, Kaufmann M, Douboyas J. Apparent increased prevalence of high-level aminoglycoside-resistant Enterococcus durans resulting from false identification by a semiautomated software system. J Clin Microbiol. 1998;36:1419–1421. doi: 10.1128/jcm.36.5.1419-1421.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyrrell G J, Rethune R N, Willey B, Low D E. Species identification of enterococci via intergenic ribosomal PCR. J Clin Microbiol. 1997;35:1054–1060. doi: 10.1128/jcm.35.5.1054-1060.1997. . (Erratum, 35:2434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wetzstein M, Volker U, Dedio J, Lobau S, Zuber U, Scheisswohl M, Hecker M, Schumann W. Cloning, sequencing, and molecular analysis of dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willey B M, Jones R N, McGeer A, Witte W, French G, Roberts R B, Jenkins S G, Nadler H, Low D E. Practical approach to the identification of clinically relevant Enterococcus species. Diagn Microbiol Infect Dis. 1999;34:165–171. doi: 10.1016/s0732-8893(99)00032-2. [DOI] [PubMed] [Google Scholar]
- 33.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]