Abstract
Guillain-Barré syndrome (GBS) and Miller-Fisher syndrome (MFS) are correlated with prior infection by Campylobacter jejuni in up to 40% of cases. Nucleotide sequence-based typing of 25 C. jejuni isolates associated with neuropathy permitted robust comparisons with equivalent data from approximately 800 C. jejuni isolates not associated with neuropathy. A total of 13 genetic lineages and 20 flaA short variable region nucleotide sequences were present among the 25 isolates. A minority of isolates (4 of 25) had the flaA short variable region nucleotide sequences that were previously proposed as a marker for GBS-associated isolates. These 4 isolates probably represented the Penner serotype 19 lineage, which has been proposed to have an association with GBS.
Guillain-Barré Syndrome (GBS) and Miller-Fisher Syndrome (MFS) are autoimmune disorders of the peripheral nervous system which cause acute flaccid paralysis (11). Both syndromes occur worldwide, with incidence rates of 0.5 to 4 per 100,000 for GBS, the more common of the two diseases (6). Up to 40% of cases are preceded by enteritis caused by the gram-negative organism Campylobacter jejuni 1 to 3 weeks before the onset of neurological symptoms (10, 14). The mechanisms by which C. jejuni infections cause neuropathy are yet to be fully elucidated; however, GBS and MFS are probably a consequence of immunological cross-reactivities of antibodies stimulated against bacterial cell surface carbohydrates with human gangliosides (5, 13). Relative to the isolates generally recovered from patients with C. jejuni enteritis, certain serotypes, especially Penner serotype 19 (7), are overrepresented among isolates from patients who develop GBS and MFS. It has been suggested that the Penner 19 serotype is associated with a C. jejuni clone which has a higher probability of association with neuropathy (4). A particular variant of the short variable region (SVR) of the flagellum-encoding gene, flaA, has been proposed as a marker for C. jejuni strains which are likely to cause GBS or MFS (12).
Developments in the molecular characterization of C. jejuni isolates, specifically the application of multilocus sequence typing (MLST) (1) and nucleotide sequence typing of the flaA SVR (9), enable precise characterization and robust comparisons of the genetic relatedness of isolates via the Internet regardless of where the isolate characterization is done (8). Nucleotide sequences are especially amenable to phylogenetic approaches for the investigation of bacterial population structure and the forces which influence it. A collection of 25 GBS- and MFS-associated C. jejuni isolates was analyzed by MLST and flaA gene sequencing, and Penner and heat-labile (HL) serotyping data were obtained. The data were compared with equivalent information for more than 800 C. jejuni isolates held in a World Wide Web-accessible database (1). Eighteen of the twenty five isolates were previously described and had been shown to be diverse by a range of serological and genetic fingerprinting methods (2, 3), but the nature of the techniques used precluded the precise genetic comparisons of isolates which are possible with nucleotide sequence data. The remaining seven isolates were previously undescribed and comprised three from GBS patients from The Netherlands (isolates 23, 24, and 25), two siblings of a Dutch GBS patient (isolates 26 and 27), and two derived from patients with GBS from Curaçao and Bonaire (isolates 22 and 28, respectively).
Nucleotide sequence accession number.
The nucleotide sequences of the flaA SVRs of the isolates analyzed in this study are available under the GenBank accession numbers listed in Table 1.
TABLE 1.
