Abstract
A biprobe assay utilizing LightCycler technology was developed to detect rifampin resistance-associated gene mutations in the Mycobacterium tuberculosis rpoB gene. Three biprobes detected all mutations present in the 46 rifampin-resistant isolates. Wild-type sequences were correctly identified in each case. The method was reproducible, accurate, and easy to use.
Mycobacterium tuberculosis is responsible for three million deaths a year worldwide (10). Recommended treatment for tuberculosis (TB) comprises a combination of four drugs: rifampin, isoniazid, pyrazinamide, and ethambutol, with or without streptomycin (5). Resistance has emerged to all of these drugs, and multidrug resistance (MDR), in which the isolate is resistant to rifampin and to at least one other drug, is becoming more common (2). As most of the MDR strains are resistant to rifampin, the rapid detection of mutations within the rpoB gene may be used for presumptive identification of MDR-TB (14), allowing the correct treatment to be given promptly. Real-time PCR and probe analysis are ideally suited to the analysis of these mutations, allowing a rapid and accurate identification of drug resistance to be made.
Our approach utilizes biprobes coupled with real-time PCR on the LightCycler. The LightCycler is a real-time PCR machine that allows both rapid PCR cycling and continuous monitoring of product formation (15). Sybr Green I, an intercalating dye that fluoresces strongly when bound to double-stranded DNA, is included in the reactions so that when PCR products are formed, fluorescence increases (11). After PCR amplification the LightCycler can monitor the melting of the DNA as temperature increases by measuring a decrease in fluorescence as the Sybr Green I is released. The negative derivative of fluorescence is plotted against temperature to give a discrete melting peak.
Sequence-specific detection of the amplicons can be achieved with a fluorescently labeled biprobe. Biprobes are sequence-specific probes labeled with the fluorophore Cy5 and blocked with biotin at the 3′ end (1). When these probes are included in the reaction after the melt cycle, a peak corresponding to a decrease in fluorescence due to the release of the probe and a peak due to melting of the PCR product can be observed. When the probe binds to the complementary sequence in the PCR product, the Cy5 label is excited by the energy transfer from Sybr Green I, resulting in an increase of light emitted by Cy5. This fluorescence is measured at a wavelength different from that emitted by Sybr Green I and so can be distinguished. Biprobes, as well as hybridizing to perfectly matched sequences, will also bind when a mismatch (mutation) is present. When the biprobe binds to a mismatched sequence, a melting temperature lower than that of a perfectly matched sequence can be distinguished. We have designed three biprobes from the 81-bp region of the rpoB gene to detect mutations in four codons.
We examined 47 rifampin-resistant isolates and a rifampin-sensitive isolate of M. tuberculosis. These had been submitted to the Public Health Laboratory Service Mycobacterium Reference Unit and identified by standard methods. DNA was extracted from cells harvested from Lowenstein-Jensen slopes as described previously (9). A 270-bp fragment was amplified with primers rpob4 (5′-CCGCAGACGTTGATCAACA-3′) and rpob5 (5′-TACGGCGTTTCGATGAACC-3′) from all of the isolates and was sequenced using these primers according to the manufacturer's instructions. Sequence analysis identified nine different mutations affecting four codons within the rpoB gene (Table 1). Of the 48 isolates sequenced, 27 had a mutation affecting codon 531, 11 had a mutation affecting codon 526, 7 had a mutation affecting codon 516, and 1 had a mutation affecting codon 511. When compared to the published sequence, the rifampin-sensitive isolate and one of the rifampin-resistant isolates showed no mutations.
TABLE 1.
