Abstract
Mycotoxins are highly toxic metabolites produced by fungi that pose a huge threat to human and animal health. Contamination of food and feed with mycotoxins is a worldwide issue, which leads to huge financial losses, annually. Decades of research have developed various approaches to degrade mycotoxins, among which the biological methods have been proved to have great potential and advantages. This review provides an overview on the important advances in the biological removal of mycotoxins over the last decade. Here, we provided further insight into the chemical structures and the toxicity of the main mycotoxins. The innovative strategies including mycotoxin degradation by novel probiotics are summarized in an in-depth discussion on potentialities and limitations. We prospected the promising future for the development of multifunctional approaches using recombinant enzymes and microbial consortia for the simultaneous removal of multiple mycotoxins.
Keywords: mycotoxins, biodegradation, probiotics, recombinant enzyme, microbial consortia
1. Introduction
The continuous occurrence of food contaminants worldwide poses a critical threat to the health of human and livestock. One of the major contaminants in food and feed products are mycotoxins, the secondary metabolites synthesized by toxigenic fungi strains, mainly those belonging to Penicillium, Aspergillus, Alternaria and Fusarium genera. Both acute and chronic exposure to mycotoxin-contaminated food may cause deleterious health effects including retarded growth, suppression of the immune response, vomiting, infertility and gastrointestinal and carcinogenic diseases [1]. These mycotoxins occur in various products, from raw agricultural products such as corn, barley, oats, fruits and herbs, to commercial commodities including aquafeeds, beverages, fruit and vegetable-derived products [2,3,4]. The contamination of mycotoxins can occur during any part of the complex food chain, including harvest, industry processing, transportation and/or storage, imposing social burdens on the food industry due to the waste created by contaminated products [5]. This creates an urgent demand for mycotoxin removal methods to minimize economic loss and hazards to consumers.
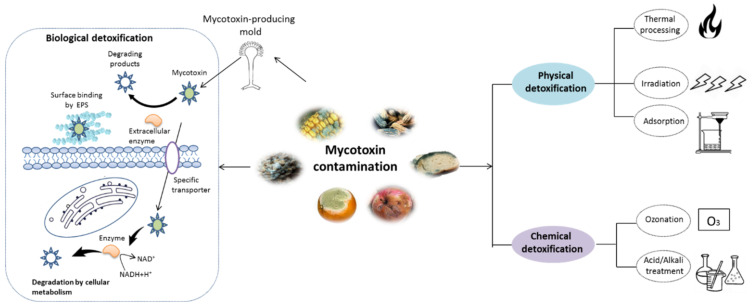
As shown in Figure 1, various strategies have been reported for mycotoxin removal, which can be roughly categorized into physical [6], chemical [7], and biological methods [8]. Although some physical and chemical approaches can remove mycotoxin with various degree of success, some problems, such as potential safety issues, loss of important nutrients and high costs, still hamper their application in the food industry [9]. In recent years, various biological detoxification approaches, particularly those using microbial cells or enzymes, have been demonstrated to be highly effective in degrading mycotoxins into less toxic products with high specificity [1,10]. Compared with the physical and chemical methods, the detoxification processes by microbes or enzyme can be handled under mild working conditions with fewer losses to the nutritional quality of food products [11,12]. To date, various microorganisms including bacteria and yeast have been found to be capable of degrading mycotoxins [13,14,15,16]. Additionally, mycotoxin-degrading enzymes obtained by direct extraction and purification from biomaterials or expression in mature microbial hosts by genetic engineering technique have been well documented [17,18]. Nonetheless, a limited number of studies have been developed to evaluate the potential safety derivatives or metabolites during the detoxification process, although the byproducts can be more harmful than the initial product [19]. Moreover, the mechanisms involved in mycotoxins detoxifications are not straightforward. Therefore, it is essential to address these questions before the real applications of biological agents.
Figure 1.
Schematic representation of mycotoxin detoxification methods. The physical methods mainly include thermal process, irradiation, and adsorption techniques, while the chemical methods involve in the treatment with acid/alkali solution and ozonation. The major mechanism of biological detoxification involves in the surface binding by extracellular polymeric substances (EPS), degradation by enzyme and cellular metabolism.