MLST allelic profiles, Penner serotypes, HL serotypes, and clonal complexes
| Isolate | STa | Allelic profilea
|
Complexa | Serotype(s) (Penner)b | Serotype (HL)b | flaA SVR accession no.c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | ||||||
| 2248d | 21 | 2 | 1 | 1 | 3 | 2 | 1 | 5 | ST-21 | 2 | ND | |
| GB 13 | 53 | 2 | 1 | 21 | 3 | 2 | 1 | 5 | ST-21 | 2 | 4 | AF354556 |
| GB 14 | 19 | 2 | 1 | 5 | 3 | 2 | 1 | 5 | ST-21 | 2 | 4 | As GB13 |
| GB 11 | 148 | 2 | 1 | 6 | 3 | 2 | 1 | 5 | ST-21 | 2 | 4 | AF354548 |
| GB 5 | 185 | 2 | 1 | 1 | 3 | 1 | 1 | 5 | ST-21 | 4, 64 | 1 | AF354547 |
| GB 26 | 186 | 2 | 1 | 1 | 3 | 1 | 1 | 3 | ST-21 | 1, 44 | UT | AF354546 |
| GB 27 | 186 | 2 | 1 | 1 | 3 | 1 | 1 | 3 | ST-21 | 1, 44 | UT | As GB26 |
| GB 23 | 122 | 6 | 4 | 5 | 2 | 2 | 1 | 5 | ST-206 | 4, 13, 43 | 90 | As AF050196 |
| MF 6 | 48 | 2 | 4 | 1 | 2 | 7 | 1 | 5 | ST-48 | 4, 64 | 1 | As GB13 |
| GB 17 | 416 | 2 | 4 | 1 | 2 | 73 | 1 | 5 | ST-48 | 4, 13, 64 | UT | AF354549 |
| GB 2 | 22 | 1 | 3 | 6 | 4 | 3 | 3 | 3 | ST-22 | UT | 36 | As GB18 |
| GB 3 | 22 | 1 | 3 | 6 | 4 | 3 | 3 | 3 | ST-22 | 19, 24 | 77 | AF354560 |
| GB 18 | 22 | 1 | 3 | 6 | 4 | 3 | 3 | 3 | ST-22 | 19 | 77 | AF354559 |
| GB 28 | 149 | 1 | 3 | 33 | 4 | 54 | 3 | 3 | ST-22 | 19 | ND | AF355599 |
| GB 19 | 61 | 1 | 4 | 2 | 2 | 6 | 3 | 17 | ST-61 | 4, 50 | 7 | AF354561 |
| GB 16 | 61 | 1 | 4 | 2 | 2 | 6 | 3 | 17 | ST-61 | 13, 65 | UT | AF354558 |
| MF 8 | 42 | 1 | 2 | 3 | 4 | 5 | 9 | 3 | ST-42 | 23, 36 | 5 | As AF050195 |
| GB 15 | 63 | 9 | 17 | 2 | 10 | 22 | 3 | 6 | ST-52 | 5, 34 | UT | AF354554 |
| MF 7 | 83 | 10 | 27 | 43 | 7 | 6 | 18 | 7 | ST-403 | 35 | UT | AF354553 |
| GB 4 | 147 | 7 | 40 | 1 | 15 | 56 | 3 | 12 | ST-51 | 37 | 28 | AF354551 |
| MF 20 | 131 | 7 | 2 | 6 | 10 | 10 | 37 | 1 | UNe | 2 | 4 | AF354552 |
| GB 25 | 131 | 7 | 2 | 6 | 10 | 10 | 37 | 1 | UN | 2 | 4 | As MF20 |
| GB 22 | 64 | 1 | 6 | 15 | 24 | 12 | 28 | 1 | UN | 13, 64 | UT | AF354557 |
| GB 21 | 132 | 1 | 6 | 22 | 24 | 12 | 28 | 1 | UN | 13, 65 | 7 | AF354556 |
| GB 1 | 29 | 8 | 1 | 17 | 3 | 16 | 1 | 1 | UN | 1 | UT | As AF050186 |
| GB 24 | 39 | 2 | 3 | 2 | 11 | 19 | 3 | 6 | UN | 31 | 32 | AF354555 |
Sequence type, allele, and complex designations follow the MLST website (http://mlst.zoo.ox.ac.uk) (1).
UT, untypeable; ND, not determined.
GenBank designations of nucleotide sequences. Entries marked “as” indicate that the sequence present in that isolate was identical to a sequence preset in the isolate number given.
