Abstract
Molecular typing of isolates revealed that neonatal coagulase-negative staphylococcal (CONS) septicemia is most frequently caused by predominant, antibiotic-resistant CONS types, which are widely distributed among both neonates and staff of the neonatal unit, suggesting cross-contamination. Therefore, infection control measures may be valuable in the prevention of this common nosocomial septicemia.
Increasing methicillin resistance has been observed among coagulase-negative staphylococcal (CONS) blood isolates in neonates (1, 4, 5, 10). Specific strains of CONS circulate in neonatal intensive care units (NICUs) for prolonged periods of time (6, 7, 12). In a previous study in our NICU, molecular typing revealed that 45% of the CONS strains causing septicemia belonged to a single type (11). Persisting CONS strains may possess specific characteristics, like resistance factors which enable them to survive and persist in the NICU, colonizing and subsequently causing septicemia, rather than being incidentally occurring strains. We studied the distribution of CONS strains in an NICU among neonates and hospital staff and also in the NICU environment (air) during a restricted period of 10 weeks to assess the relatedness of strains and to detect niches that are more likely to harbor strains causing infection.
During a period of 10 weeks, CONS strains were collected weekly from colonization sites of all admitted neonates (throat and skin), attending hospital staff (nose and hands), and from the NICU environment (air, by using settling plates on two locations during 2-h periods) (8). Blood isolates from cases of neonatal septicemia were included. Identification as CONS was based on Gram staining, production of catalase, and the absence of the coagulase gene. Detection of coagulase and mecA genes was performed by multiplex PCR (9). CONS strains were typed by randomly amplified polymorphic DNA (RAPD) analysis by using 0.5 μg of genomic DNA prepared by the Miniprep method (2) and 50 pmol of primer ERIC2 (5′AGTAAGTGACTGGGGTGAGCG-3′) in each PCR amplification (12). The resulting banding patterns were measured, compared, and assigned type names. Patterns differing by one band were considered different types. Differences in band intensity were not taken into consideration (3).
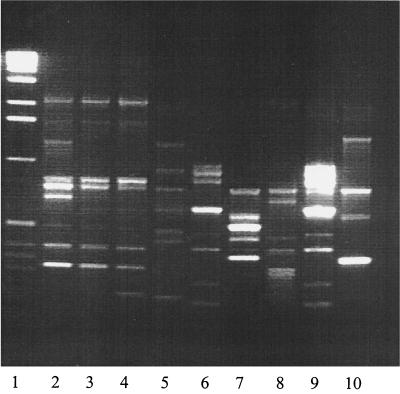
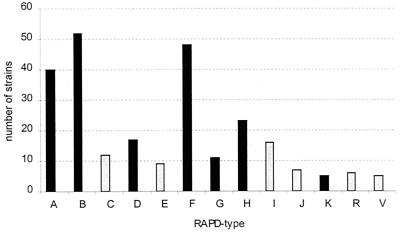
A selection of 282 out of a total of 539 CONS strains were included, comprising all blood isolates from 8 neonates with septicemia (9 strains), a selection of 163 strains from the colonization sites of the 53 neonates (62 strains), at least 1 set of strains (1 from hand, 1 from nose) from 56 members of the staff (142 strains), and all strains from the air (69). The mecA gene was detected in 127 strains (45%), comprising 18 of the 69 air samples (26%), 55 of the 142 staff samples (39%), 47 of the 62 neonatal colonization site samples (76%) and 7 of 9 neonatal blood strains (78%). RAPD typing revealed 41 different molecular types. Among the strains from the neonatal colonizing sites, 14 different types were identified; from the staff, 27 types were identified; and from the NICU air samples, 25 types were identified. RAPD typing of the nine blood isolates yielded seven different banding patterns (Fig. 1). Seventeen of the 41 RAPD types were unique and 13 of the other 24 were more prevalent, being isolated ≥5 times, whereas 8 of these were isolated ≥10 times (types A, B, C, D, F, G, H, and I). RAPD types A, B, and F were most frequently identified (40, 51, and 47 times, respectively) (Fig. 2). Eight of the nine septicemia strains belonged to the eight RAPD types isolated ≥10 times. Four of the blood isolates belonged to the three types isolated most frequently (≥40 times): RAPD types A, B, and F. The distribution of the RAPD types of CONS strains among neonates, staff, and NICU air and of mecA gene carriage is shown in Table 1. RAPD type D was predominant among neonates (14 of 17), RAPD type A (22 of 40) and particularly types B (38 of 52) and F (42 of 47) predominated among the staff, and RAPD type H was predominant among the NICU air strains (14 of 23). The highest mecA gene carriage was found in neonatal CONS strains: 7 of 8 blood strains and 44 of 50 neonatal colonizing strains (both 88%) were mecA positive (Table 1). Fifty-three of 120 CONS strains from the staff (44%) were mecA positive and 9 of 38 strains (24%) from the NICU air were positive. Although the numbers are low, this distribution of mecA gene carriage seems to be independent of molecular type and more related to the site of isolation, which is illustrated in the case of RAPD type A and B strains: 16 of 16 (100%) neonatal RAPD type A strains were mecA positive, as opposed to 18 of 22 (82%) positive strains from the staff and none of the 2 NICU air strains. Eight of 9 neonatal RAPD type B strains (89%) were mecA positive, as opposed to 10 of the 38 (26%) staff strains and 2 of the 4 (50%) NICU air strains.
