Abstract
This report describes the application of PCR fingerprinting for the identification of species and varieties of common dermatophytes and related fungi utilizing as a single primer the simple repetitive oligonucleotide (GACA)4. The primer was able to amplify all the strains, producing species-specific profiles for Microsporum canis, Microsporum gypseum, Trichophyton rubrum, Trichophyton ajelloi, and Epidermophyton floccosum. Intraspecific variability was not observed for these species. Instead, three different profiles were observed in the Trichophyton mentagrophytes group.
Routine procedures for dermatophyte species identification rely on examination of the colony (pigmentation of the surface and reverse sides, topography, texture, and rate of growth) and microscopic morphology (size and shape of macroconidia and microconidia, spirals, nodular organs, and pectinate branches). Further identification characteristics include nutritional requirements (vitamins and amino acids) and temperature tolerance, as well as urease production, alkaline production of bromocresol purple medium, in vitro hair perforation, etc. (9, 16). Morphological and physiological characteristics can frequently vary; in fact, the phenotypic features can be easily influenced by outside factors such as temperature variation, medium, and chemotherapy (11) and therefore strain identification is often difficult.
In the last few years genotypic approaches have proven to be useful for solving taxonomic problems regarding dermatophytes; in fact, genotypic differences are considered more stable and more precise than phenotypic characteristics (2, 11).
Molecular methods, such as restriction fragment length polymorphism analysis of mitochondrial DNA (1, 7, 8), sequencing of the internal transcribed spacer (ITS) region of the ribosomal DNA (3, 4), sequencing of protein-encoding genes (5, 6), and PCR (random amplification of polymorphic DNA [RAPD] [15], arbitrarily primed PCR [AP-PCR] [10, 11], and PCR fingerprinting [2]), have brought important progress in distinguishing between species and strains. However, most of these techniques (e.g., restriction fragment length polymorphism analysis, sequencing) are complex, laborious, time-consuming, and not easily employable for routine identification of dermatophytes; in contrast, PCR technology is simple, rapid, and, in the absence of specific nucleotide sequence information for the many dermatophyte species, able to generate species-specific or strain-specific DNA polymorphisms on the basis of characteristic band patterns detected by agarose gel electrophoresis (2, 11).
This report describes the application of PCR fingerprinting for the identification of species and varieties of common dermatophytes and related fungi utilizing as a single primer the simple repetitive oligonucleotide (GACA)4 previously used by Meyer and others to distinguish strains of Cryptococcus neoformans (12, 13) and species of the genus Candida (14).
The species and varieties we have studied are Microsporum canis, Microsporum gypseum, Trichophyton ajelloi, Trichophyton mentagrophytes var. asteroides, T. mentagrophytes var. granulosum, T. mentagrophytes var. lacticolor, T. mentagrophytes var. radians, T. mentagrophytes of undetermined variety, Trichophyton interdigitale, Trichophyton rubrum, and Epidermophyton floccosum.
Furthermore, the study was conducted on various strains both from collections and from clinical isolation with the aim of finding the presence of an intraspecific variability.
Strains.
Out of a total of 140 strains selected for the study, 29 were obtained from the collection of the Institut Pasteur of Paris, France. One hundred eleven clinical isolates were recovered from humans with dermatophytosis as well as from cats and dogs with or without visible lesions. The clinical strains were isolated in Florence (Department of Public Health—Microbiology Unit and Department of Dermatological Sciences) and Pisa (Department of Animal Pathology), Italy, during 1997 and 1998 and identified using conventional culture and microscopic techniques.
The origins of the strains are listed in Tables 1 and 2.
TABLE 1.
