Abstract
We used PCR to detect hepatitis C virus (HCV) RNA among supernatants of ground Culex quinquefasciatus mosquitoes that (i) had been fed HCV-positive blood, (ii) had been intrathoracically inoculated with HCV-positive blood, or (iii) were from homes of hepatitis C patients. HCV RNA was detectable under all three conditions, but it did not replicate in mosquitoes and was not detectably transmitted during feeding.
Hepatitis C virus (HCV) is an RNA virus causing sporadic non-A, non-B hepatitis. Other HCV transmission routes may exist in addition to the obvious parenteral routes of transmission. HCV, pestivirus, and flavivirus are of the family Flaviviridae, containing over 60 arthropod-borne viruses. Many flaviviruses (causing yellow fever, Japanese encephalitis, dengue) are transmitted by mosquitoes. HCV has been proven to replicate in mosquito cells (R. Germi, J. M. Crance, and D. Garin, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. A-16790, 2000). Therefore, we investigated possible mosquito-borne HCV replication and transmission. Taiwan is subtropical. Culex quinquefasciatus is common in tropical and subtropical areas of Asia, Africa, and America. Because our preliminary work detected HCV RNA in C. quinquefasciatus more than in Aedes aegypti or Aedes albopictus, C. quinquefasciatus was studied and is described herein.
A continuous laboratory colony of C. quinquefasciatus (Jin-Men strain) was maintained. Four-day-old females, deprived of 10% sucrose solution meal for 24 h, were tested. The feeding apparatus was from Wade (6). HCV-positive whole blood, confirmed through nested PCR, was stored at 4°C and heated to 37°C for feedings.
Twenty 4-day-old female mosquitoes were ground separately, immediately (day zero) after feeding on HCV-positive blood. Supernatant solutions were prepared. An additional 260 mosquitoes were similarly tested from days 1 to 30 after feeding. Another group of mosquitoes (n = 90) was infected by intrathoracic inoculation (0.96 μl) using pulled disposable pipette needles (4). Supernatants were prepared and tested as described above for a similar number of days. A total of 49 female C. quinquefasciatus mosquitoes, collected at three homes of patients with chronic HCV infection, were tested as described above. Of these 49 mosquitoes, 27 had recently fed. Supernatants of all mosquitoes were tested for both positive- and negative-stranded HCV RNA by PCR.
Oral transmission was tested by inserting mosquito proboscises into capillary pipettes containing 3.5 μl of a solution (10% fetal bovine serum, 10% sucrose) for 60 to 90 min (5). Pipette contents were tested for HCV RNA. Groups of 15 mosquitoes were tested at 9, 11, 17, 21, and 25 days (75 mosquitoes in total) after HCV-positive-blood feeding, for biological oral transmission. Groups of 30 mosquitoes were tested, immediately after and 10 and 30 min after HCV-positive-blood feeding, for mechanical oral transmission via small quantities of blood on mosquito proboscises.
The mosquito supernatant RNA extraction was performed using a modified single-step RNA isolation method. Positive-stranded HCV RNA cDNA was synthesized at 37°C, for 3 h, with antisense primer 5′-AACACTACTCGGCTAGCAGT-3′ and moloney murine leukemia virus reverse transcriptase. Negative-stranded HCV RNA cDNA was synthesized similarly using sense primer 5′-ACTCCACCATAGATCATCCC-3′. Products were boiled to inactivate reverse transcriptase activity and then treated with RNase A to digest all RNA molecules and avoid possible amplification of positive-stranded HCV RNA (1). Appropriate controls were included with each assay. Nested PCR amplification from the 5′ noncoding region was performed with corresponding primer pairs (outer primers: 5′-ACTCCACCATAGATCATCCC-3′, nucleotides [nt] 23 to 42, and 5′-AACACTACTCGGCTAGCAGT-3′, nt 264 to 245; inner primers: 5′-TTCACGCAGAAAGCGTCTAG-3′, nt 62 to 81, and 5′-GTTGATCCAAGAAAGGACCC-3′, nt 206 to 187). Aliquots (15 μl) of the PCR products were analyzed by agarose gel electrophoresis. The detection sensitivity of the nested PCR was about 10 to 100 HCV genome equivalents.