Position of mutations in isolates as determined by DNA sequencing
| Mutation position | Nucleotide change | Amino acid change | No. of mutations in this study |
|---|---|---|---|
| 531 | TCG→TTG | Ser→Leu | 23 |
| 531 | TCG→TGG | Ser→Trp | 4 |
| 526 | CAC→TAC | His→Tyr | 6 |
| 526 | CAC→GAC | His→Asp | 3 |
| 526 | CAC→CTC | His→Leu | 1 |
| 526 | CAC→TGC | His→Cys | 1 |
| 516 | GAC→GTC | Asp→Val | 6 |
| 516 | GAC→TTC | Asp→Phe | 1 |
| 511 | CTG→CCG | Leu→Pro | 1 |
A 183-bp amplicon was amplified with primers rpob1 (5′-AGGAGTTCTTCGGCACCAG-3′) and rpob2 (5′-GGGTTTCGATCGGGCACAT-3′) in a reaction mixture containing 1× PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl, and 2 mM magnesium chloride), a 200 μM concentration of each deoxynucleoside triphosphate, a 200 nM concentration of each primer, and 1 U of Taq DNA polymerase. After an initial denaturation step of 5 min at 95°C, PCR cycling was performed for 30 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The three biprobes were tested in a second-round, asymmetric PCR on the LightCycler using strains representing the different mutations and a rifampin-sensitive isolate. A second-round, asymmetric PCR was performed to preferentially amplify the DNA strand to which the biprobe would bind. PCRs (10 μl) contained 1 μl of the symmetric PCR product, Tris-HCl (pH 8.3) (500 mM), MgCl2 (5 mM), bovine serum albumin (1 mg/ml), deoxynucleoside triphosphates (a 200 μM concentration of each) (Gibco BRL), rpoB reverse primer (5′-G GCACGCTCGCGTGACA-3′) (5 pmol/μl), Platinum Taq polymerase (0.4 U) (Gibco BRL), Sybr Green I (Bio/Gene Ltd.) diluted 0.01%, and a biprobe (5 pmol/μl). The cycling parameters used were denaturation at 95°C for 20 s and 40 amplification cycles (temperature transition rate of 20°C/s) at 95°C for 2 s, 55°C for 1 s, 60°C for 15 s, and 74°C for 10 s. PCR cycling was followed by melting-curve analysis at 40 to 99°C (temperature transition rate of 0.2°C/s) with continuous fluorescence readings.
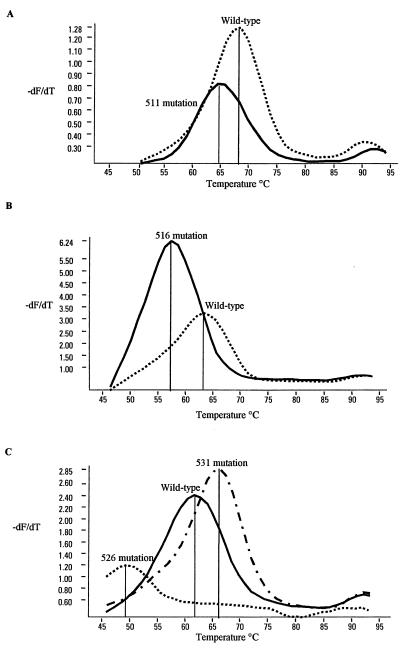
Biprobe A (5′ Cy5-CACCCAGCTGAGCCAATTC-biotin 3′) spanned the mutations at codon 511. The melt temperature of the normal sequence was determined to be 69°C, and when a mutation was present the melt temperature was found to be 65°C (Fig. 1A). Biprobe B (5′ Cy5-TTCATGGACCAGAACAACCC-biotin 3′) spanned codon 516, and when a mutation was present at codon 516, the melt temperature was found to be 57°C, compared to 64°C in a wild-type sequence (Fig. 1B). Biprobe C (5′ Cy5-GTTGACCCACAAGCGCC-biotin 3′) spanned the mutations at position 526, and three peaks were observed with this probe. The wild-type sequence melted at 61°C, the sequence containing a mutation at position 526 melted at 49°C, and a third melting peak which corresponded to a mutation at codon 531 was observed at 66°C (Fig. 1C). This third melting peak was confirmed by testing more strains known to have this mutation and was reliably reproduced.
FIG. 1.
Biprobe peaks observed with M. tuberculosis strains that have mutations at codons 511 (A) 516 (B), and 526 and 531 (C). −dF/dT is the first negative derivative of the change in fluorescence (dF) divided by the change in temperature (dT).