Reference strains (collection of Institut Pasteur) investigated in this study
| Species | Institut Pasteur collection no. | Origin |
|---|---|---|
| Microsporum spp. | ||
| M. canis | 1687-87 | Human |
| 2144-93 | Cat | |
| 2145-93 | Cat | |
| 2289-94 | Human | |
| M. gypseum | 1463-83 | Human |
| 2143-93 | Dog | |
| Trichophyton spp. | ||
| T. ajelloi | 1469-83 | Unspecified |
| 2253-94 | Unspecified | |
| T. mentagrophytesa | 401-69 | Human |
| 404-56 | Human | |
| 407-74 | Dog | |
| 877-71 | Human | |
| 1468-83 | Human | |
| T. mentagrophytes var. asteroides | 402-69 | Human |
| T. mentagrophytes var. granulosum | 1182-79 | Human |
| 1711-88 | Human | |
| T. mentagrophytes var. lacticolor | 165-53 | Unspecified |
| T. mentagrophytes var. radians | 409-60 | Human |
| T. interdigitale | 102-77 | Human |
| 406-72 | Human | |
| 447-74 | Human | |
| 1465-83 | Human | |
| 2189-93 | Human | |
| 2190-93 | Human | |
| 2191-93 | Human | |
| T. rubrum | 2073-92 | Human |
| 2360-96 | Human | |
| Epidermophyton sp. | ||
| E. floccosum | 1454-83 | Human |
| 1559-84 | Human |
Unspecified variety.
TABLE 2.
Clinical isolates used in this study
| Species | No. of strains investigated | Origin | Symptomatology |
|---|---|---|---|
| Microsporum spp. | |||
| M. canis | 12 | Human | Present |
| 5 | Cat | Present | |
| 15 | Cat | Absent | |
| 5 | Dog | Present | |
| 12 | Dog | Absent | |
| M. gypseum | 1 | Human | Present |
| 3 | Cat | Absent | |
| 2 | Cat | Present | |
| 3 | Dog | Absent | |
| 5 | Dog | Present | |
| Trichophyton spp. | |||
| T. ajelloi | 3 | Dog | Absent |
| T. mentagrophytes var. granulosum | 1 | Human | Present |
| T. mentagrophytesa | 11 | Human | Present |
| 2 | Dog | Present | |
| 1 | Rabbit | Present | |
| 1 | Chinchilla | Present | |
| T. interdigitale | 10 | Human | Present |
| T. rubrum | 12 | Human | Present |
| Epidermophyton sp. | |||
| E. floccosum | 7 | Human | Present |
Unspecified variety.
DNA extraction.
The strains were grown in Sabouraud's dextrose agar at 25°C; after 2 weeks some mycelium was cut from the agar and transferred to Sabouraud's dextrose broth. After 2 weeks at 25°C, superficial mycelial growth was transferred to a mortar, washed with distilled water, and pestled. For rapid DNA extraction we used the Dynabeads DNA Direct System I (Dynal) based on biomagnetic separation.
In brief, about 20 μl of pestled mycelium was transferred to an Eppendorf tube and incubated with 200 μl of Dynabeads (paramagnetic polystyrene beads in lysis buffer) for 10 min at 65°C so as to obtain cell lysis and the adsorption of the released DNA to the Dynabead surface. This step was followed by magnetic separation of the DNA-Dynabeads complex and by two or three subsequent washings that removed any residual contaminant and eliminated potential PCR inhibitors. The DNA-Dynabeads complex was resuspended in TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA), and DNA was eluted for 5 min at 65°C; it was then ready for PCR or storage at −20°C.
For some strains several cultures and extractions were performed.
PCR fingerprinting.
The simple repeat sequence (GACA)4 was used as a single primer (12–14) in the PCR amplification.
Amplification reactions were performed in volumes of 50 μl containing 25 ng of template DNA, reaction buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl), 2.5 mM MgCl2, 200 μM (each) dATP, dCTP, dGTP, and dTTP, 160 ng of primer, and 2.5 U of Taq DNA polymerase (GeneAmp PCR Core reagents; Perkin-Elmer Cetus, Norwalk, Conn.).
The samples were overlaid with sterile paraffin oil (Carlo Erba) and PCR was performed for 39 cycles in a DNA Thermal Cycler (Perkin-Elmer Cetus) with 1 min of denaturation at 93°C, 1 min of annealing at 50°C, and 1 min of extension at 72°C and then a final extension for 7 min at 72°C.
PCR products (20 μl/sample) were separated by electrophoresis in 1% agarose gels for 2 h at 5V/cm in 0.5× TBE buffer (0.045 M Tris-borate [pH 8.3], 1 mM EDTA). Amplification products were detected by staining with ethidium bromide and were visualized under UV light.
