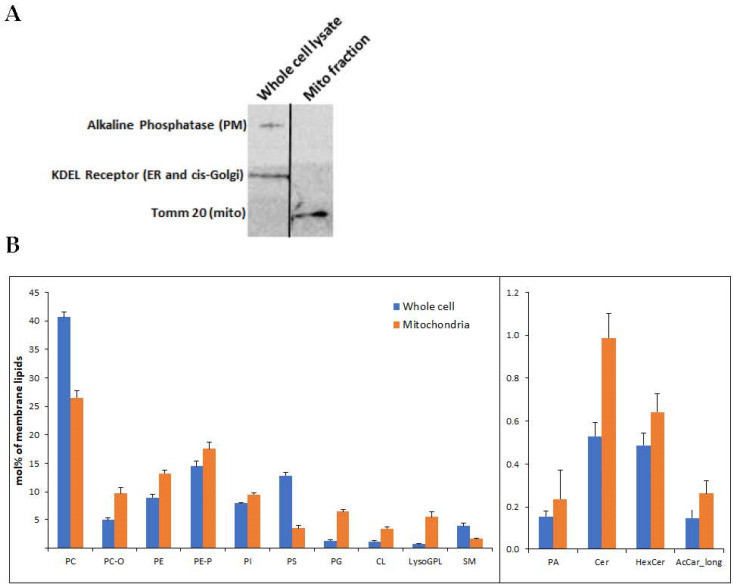
Figure 1.
(A) Mitochondria were isolated from U118MG cells according to Kappler et al. The mitochondrial isolate and an identical amount (20 μg) of the whole cell lysate were subjected to SDS-PAGE and immunoblotting using antibodies against the PM marker alkaline phosphatase, the endoplasmic reticulum marker KDEL sequence and the mitochondrial marker Tom20. Representative results from 3 different experiments are shown. (B) Lipidomics of whole cell input and isolated mitochondrial fraction. The enrichment of CL (p < 3.54 × 10−9) and PG (p < 2.68 × 10−6) can be observed in the mitochondrial fraction, which is also depleted of several PM markers, such as PC (p < 2.17 × 10−8) and PS (p < 2.91 × 10−12). The levels of the other lipids were also different between whole cells and isolated mitochondria: PC-O (p < 1.47 × 10−7), PE (p < 5.45 × 10−7), PE-P (p < 0.003), PI (p < 4.00 × 10−5), LysoGPL (p < 0.0004), SM (p < 0.001), PA (p < 0.01), Cer (p < 1.51 × 10−6), HexCer (p < 0.0004), AcCar_long (p < 0.101). Shown are mean ± SEM, n = 6. Differences determined by Student’s t-tests. PC and PC-O, phosphatidylcholine (diacyl and alkyl-acyl); PE and PE-P, phosphatidylethanolamine (diacyl and alkenyl-acyl); PI, phosphatidylinositol; PS, phosphatidylserine; PG, phosphatidylglycerol; CL, cardiolipin; LysoGPL, Lyso glycerophospholipids; SM, sphingomyelin; PA, phosphatidic acid; Cer, ceramide; HexCer, hexosyl ceramide; AcCar_long, long-chain acylcarnitine.