FIG. 1.
Agarose gel containing the amplification products of all neonatal blood CONS strains. Lane 1, molecular weight marker; lanes 2 to 10, neonatal blood isolates. The banding patterns of lanes 3 and 4 are identical (RAPD type A), as are those of lanes 6 and 9 (RAPD type D).
FIG. 2.
Frequency of the 13 most prevalent RAPD types of CONS strains distinguished among 282 strains derived from neonates, hospital staff, and the environment (NICU air). Types A, B, D, F, G, H, and K were also found in neonatal blood strains (dark bars).
TABLE 1.
Distribution of the eight most frequent RAPD types of CONS strains, isolated ≥10 times, and their mecA gene carriage
| RAPD type | No. of mecA-positive strains/total no. (%) from:
|
|||
|---|---|---|---|---|
| Neonate colonization sitesa | Blood of neonates with septicemia | Medical staff colonization sitesb | NICU air | |
| A | 14/14 (100) | 2/2 (100) | 18/22 (82) | 0/2 (0) |
| B | 7/8 (88) | 1/1 (100) | 10/38 (26) | 2/4 (50) |
| C | 5/5 (100) | 2/2 (100) | 2/5 (40) | |
| D | 11/12 (92) | 2/2 (100) | 0/1 (0) | 2/2 (100) |
| F | 1/1 (100) | 18/42 (43) | 1/4 (25) | |
| G | 3/5 (60) | 0/1 (0) | 2/3 (67) | 1/2 (50) |
| H | 1/1 (100) | 1/1 (100) | 0/7 (0) | 1/14 (7) |
| I | 3/5 (60) | 3/5 (60) | 0/5 (0) | |
| Total | 44/50 (88) | 7/8 (88) | 53/120 (44) | 9/38 (24) |
Skin and throat.
Hand and nose.
In the present study, we have examined the molecular type and mecA gene carriage of CONS strains isolated from neonates, hospital staff, and the NICU environment in an attempt to determine whether there is any relation between the circulation of specific CONS strains, the propensity to cause septicemia, and methicillin resistance. Although a large number of molecular types of CONS strains circulated in the NICU among neonates, staff, and in the air, clearly a small number of types predominated. The eight most prevalent types (isolated ≥10 times) comprised 55% (38 of 69) of the NICU air strains, 85% (120 of 142) of the staff colonizing strains, 81% (50 of 62) of the neonatal colonizing strains, and 89% (8 of 9) of the neonatal blood strains. The persistence of these strains in the NICU could not be adequately assessed because the study period was restricted to 10 weeks. It has been demonstrated that strains can persist for a year or longer (6, 7, 11, 12).