The aim of the current review is to address some of the developments on (a). Novel mycotoxin-removal approaches, notably based on the promising probiotics, recombinant enzyme and microbial consortia, and the main mechanisms of action underlying the degradation process; (b). New strategies for the elucidation of the degradation pathway and a risk assessments of the degrading by-products; (c). Challenges and limitations in the application of biological mycotoxin removal.
2. Chemical Structure and Toxic Effects of Mycotoxins
Since 1960, more than 400 forms of mycotoxins have been identified and reported. The most common mycotoxins in food include aflatoxins (AFs), trichothecenes, ochra-toxins, zearalenone (ZEA), fumonisins, and patulin, which are highly toxic [20]. These notable contaminants can be produced by various toxigenic fungi due to improper harvesting, storage or transport conditions [21]. Most of them have complex chemical structures containing specific groups that confer the toxic effects. Basic information of the main mycotoxins including chemical structure, biological effect and producing fungi is summarized in Table 1 [21,22,23,24,25].
Table 1.
Chemical structure, toxic groups, biological effects of main mycotoxins and producing fungi.
| Mycotoxins | Chemical Structure | Main Toxic Groups | Main Degradation Products | Organ/System Affected | Main Clinical Signs | Producing Fungi |
|---|---|---|---|---|---|---|
| Aflatoxins (B1, B2, G1, G2) |
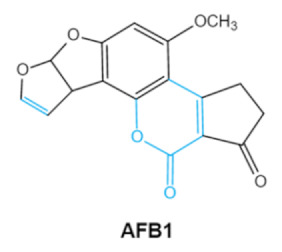
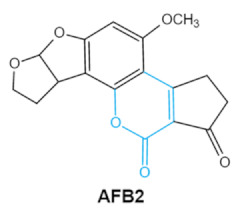
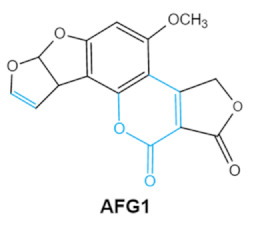
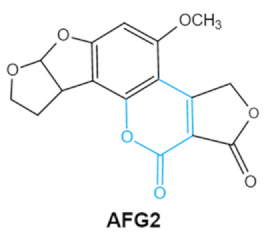
|
Lactone ring Double bond in difuran ring moiety |
AFB1-8,9dihydrodiol, AFB1-8,9-epoxide, dihydrohydroxyaflatoxin B1, | Liver, kidney, immune system | Hepatitis, carcinogenic, abdominal pain, vomiting, increased susceptibility to disease, immunosuppressive and carcinogenic effects |
Aspergillus. Flavus
A. Parasiticus A. nomius |
| Zearalenones (ZEA) |
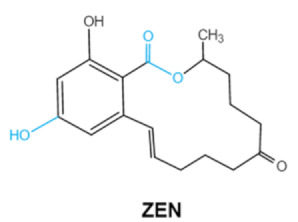
|
Lactone ring C-4 hydroxyl group |
α-/β-zearalenol, α-/β-zearalanol,zearalenone-4-sulfate, 1-(3,5-dihydroxyphenyl)-6′-hydroxy-l’-undecen-l0′-one, (5S)-5-({2,4-dihydroxy-6-[(1E)-5-hydroxypent-1-en-1-yl]benzoyl}oxy)hexanoic acid |
Reproductive tract, mainly female | Hyperestrogenism, Reproductive disorders |
Fusarium graminearum
(F. roseum)
F. culmorum F. equiseti F. cerealis F. verticillioides F. incarnatum |
| Ochratoxins (A,B,C) (OTs) |