Each sample of genomic DNA was amplified in duplicate in the same PCR and in repeated PCRs at different times.
The genomic analysis of all the strains was done utilizing rigorously standardized concentrations of the reagents, the same thermal cycler, and the same cycling conditions.
The primer we used [(GACA)4] was able to amplify all the strains, and the method used was shown to be a simple, rapid, and reproducible technique. In fact, multiple extractions of the same strain starting with cultures grown for different times produced PCR fingerprinting profiles showing the same distribution of bands of both strong and weak intensity.
Each strain produced the same genomic profiles whether in the same PCR (double amplified sample) or in PCRs repeated at different times. Occasional changes in band intensities were observed which could be attributed to slight variations in the reaction conditions (13).
To confirm that the observed bands were really amplified genomic DNA and not primer artifacts, genomic DNA was omitted from the control reaction mixture for each PCR. We did not observe amplification products in any control reaction.
The PCR fingerprints of all strains yielded up to 11 bands, ranging from approximately 394 to 2,399 bp in length; the number of brightly colored fragments varied from 2 to 5 according to the species, while that of the weakly colored ones varied from 2 to 8.
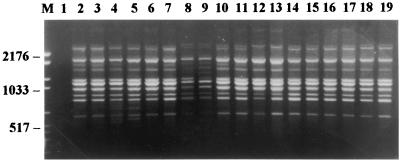
All 53 strains of M. canis produced profiles which are perfectly superimposable without distinction between the collection strains and the strains isolated from humans, cats, and dogs, with or without the presence of clinical lesions (Fig. 1).
FIG. 1.
PCR fingerprints of M. canis strains (each strain was amplified in duplicate). Lanes: M, molecular weight marker VI (Boehringer Mannheim), size range, 154 to 2,176 bp; 1, control reaction without template DNA; 2 to 7, three strains from humans; 8 to 15, four strains from dogs; 16 to 19, two strains from cats.
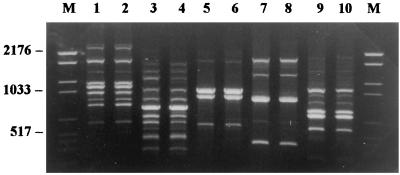
Species-specific profiles were also observed for the species M. gypseum (16 strains), T. rubrum (14 strains), T. ajelloi (5 strains), and E. floccosum (9 strains) for which no intraspecific variability was noted; the complexity of the profiles of M. canis, M. gypseum, and E. floccosum was contrasted by the simplicity of those presented by T. rubrum and T. ajelloi, which for their limited number of bands and their distribution are easily distinguished from those of the other species examined (Fig. 2).
FIG. 2.
PCR fingerprints of five different dermatophyte species (two different strains from each species). Lanes: M, molecular weight marker VI (Boehringer Mannheim), size range, 154 to 2,176 bp; 1 and 2, M. canis; 3 and 4, M. gypseum; 5 and 6, T. rubrum; 7 and 8, T. ajelloi; 9 and 10, E. floccosum.
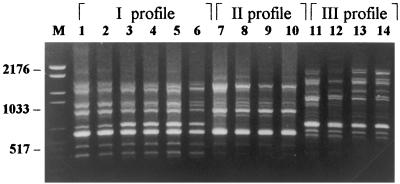
The profiles of T. mentagrophytes and T. interdigitale were also complex and we noted in them a discrete variability which allowed us to group the strains examined into three different profiles. The first and second profiles differ by only a fragment (approximately 653 bp) while the third profile is very different from the other two (Fig. 3).
FIG. 3.
PCR fingerprints showing three different profiles of T. mentagrophytes group strains (each strain amplified in duplicate). Lanes: M, molecular weight marker VI (Boehringer Mannheim), size range, 154 to 2,176 bp; 1 and 2, T. mentagrophytes var. granulosum; 3 to 6, T. interdigitale (two strains); 7 and 8, T. mentagrophytes var. asteroides; 9 and 10, T. mentagrophytes var. radians; 11 and 12, T. mentagrophytes var. lacticolor; 13 and 14, T. mentagrophytes of unspecified variety.