Previously published ST-21 isolate included for comparative purposes.
UN, currently unassigned to a lineage.
The MLST profiles demonstrated that the 25 isolates were genetically diverse, with 20 different sequence types (STs); these were assigned names and grouped into 13 distinct clonal complexes, which are thought to correspond to genetic lineages, using the previously published definitions (1) (Table 1). The high level of genetic diversity among these isolates was also reflected phenotypically in their large range of Penner and HL serotypes (Table 1). The level of diversity observed among the 25 isolates was similar to that present in the C. jejuni MLST database (http://mlst.zoo.ox.ac.uk/), which contained 814 isolates of human, animal, and environmental origin at the time of comparison. The most common lineage or clonal complex present in the MLST database was the ST-21 complex, which made up 260 of the 814 isolates (32%) at the time of comparison. While no representatives of ST-21 itself were present among the collection of 25 isolates, 6 of the 25 isolates were considered to be variants of this sequence type (Table 1). Of these 6 variants, 2 STs, ST-19 and ST-53, were frequently found in the database, with 30 and 19 isolates, respectively. A total of 4 isolates belonged to the ST-22 complex, which is associated with Penner serotype 19 and HL 36 and 77 (Table 1); this lineage is comparatively poorly represented in the database (12 of 814 isolates [1.5%]). The ST-61 and ST-48 complexes were represented twice, and the ST-42, ST-51, ST-52, ST-206, and ST-403 complexes once each—in all of these examples the abundance of these lineages among the neuropathy-associated isolates was consistent with their abundance in the database as a whole. The absence of members of the ST-45 complex, which was the second-most-common lineage represented in the database (74 of 814 isolates), may indicate that members of this complex have a low probability of association with GBS or MFS. The MLST data were compatible with previous analyses of these isolates (2). Isolates 13 and 14, which were previously reported to exhibit whole genome homology (4), belonged to distinct STs of the ST-21 complex, differing by only one nucleotide in the gltA locus, probably corresponding to a point mutation. Isolates MF20 and GB25 had identical STs and may both belong to an as yet undefined clonal complex. The same may be true for isolates GB22 and GB21, which shared six of the seven MLST loci.
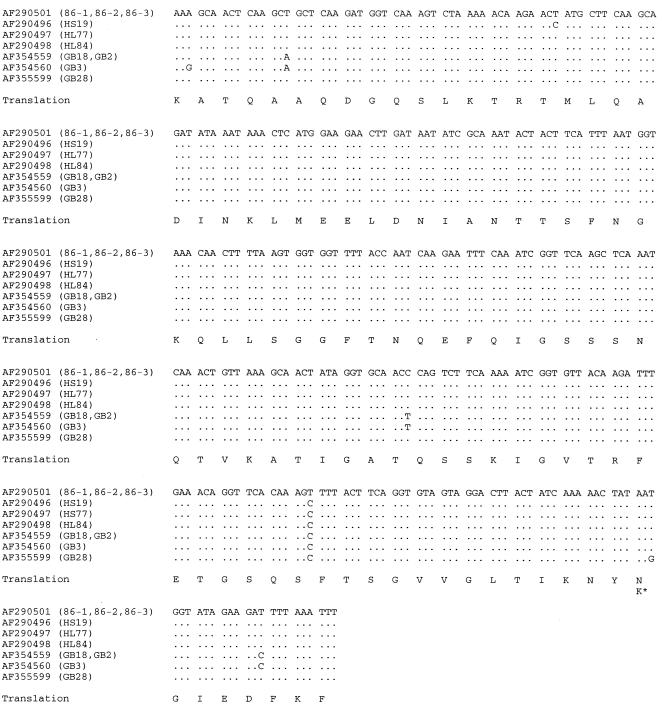
The nucleotide sequences of the flaA SVR present among the 25 isolates were diverse, with 20 distinct SVR nucleotide sequences (Table 1). The two most diverse sequences differed at 63 of 321 nucleotide sites (19.6%), corresponding to 16 differences (15.0%) among the 107 amino acids. The flaA sequences from the four ST-22 complex isolates were similar to each other and to the flaA sequences reported to be associated with GBS previously (0.3 to 1.6% nucleotide sequence differences with only one amino acid change [Fig. 1]) (12). These SVR sequences differed at 17 to 62 nucleotide sites (5.3 to 19.3%) and 6 to 16 amino acids (5.6 to 15.0%) from the remaining SVR sequences obtained for the isolates described here. The flaA SVR nucleotide sequence data provided slightly more discrimination than PCR-restriction fragment length polymorphism (RFLP) fingerprint patterns, with 14 unique sequences compared to 12 patterns by PCR-RFLP among the 18 previously described isolates (3). Isolates with identical PCR-RFLP patterns either had identical SVR sequences or differed at one nucleotide. Identical fla SVR sequences have been described for isolates which were not associated with GBS or MFS (data not shown).