The presence and codon position of mutations in 48 M. tuberculosis strains determined by melting-peak analysis using the three biprobes correlated with the DNA sequence analysis. Each of the 46 resistant isolates showed an altered melting peak with only one of the three biprobes. Wild-type melting peaks were obtained with the other two biprobes for each of the 46 resistant isolates. For the rifampin-sensitive isolate and one rifampin-resistant isolate, wild-type melting peaks were observed with all three biprobes.
We have described an accurate and reproducible test for identification of the main rpoB mutations that cause rifampin resistance in M. tuberculosis. The real-time PCR assays can be completed in just 30 min, and all three probes can be run under the same cycling conditions. MDR-TB isolates are being isolated more frequently, and it is increasingly important to identify rifampin-resistant isolates rapidly. A previous study by Torres et al. (13) utilized real-time PCR and hybridization probes designed to bind to the mismatched sequence to detect two of the four mutations in the rpoB gene. Biprobes are very flexible in that they are designed to bind to perfectly matched sequences but will also bind when a mismatch is present. This allows unknown mutations to be detected. Sequencing data from other studies (3, 4, 6, 7, 8, 12) and our own data indicate that approximately 98% of mutations that give the resistant phenotype would be detected by our biprobe system. This system could also be readily applied to examination of clinical material, for example, by analyzing samples from rapid liquid cultures. LightCycler technology is also amenable to automation, making it useful for routine clinical use.
Acknowledgments
This work was partially funded by the Biotechnology and Biology Research Scientific Council (BBSRC).
REFERENCES
- 1.Bio/Gene Ltd. and the Secretary of State for Defense. July 1999. Nucleic acid detection system. British patent GB2333359A.
- 2.Brown T J, Tansel O, French G L. Simultaneous identification and typing of multi-drug-resistant Mycobacterium tuberculosis isolates by analysis of pncA and rpoB. J Med Microbiol. 2000;49:651–656. doi: 10.1099/0022-1317-49-7-651. [DOI] [PubMed] [Google Scholar]
- 3.Caugant D A, Sandven P, Eng J, Jeque J T, Tonjum T. Detection of rifampicin resistance among isolates of Mycobacterium tuberculosis from Mozambique. Microb Drug Resist. 1995;1:321–326. doi: 10.1089/mdr.1995.1.321. [DOI] [PubMed] [Google Scholar]
- 4.De Beenhouwer H, Lhiang Z, Jannes G, Mijs W, Machtelinckx L, Rossau R, Traore H, Portaels F. Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 5.Joint Tuberculosis Committee of the British Thoracic Society. Chemotherapy and management of tuberculosis in the United Kingdom: recommendations. Thorax. 1998;53:536–548. [PMC free article] [PubMed] [Google Scholar]
- 6.Kapur V, Li L-L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Mussen J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapur V, Li L L, Hamrick M R, Plikaytis B B, Shinnick T M. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch Pathol Lab Med. 1995;119:131–138. [PubMed] [Google Scholar]
- 8.Morris S, Bai G H, Suffys P, Portillo-Gomez L, Fairchok M. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 9.Patel S, Wall S, Saunders N A. Heminested inverse PCR for IS6110 fingerprinting of Mycobacterium tuberculosis strains. J Clin Microbiol. 1996;34:1686–1690. doi: 10.1128/jcm.34.7.1686-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raviglione M C, Snider D E, Kochi A. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 11.Ririe K M, Rasmussen R P, Wittwer C T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 12.Telenti A, Imboden P, Marchesi F, Schmidheini T, Bodmer T. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob Agents Chemother. 1993;37:2054–2058. doi: 10.1128/aac.37.10.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres M J, Criado A, Palomares J C, Aznar J. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:3194–3199. doi: 10.1128/jcm.38.9.3194-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vareldzis B P, Grosset J, Crofton J, Laszlo A. Drug-resistant tuberculosis: laboratory issues. World Health Organization recommendations. Tuber Lung Dis. 1994;75:1–7. doi: 10.1016/0962-8479(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 15.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]