In the first profile we find all the strains of T. mentagrophytes var. granulosum and of T. interdigitale, in the second T. mentagrophytes var. asteroides and T. mentagrophytes var. radians, and in the third T. mentagrophytes var. lacticolor; the strains of T. mentagrophytes of unspecified variety are distributed as follows in all three profiles: in the first we find strains isolated from both humans and animals and in the second and third we find only strains isolated from humans (Table 3).
TABLE 3.
Strain profiles and origins of T. mentagrophytes group isolates
| Profile | Species | No. of isolates from Institut Pasteur collection
|
No. of clinical isolates
|
Total no. of isolates | ||
|---|---|---|---|---|---|---|
| Human | Animal | Human | Animal | |||
| I | T. mentagrophytes var. granulosum | 2 | 1 | 3 | ||
| T. interdigitale | 7 | 10 | 17 | |||
| T. mentagrophytesa | 1 | 1 | 6 | 4 | 12 | |
| II | T. mentagrophytes var. asteroides | 1 | 1 | |||
| T. mentagrophytes var. radians | 1 | 1 | ||||
| T. mentagrophytesa | 3 | 3 | ||||
| III | T. mentagrophytes var. lacticolorb | 1 | ||||
| T. mentagrophytesa | 5 | 5 | ||||
Unspecified variety.
From the Institut Pasteur collection, with unspecified origin.
PCR fingerprinting has proven to be a simple and reproducible method. In fact, by strictly maintaining the experimental conditions we have had superimposable profiles.
PCR fingerprinting, along with a fast DNA extraction method, proved to be a rapid method in comparison to other techniques of molecular biology (restriction fragment length polymorphism analysis, sequencing of the ITS region, and sequencing of protein-encoding genes).
In fact, in about 6 h we can obtain electrophoretic profiles starting from cultures, as the DNA extraction technique we used does not require more than half an hour and the amplification requires about 5 h.
The primer we used produced species-specific profiles for M. canis, M. gypseum, T. rubrum, T. ajelloi, and E. floccosum and allowed us to detect three different profiles for T. mentagrophytes in relation to the varieties studied. The fact that all the strains of T. interdigitale belong to one of these profiles confirms the notion that this species is closely related to or a variety of T. mentagrophytes.
The great diversity of profiles between the T. mentagrophytes group and T. rubrum seems particularly interesting and makes the two species quite distinguishable, while with the classical technique their identification is often difficult.
These species-specific profiles could be used for identifying strains that do not present typical morphological characteristics and therefore cannot be identified in the classical way.
In our experience we isolated some strains from dogs and cats that did not produce conidia but presented a characteristic orange pigmentation on the reverse side of the colony. PCR fingerprinting of these strains has produced electrophoretic profiles that are superimposable with those produced by the strains of M. canis reported in this work, causing us to hypothesize that they belong to this species (unpublished data).
Dermatophytes in culture easily lose their typical morphological characteristics, so PCR fingerprinting could be used for reidentifying the collection strains. In our experience we were able to reidentify numerous strains of our collection which had lost their typical morphological characteristics (unpublished data).
Since genomic research does not necessarily imply the use of live organisms, PCR fingerprinting could also be used for studying dead strains. We have no experience in this regard but Liu et al. (11) have been able to confirm the identities of 20-year-old nonviable dermatophyte isolates by AP-PCR.
In conclusion, while the only disadvantage of the use of PCR fingerprinting for identifying dermatophytes is the relatively higher cost in comparison to the classical method, the advantages of its use are many.
It is a technique of simple execution, it is rapid in comparison with other techniques of molecular biology, especially thanks to the speed of the DNA extraction, and it can be applied in cases where it is necessary to identify strains not presenting typical morphological characteristics.
Therefore, this method can be of great utility when it is not possible to use, for the above-specified reasons, the classical method, which is still valid and advisable for identifying strains with well-characterized morphological aspects.