FIG. 1.
Comparison of the unique flaA SVR nucleotide sequences present in the ST-22 complex isolates with SVR sequences proposed to be associated with GBS (12). Sequences are identified by their GenBank number, and the name of the isolate(s) in which the sequences have been described is given in brackets. The sequences are compared to the sequence under accession no. AF290501: a period (.) indicates identity with this sequence, a nucleotide substitution being represented by the appropriate letter. The amino acid translations of these sequences are shown by single-letter amino acid code, and the single nonsynonymous base change (asparagine to lysine) is indicated thus: K∗. The identical sequences for isolates 86-1, 86-2, and 86-3 are separately stored in GenBank under numbers AF290501, AF290502, and AF290503, respectively.
There was no detectable allele bias among the GBS- and MFS-associated isolates for MLST loci when they were compared with the database of more than 800 isolates (data not shown). Isolates GB 19 and GB 16 contained an uncA allele (uncA-17) which exhibited ∼90% nucleotide sequence identity with the other 11 uncA alleles present in the isolate collection, which shared ∼97% nucleotide sequence identity. This was likely to be due to the recruitment of the allele sequence from another related species, as has been observed previously with glyA alleles with high homology to Campylobacter coli sequences being found in some C. jejuni isolates (1). The allele uncA-17 has been identified in two C. coli isolates (data not shown). The allele sequences found at each locus of the GBS- and MFS-associated isolates were distributed among the C. jejuni alleles present in the MLST database in phylogenetic reconstructions made by several techniques (data not shown).
These data support the view that diverse C. jejuni genotypes, as measured by MLST and flaA SVR nucleotide sequences, and serotypes reflecting capsular (Penner) and flagellar (HL) antigens can be associated with neuropathy. The most common lineage was the ST-21 complex, which was also the most common lineage in the collection of mainly human enteritis-associated isolates available in the MLST database (http://mlst.zoo.ox.ac.uk). The next-most-common lineage among the isolates examined was the ST-22 complex, and, although the numbers are small, this complex was apparently overrepresented in the neuropathy-associated isolate collection relative to the isolates which are described in the database as a whole. It is possible from these data that the ST-45 complex is relatively underrepresented among neuropathy-associated isolates. These observations are consistent with the ST-22 complex being the Penner serotype 19 virulent clone proposed to be associated with GBS (4). It appears likely that the flaA SVR sequence that was proposed as a marker for GBS-associated C. jejuni strains may be a marker for this clonal complex; however, since this sequence was present in only 4 of the 25 neuropathy-associated isolates examined here, it is not a useful marker for all C. jejuni strains likely to cause GBS.
Acknowledgments
This work was funded by the United Kingdom Ministry of Agriculture Fisheries and Food, contract number OZ0604. This publication made use of the Multi Locus Sequence Typing website (http://mlst.zoo.ox.ac.uk) developed by Man-Suen Chan and sited at the University of Oxford. The development of this site is funded by the Wellcome Trust.