A striking pattern in mecA gene carriage of the CONS strains was found, with the highest carriage occurring among the neonatal strains (76% of the colonizing strains and 78% of the septicemia strains) and lower carriage occurring among the colonizing strains of the staff (39%) and from the NICU environment (26%). The high mecA gene carriage of the neonatal CONS strains and particularly of the blood strains is probably an effect of selection by antibiotic usage, which is usually high in NICUs. Although appropriate assessment was impossible due to the low sample numbers, the distribution of mecA gene carriage seemed independent of molecular type, as illustrated by the distribution of mecA gene carriage in RAPD types A and B, both of which are frequently found on neonates and staff, but with the highest mecA gene carriage in neonates. It is likely that predominant RAPD types of CONS strains with high mecA gene carriage persist in the NICU. In a recent study among CONS strains isolated from clinical specimens from neonates during periods in 1996, 1997, and 1998, pulsed-field gel electrophoretic typing showed four main clones of CONS which were more antibiotic resistant than the sporadic strains and appeared to persist in the NICU, indicating transmission among patients (13). These findings were confirmed in the present study. Moreover, we found evidence for transmission via hospital staff and persistence in the NICU air. In addition, we showed that of a specific type, the highest proportion of mecA-positive CONS strains was found among those strains isolated from neonates (blood or colonizing), suggesting selection of resistant strains by antibiotic pressure. Both factors, cross-contamination with predominant, resistant strains and selection by antibiotic pressure, are potentially preventable by strict adherence to hospital infection control measures and restrictive antibiotic use.
In conclusion, neonatal septicemia is most frequently caused by predominant CONS strains, with high mecA gene carriage, probably due to antibiotic selection. These strains were frequently distributed among neonates and staff, suggesting cross-contamination of resistant strains. Strict adherence to hospital infection control measures and restricted antibiotic usage are potential strategies in the prevention of nosocomial CONS septicemia in neonatal intensive care units.
REFERENCES
- 1.Archer G L, Climo M W. Antimicrobial susceptibility of coagulase-negative staphylococci. Antimicrob Agents Chemother. 1994;38:2231–2237. doi: 10.1128/aac.38.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Boston, Mass: John Wiley & Sons; 1995. p. 2.4.1. [Google Scholar]
- 3.Bingen E, Barc M C, Brahimi N, Vilmer E, Beaufils F. Randomly amplified polymorphic DNA analysis provides rapid differentiation of methicillin-resistant coagulase-negative staphylococcus bacteremia isolates in pediatric hospital. J Clin Microbiol. 1995;33:1657–1659. doi: 10.1128/jcm.33.6.1657-1659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers H F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackbarth C J, Chambers H F. Methicillin-resistant staphylococci: detection methods and treatment of infections. Antimicrob Agents Chemother. 1989;33:995–999. doi: 10.1128/aac.33.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huebner J, Pier G B, Maslow J N, Muller E, Shiro H, Parent M, Kropec A, Arbeit R D, Goldmann D A. Endemic nosocomial transmission of Staphylococcus epidermidis bacteremia isolates in a neonatal intensive care unit over 10 years. J Infect Dis. 1994;169:526–531. doi: 10.1093/infdis/169.3.526. [DOI] [PubMed] [Google Scholar]
- 7.Low D E, Schmidt B K, Kirpalani H M, Moodie R, Kreiswirth B, Matlow A, Ford-Jones E L. An endemic strain of Staphylococcus haemolyticus colonizing and causing bacteremia in neonatal intensive care unit patients. Pediatrics. 1992;89:696–700. [PubMed] [Google Scholar]
- 8.McGowan J E, Jr, Weinstein R A. The role of the laboratory in control of nosocomial infection, p 187–220. In: Bennett J V, Brachman P S, editors. Hospital infection. Boston, Mass: Little, Brown and Company; 1992. [Google Scholar]
- 9.Schmitz F J, Hofmann B, Verhoef J, Finken-Eigen M, Heinz H P, Kohrer K. Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by a multiplex PCR. J Med Microbiol. 1997;46:773–778. doi: 10.1099/00222615-46-9-773. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki E, Hiramatsu K, Yokota T. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob Agents Chemother. 1992;36:429–434. doi: 10.1128/aac.36.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermont C L, Hartwig N G, Fleer A, de Man P, Verbrugh H, van den Anker J, de Groot R, van Belkum A. Persistence of clones of coagulase-negative staphylococci among premature neonates in neonatal intensive care units: two-center study of bacterial genotyping and patient risk factors. J Clin Microbiol. 1998;36:2485–2490. doi: 10.1128/jcm.36.9.2485-2490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villari P, Sarnataro C, Iacuzio L. Molecular epidemiology of Staphylococcus epidermidis in a neonatal intensive care unit over a three-year period. J Clin Microbiol. 2000;38:1740–1746. doi: 10.1128/jcm.38.5.1740-1746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]