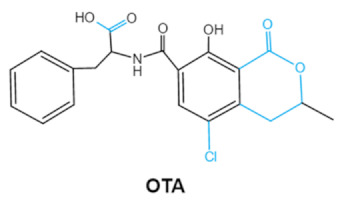
|
Isocoumarin moiety Carboxyl group of the phenylalanine moietyCl group |
L-βphenylalanine, OTα | Liver, kidney, immune system, inhibit RNA, DNA and protein synthesis in kidney | Nephritis, enlargement of kidney and hepatitis |
A. ochraceus
A. carbonarius A. niger P. verrucosum P. nordicum |
| Fumonisins FBs (B1, B2) |
|
Two tricarballylic acid side chains Free amino group |
2-oxo-12,16-dimethyl-3,5,10,14,15-icosanepentol hemiketal, NacetylAP1, |
Lungs and heart (pig), central nervous system (horse), liver, immune system | Porcine pulmonary edema (PPE), equine leukoencephalomalacia | Fusarium section Liseola |
| Trichothecenes (DON, T-2, HT-2) TCNs |
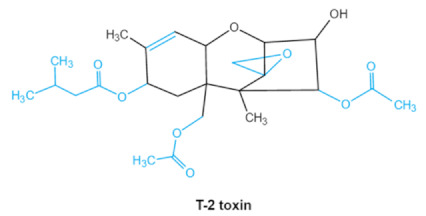
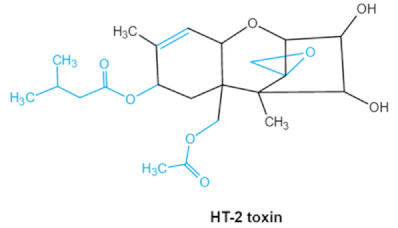
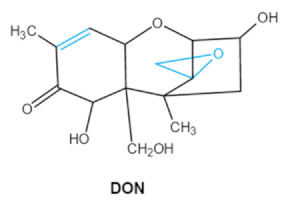
|
Epoxide group Acylated side groups C9-10 double bond |
HT-2 toxin, T-2 triol, T-2 tetraol, de-epoxy T-2 tetraol, 3α,7α,15α-triacetoxy-deoxynivalenol, de-epoxy deoxynivalenol, 3-acetyldeoxynivalenol, diaacetoxydeoxynivalenol. Epoxymonoacetoxyscirpenol, de-epoxyscirpentrio |
Central nervous system, gastrointestinal tract, liver, immune system | Anorexia, vomiting, abdominal pains, cardiovascular dysfunction |
F. acuminatum F. sporotrichioides F. langsethiae Fusariumgraminearum, F. culmorum F. cerealis F. culmorum F. graminearum F. sporotrichioides F. poae |
| Patulin PAT |
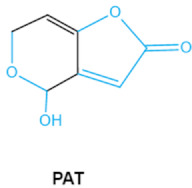
|
Furan, pyran or lactone ring Hemiacetal |
Ascladiol, hydroascladiol, desoxypatulinic acid, 3-keto-5-hydroxypentanal, glyoxylic acid |
Gut epithelium, liver, kidney, immune system | Oral and epithelial lesion, loss of appetite Nausea, vomitin, gastric ulcers |
Penicillim expansum Bysochlamis nívea
Aspergillus clavatus P. griseofulvum |
2.1. Aflatoxins (AFs)
Aflatoxin (AFs) is the most widely studied mycotoxin encountered in agricultural commodities. They are mainly produced by Aspergillus species, Aspergillus flavus, A. parasiticus and A. nomius. AFs shave been reported to induce hepatic toxicity, nephron toxicity and immune suppression in humans [26]. Of the AFs, AFB1 is extremely hazardous and has been considered as a Group I of naturally occurring carcinogens [27]. AFs are furanocoumarins associated with a bisdihydrodifuran or tetrahydrobisfuran united to a coumarin, replaced by a cyclopentanone or a lactone [28]. Lactone ring and the double bond in the terminal furan ring of AFs are responsible for the toxicity and carcinogenic activity [23,29].