REFERENCES
- 1.Dingle K E, Colles F M, Wareing D R A, Ure R, Fox A J, Bolton F E, Bootsma H J, Willems R J L, Urwin R, Maiden M C J. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duim B, Ang C W, van Belkum A, Rigter A, van Leeuwen N W J, Endtz H P, Wagenaar P A. Amplified fragment length polymorphism analysis of Campylobacter jejuni strains isolated from chickens and from patients with gastroenteritis or Guillain-Barré or Miller Fisher syndrome. Appl Environ Microbiol. 2000;66:3917–3923. doi: 10.1128/aem.66.9.3917-3923.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endtz H P, Ang C W, van den Braak N, Duim B, Rigter A, Price L J, Woodward D L, Rodgers F G, Johnson W M, Wagenaar J A, Jacobs B C, Verbrugh H A, van Belkum A. Molecular characterization of Campylobacter jejuni from patients with Guillain-Barré and Miller Fisher syndromes. J Clin Microbiol. 2000;38:2297–2301. doi: 10.1128/jcm.38.6.2297-2301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujimoto S, Allos B M, Misawa N, Patton C M, Blaser M J. Restriction fragment length polymorphism analysis and random amplified polymorphic DNA analysis of Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome. J Infect Dis. 1997;176:1105–1108. doi: 10.1086/516522. [DOI] [PubMed] [Google Scholar]
- 5.Hughes R A, Hadden R D, Gregson N A, Smith K J. Pathogenesis of Guillain-Barré syndrome. J Neuroimmunol. 1999;100:74–97. doi: 10.1016/s0165-5728(99)00195-2. [DOI] [PubMed] [Google Scholar]
- 6.Hughes R A, Rees J H. Clinical and epidemiologic features of Guillain-Barré syndrome. J Infect Dis. 1997;176(Suppl. 2):S92–S98. doi: 10.1086/513793. [DOI] [PubMed] [Google Scholar]
- 7.Kuroki S, Saida T, Nukina M, Haruta T, Yoshioka M, Kobayashi Y, Nakanishi H. Campylobacter jejuni, strains from patients with Guillain-Barré syndrome belong mostly to Penner serogroup 19 and contain beta-N-acetylglucosamine residues. Ann Neurol. 1993;33:243–247. doi: 10.1002/ana.410330304. [DOI] [PubMed] [Google Scholar]
- 8.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meinersmann R J, Helsel L O, Fields P I, Hiett K L. Discrimination of Campylobacter jejuni by fla gene sequencing. J Clin Microbiol. 1997;35:2810–2814. doi: 10.1128/jcm.35.11.2810-2814.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molnar G K, Mertsola J, Erkko M. Guillain-Barré syndrome associated with Campylobacter infection. Br Med J. 1982;285:652. doi: 10.1136/bmj.285.6342.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachamkin I, Allos B M, Ho T. Campylobacter species and Guillain-Barré syndrome. Clin Microbiol Rev. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang R S, Figueroa G, Bryden L, Ng L K. Flagella as a potential marker for Campylobacter jejuni strains associated with Guillain-Barré syndrome. J Clin Microbiol. 2001;39:762–764. doi: 10.1128/JCM.39.2.762-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willison H J, O'Hanlon G M. The immunopathogenesis of Miller Fisher syndrome. J Neuroimmunol. 1999;100:3–12. doi: 10.1016/s0165-5728(99)00213-1. [DOI] [PubMed] [Google Scholar]
- 14.Yuki N, Yoshino H, Sato S, Miyatake T. Acute axonal poly-neuropathy associated with anti-GM1 antibodies following Campylobacter enteritis. Neurology. 1990;40:642–647. doi: 10.1212/wnl.40.12.1900. [DOI] [PubMed] [Google Scholar]