To confirm PCR products as specific and full-length HCV genomes, extracted RNA was tested with four other primer sets specific for the core (outer primers: 5′-CGCGCGACTAGGAAGACTTC-3′, nt 480 to 499, and 5′-ATGTACCCCATGAGGTCGGC-3′, nt 751 to 732; inner primers: 5′-AGGAAGACTTCCGAGCGGTC-3′, nt 489 to 508, and 5′-GAGCCATCCTGCCCACCCCA-3′, nt 632 to 613), E1/E2 (outer primers: 5′-TGGCTTGGGACATGATGATG-3′, nt 1294 to 1313, and 5′-TCACAACGCTCTCCTCGGGT-3′, nt 2299 to 2280; inner primers: 5′-TGCTCCGGATCCCTCAAGC-3′, nt 1351 to 1369, and 5′-TGATGTGCCAGCTGCCATTG-3′, nt 1608 to 1589), NS3 (outer primers: 5′-GTAACACGTGTGTCAC-3′, nt 4702 to 4717, and 5′-GCATGCCATGATGTAT-3′, nt 5285 to 5270; inner primers: 5′-GACAGTCGACTTCAGCTT-3′, nt 4721 to 4738, and 5′-TGGTTATGGGGTGCGTGA-3′, nt 5268 to 5251) and NS5 (outer primers: 5′-CATGTAAACCTCTCCTACGG-3′, nt 6748 to 6767, and 5′-TAGCAAGCTCTGCCAAAGCA-3′, nt 7386 to 7367; inner primers: 5′-AATACGTGGTTGGGTCACAG-3′, nt 6798 to 6817, and 5′-CCTCTTTCTCCGTGGAGGTG-3′, nt 7334 to 7315) HCV regions. We tested two mosquitoes with each primer set, 1 and 2 days after HCV-positive-blood feeding with the blood used in the laboratory feeding, a mosquito trapped in a hepatitis C patient's home, and the patient's serum. PCR products were purified and sequenced directly using sense and nested primers with a cycle-sequencing protocol that incorporated fluorescent dye terminators via automatic sequencer analysis and PILEUP software.
As measured in a branched DNA assay, the virus titer of the whole blood used for feeding mosquitoes was 7.908 Meq/ml. The HCV RNA detection rates in the mosquitoes were 100% (40 of 40) and 13% (32 of 240) within 1 day and 2 to 30 days after HCV-positive-blood feeding, respectively, compared to 70% (14 of 20) and 11% (8 of 70) after intrathoracic inoculation. HCV RNA could be detected in some mosquitoes up to 25 days after feeding. For mosquitoes trapped in hepatitis C patients' homes, the detection rate was 11% (3 of 27). Negative-stranded HCV RNA was undetectable at any time in any case, indicating a lack of HCV RNA replication. Tests of biological and mechanical HCV transmission yielded negative results.
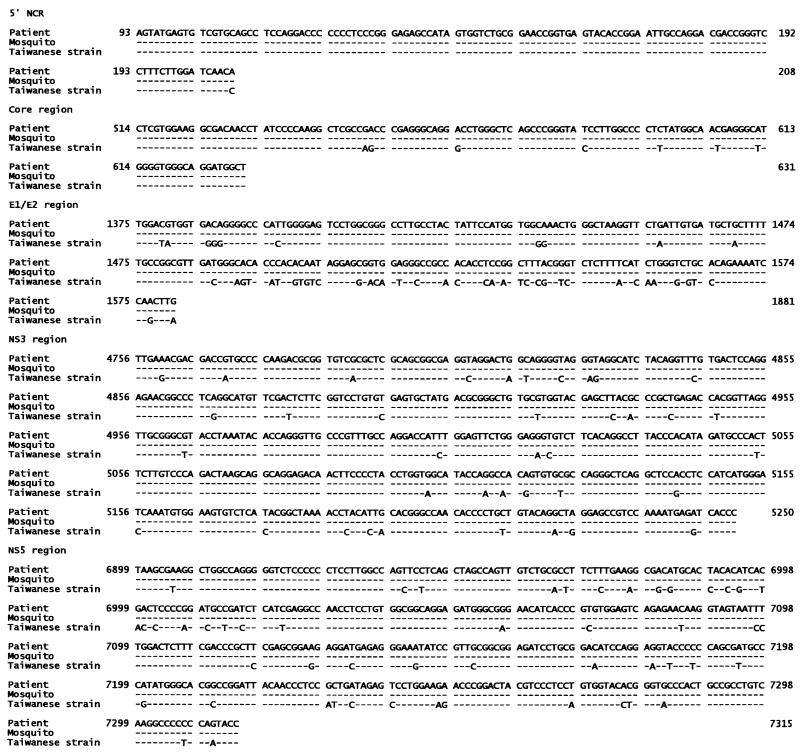
In sequence analysis and comparison of PCR products for five HCV regions, all PCR products were found to be HCV positive except for one day-2 mosquito, which tested HCV negative for the NS5 region. All HCV isolated from mosquitoes was genomically identical to HCV in the source blood. Homology with the Taiwanese strain (2) in the five regions was 99.1, 94.1, 76.8, 92.3, and 88.0%, respectively (Fig. 1), confirming that the PCR products were specific and represented the full-length HCV genome.