2.2. Trichothecenes
As one of the major classes of mycotoxins, trichothecenes (TCNs) cause a significant economic impact on the food chain each year. TCNs are formed in nature by some filamentous fungi such as Aspergillus, Penicillium, Fusarium, Spicellum and Stachybotrys, among them Fusarium is the most common toxin source in cereals, vegetables, and feedstuff [30,31]. The consumption of TCNs-contaminated food may affect the gastrointestinal tract, kidney, liver, and immune system, and cause health issues such as anorexia, vomiting, abdominal pains and cardiovascular dysfunction [32]. TCNs are tetracyclic sesquiterpenoid epoxides and form a family of over 200 toxins with an atricyclic 12,13-epoxytrichothec-9ene (EPT) core structure in common [33]. According to their structure, toxins in this family are classified into four groups (types A, B, C, and D) based on the substitution pattern of EPT, among them, type A and type B are of special interest because of their high toxicity [34]. Representative type ATCNs includes T-2 toxin (T-2), HT-2 toxin (HT-2) and neosolaniol (NEO), while type BTCNs were represented by deoxynivalenol (DON) and its acetyl derivatives [35]. The 12, 13-epoxide ring, 9–10 double bond and acylated side groups in trichothecenes are essential for imparting the toxic effects in inhibiting the synthesis of protein, RNA and DNA [36,37]. Generally, the complete detoxification of TCNs is a challenge due to their stable and complex structure [38].
2.3. Ochratoxins
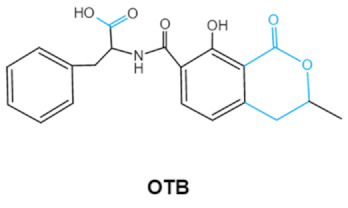
Ochratoxins (OTs) belong to a family of mycotoxins commonly found in various food and feed mainly produced by Penicillium and Aspergillus species, including P. verrucosum, P. nordicum, A. ochraceus, A. carbonarius and A. niger [39]. They comprise a group of coumarin derivatives, including ochratoxin A (OTA), ochratoxin B (OTB) and ochratoxin C (OTC), of which OTA is the most prevalent and toxic compound [40]. OTA is of neurotoxicity, carcinogenesis, immunotoxicity and hepatotoxicity, which poses a serious threatens to both human and animal health. [41]. Isocoumarin moiety is the key toxic group of OTA, and the carboxyl group of the phenylalanine moiety as well as the Cl group contribute to its toxicity [25].
2.4. Zearalenone
Zearalenone (ZEA) is a kind of nonsteroidal mycotoxin that contaminates a variety of cereals [42]. ZEA is a secondary metabolite that is synthesized by several species of mold fungi belonging to the Fusarium family that colonizes plants grown in the temperate climate zone such as corn, maize and other grain crops [43]. ZEA belongs to xenoestrogens that have a chemical structure similar to natural estrogens. Therefore, tZEA can bind to estrogen receptor sites, causing hormonal imbalance that may lead to reproductive diseasesin human and livestocks [44]. Lactone group in the chemical structure of ZEA is critical to the toxicity of ZEA. It has been reported that the toxic effects of ZEA can be significantly reduced by the cleavage of the lactone ring [45].
2.5. Fumonisins
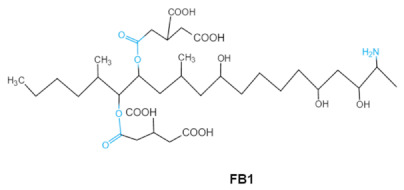
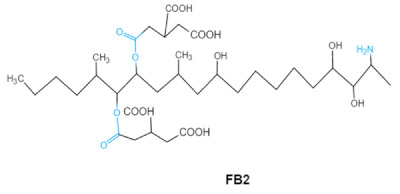
Fumonisins (FBs), polyketide-derived mycotoxins, are mainly produced by Fusarium mold species that commonly infect corn and other agricultural products [46]. Based on the structural difference, four main groups of fumonisin analogs have been characterized, and among them, FB1 is the most prevalent and toxic [47]. FBs have a chemical structure similar to sphingolipid long-chain bases, such as sphinganine (Sa) and sphingosine (So) which are intermediate compounds in the sphingolipid metabolism, which can inhibit ceramide synthase and block the biosynthesis of complex sphingolipids, resulting in a wide spectrum of pathological effects in humans and animals [48]. The tricarballylic acid (TCA) side groups and the free amino group play important roles in FBs toxicity mechanism. It has been found that the removal of these groups can significantly reduce both phytotoxicity and mammalian cytotoxicity [49,50].