REFERENCES
- 1.De Bièvre C, Dauguet C, Nguyen V H, Ibrahim-Granet O. Polymorphism in mitochondrial DNA of several Trichophyton rubrum isolates from clinical specimens. Ann Inst Pasteur Microbiol. 1987;138:719–727. doi: 10.1016/0769-2609(87)90149-9. [DOI] [PubMed] [Google Scholar]
- 2.Gräser Y, El Fari M, Presber W, Sterry W, Tietz H J. Identification of common dermatophytes (Trichophyton, Microsporum, Epidermophyton) using polymerase chain reactions. Br J Dermatol. 1998;138:576–582. doi: 10.1046/j.1365-2133.1998.02165.x. [DOI] [PubMed] [Google Scholar]
- 3.Gräser Y, El Fari M, Vilgalys R, Kuijpers A F A, De Hoog G S, Presber W, Tietz H J. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med Mycol. 1999;37:105–114. [PubMed] [Google Scholar]
- 4.Gräser Y, Kuijpers A F A, Presber W, De Hoog G S. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med Mycol. 1999;37:315–330. doi: 10.1046/j.1365-280x.1999.00234.x. [DOI] [PubMed] [Google Scholar]
- 5.Kano R, Nakamura Y, Watari T, Watanabe S, Takahashi H, Tsujimoto H, Hasegawa A. Phylogenetic analysis of 8 dermatophyte species using chitin synthase 1 gene sequences. Mycoses. 1997;40:411–414. doi: 10.1111/j.1439-0507.1997.tb00176.x. [DOI] [PubMed] [Google Scholar]
- 6.Kano R, Okabayashi K, Nakamura Y, Ooka S, Kashima M, Mizoguchi M, Watanabe S, Hasegawa A. Differences among chitin synthase 1 gene sequences in Trichophyton rubrum and T. violaceum. Med Mycol. 2000;38:47–50. doi: 10.1080/mmy.38.1.47.50. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki M, Ishizaki H, Aoki M, Watanabe S. Phylogeny of Nannizzia incurvata, N. gypsea, N. fulva, and N. otae by restriction enzyme analysis of mitochondrial DNA. Mycopathologia. 1990;112:173–177. doi: 10.1007/BF00436650. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki M, Aoki M, Ishizaki H, Nishimura K, Miyaji M. Phylogeny of Epidermophyton floccosum and other dermatophytes. Mycopathologia. 1996;134:121–128. doi: 10.1007/BF00436718. [DOI] [PubMed] [Google Scholar]
- 9.Kwon-Chung K J, Bennett J E. Dermatophytoses. In: Kwon-Chung K J, Bennett J E, editors. Medical mycology—1992. Philadelphia, Pa: Lea & Febiger; 1992. pp. 105–161. [Google Scholar]
- 10.Liu D, Coloe S, Baird R, Pedersen J. PCR identification of Trichophyton mentagrophytes var. interdigitale and T. mentagrophytes var. mentagrophytes dermatophytes with a random primer. J Med Microbiol. 1997;46:1043–1046. doi: 10.1099/00222615-46-12-1043. [DOI] [PubMed] [Google Scholar]
- 11.Liu D, Coloe S, Baird R, Pedersen J. Application of PCR to the identification of dermatophyte fungi. J Med Microbiol. 2000;49:493–497. doi: 10.1099/0022-1317-49-6-493. [DOI] [PubMed] [Google Scholar]
- 12.Meyer W, Mitchell T G, Freedman E Z, Vilgalys R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2274–2280. doi: 10.1128/jcm.31.9.2274-2280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer W, Mitchell T. Polymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequences: strain variation in Cryptococcus neoformans. Electrophoresis. 1995;16:1648–1656. doi: 10.1002/elps.11501601273. [DOI] [PubMed] [Google Scholar]
- 14.Meyer W, Latouche G N, Daniel H M, Thanos M, Mitchell T G, Yarrow D, Schönian G, Sorrell T. Identification of pathogenic yeasts of the imperfect genus Candida by polymerase chain reaction fingerprinting. Electrophoresis. 1997;18:1548–1559. doi: 10.1002/elps.1150180911. [DOI] [PubMed] [Google Scholar]
- 15.Mochizuki T, Sugie N, Uehara M. Random amplification of polymorphic DNA is useful for the differentiation of several anthropophilic dermatophytes. Mycoses. 1997;40:405–409. doi: 10.1111/j.1439-0507.1997.tb00175.x. [DOI] [PubMed] [Google Scholar]
- 16.Weitzman I, Summerbell R C. The dermatophytes. Clin Microbiol Rev. 1995;8:240–259. doi: 10.1128/cmr.8.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]