FIG. 1.
Nucleotide sequence alignment of the 5′ noncoding region, core, E1/E2, NS3, and NS5 fragments of HCV from a patient and mosquitoes. The sequences are compared with the Taiwanese strain of HCV, as published by Chen et al. (2) (EMBL/GenBank M84754). The nucleotide numbering system is according to that of GenBank sequence number M84754.
This study showed HCV existence in the majority of C. quinquefasciatus organisms studied within 1 day after HCV-positive-blood feeding and intrathoracic inoculation, with the latter exposure route included to avoid possible midgut HCV barriers. The HCV detection rate declined rapidly, consistent with findings in mosquitoes from hepatitis C patients' homes and other reports concerning hepatitis B virus (HBV) and C. quinquefasciatus (3), although HCV could be detected in some mosquitoes for up to 25 days. Furthermore, we showed no HCV replication in these mosquitoes, even when the midgut was bypassed by employing intrathoracic inoculation. This evidence strongly weighs against mosquitoes being reservoirs of HCV.
Arthropod vectors have been suspected in mechanical HBV transmission (7). However, our tests of biological and mechanical HCV transmission by C. quinquefasciatus showed negative results. Low HCV RNA titers in patient sera and different species tropisms for HCV RNA replication are probably the reasons why mechanical and biological HCV transmission does not occur in mosquitoes. It seems C. quinquefasciatus is not an HCV risk factor.
Acknowledgments
This work was supported by grants NSC85-2331-B006-024-MH and NSC86-2315-B006-001-MH from the National Science Council.
REFERENCES
- 1.Chang T T, Young K C, Yang Y J, Lai K A, Wu H L, Wu M H, Chen M Y, Lin X Z, Lin C Y, Shin J S. Incidence of post-transfusion hepatitis in Taiwan before and after introduction of anti-HCV testing. Liver. 1996;16:201–206. doi: 10.1111/j.1600-0676.1996.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 2.Chen P J, Lin M H, Tai K F, Liu P C, Lin C J, Chen D S. The Taiwanese hepatitis C virus genome: sequence determination and mapping the 5′termini of viral genomic and antigenomic RNA. Virology. 1992;188:102–113. doi: 10.1016/0042-6822(92)90739-c. [DOI] [PubMed] [Google Scholar]
- 3.Fouche A, Crewe R M, Windsor I M, Karim S S. Persistence of hepatitis B antigen in Culex quinquefasciatus (Diptera: Culicidae) J Med Entomol. 1994;27:697–700. doi: 10.1093/jmedent/27.4.697. [DOI] [PubMed] [Google Scholar]
- 4.Rosen L, Gubler D. The use of mosquitoes to detect and propagate dengue virus. Am J Trop Med Hyg. 1974;23:1153–1160. doi: 10.4269/ajtmh.1974.23.1153. [DOI] [PubMed] [Google Scholar]
- 5.Schoepp R J, Beaty B J, Eckels K H. Dengue 3 virus infection of Aedes albopictus and Aedes aegypti: comparison of parent and progeny candidate vaccine viruses. Am J Trop Med Hyg. 1990;42:89–96. doi: 10.4269/ajtmh.1990.42.89. [DOI] [PubMed] [Google Scholar]
- 6.Wade J O. A new design of membrane feeder incorporating an electrical blood stirring device. Ann Trop Med Parasitol. 1976;70:113–120. doi: 10.1080/00034983.1976.11687102. [DOI] [PubMed] [Google Scholar]
- 7.Zebe H, Sanwald R, Ritz E. Insect vectors in serum hepatitis. Lancet. 1972;i:1117–1118. doi: 10.1016/s0140-6736(72)91452-3. [DOI] [PubMed] [Google Scholar]