2.6. Patulin
Patulin (PAT) is a tetraketide mycotoxin most commonly found in apples and apple-derived products [51]. It is produced by a variety of filamentous fungi including Penicillium, Aspergillus and Byssochlamys [52]. Acute PAT exposure damages gut epithelium, liver, kidney, and immune system, causing symptoms such as nausea, vomiting and intestinal hemorrhages [53]. PAT has also been shown to affect the activities of thiol-containing enzymes that play important roles in glycolysis, since this mycotoxin can easily form covalent sulfhydryl-containing compounds, which is the main mechanism associated with PAT toxicity [54]. The destruction of lactone, furan or the pyran ring and hemiacetal in the structure of PAT may be an indicator of toxicity reduction [8].
3. Mycotoxin Removal by Probiotics
3.1. Fast Glance to Probiotic Properties
Probiotics, defined as ‘‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’’ by the World Health Organization (WHO), have been incorporated into various kinds of products such as foods, drugs, and dietary supplements [55]. Beyond their basic nutritional value, probiotics provide numerous benefits such as the alleviation of lactose intolerance, maintaining balance of the digestive tract and modulation of inflammatory responses [56]. The most extensively studied and widely used probiotics include Lactobacillus, Bifidobacterium, yeast Saccharomyces cerevisiae as well as some Bacillus species [57]. Gaggìa et al. (2010) summarized the species of probiotics and their health-promoting characteristics based on extensive studies and an internet search of commercial products [58]. These nonpathogenic, nontoxigenic, and fermentative microorganisms are quite commonly added to “functional foods” and growth supplements for human health. The application of probiotics is also prevalent in the aquaculture field, acting as alternatives for antibiotics or disinfectants to improve water quality [59]. Moreover, specific strains of probiotic bacteria were indicated to be effective in removing contaminants such as heavy metals, pesticides as well as mycotoxins, which might act as promising bio-agents for food safety [60,61,62].
3.2. Mycotoxins Detoxification by Probiotics and the Related Mechanism
The biological detoxification of mycotoxins by probiotic bacteria was reviewed years ago [28,55,59]. Probiotics, particularly lactic acid bacteria (LAB) and yeasts such as Saccharomyces genus, can remove mycotoxins primarily through two mechanisms, surface adsorption or biodegradation (Figure 1). Toxins removal by surface adsorption of microorganisms is a fast and reversible process that does not cause any chemical changes in the substrate. In contrast, biodegradation is permanent but results in undesirable metabolites.
3.2.1. Cell-Binding of Mycotoxins
In the last three decades, numerous studies have been developed regarding toxin binding by the cell-surface of probiotics such as Lactobacillus rhamnosus, Lactobacillus amylovorus, Lactobacillus plantarum, Lactobacillus pentosus [63,64,65,66]. Most of studies in vitro have been carried out using viable cells or heat-treated cells with aqueous toxic standards. The binding capacity of mycotoxins was strain-dependent, and depends on the natural structure of the toxin as well as physicochemical conditions. In vivo studies in animal trials and human clinical trials investigating the real effect of probiotics binding capacity as antidote for mycotoxin detoxification have been reviewed [67]. Although certain probiotic strains have shown a positive role in the restoration of mycotoxins damage, studies concerning the fate of the ingested mycotoxins and probiotic-mycotoxin still need to be supplemented.
The structural components of the probiotic cell wall play a pivotal role in the binding of toxins by microorganisms. LAB has the typical cell wall structure of Gram-positive bacteria, which consists of thick and multilayered peptidoglycan (PG) sacculi decorated with other glycopolymers including teichoic acids (TAs), polysaccharides (PSs) and proteinatious Slayer [68]. The binding efficiency of different LAB species seems most closely related to the amino acid sequence of peptide bridges of the PG [69]. The cell wall of LAB contains plenty of negatively charged functional groups that might facilitate the binding capacity due to the presence of S-layer proteins [60]. The literature available has limited information on the role of teichoic acids and polysaccharide involved in the toxins binding mechanism, experimental studies of enzymatic degradation showed that deficiency of these components in cell wall leads to less toxin accumulation [70,71]. The cell wall of yeasts such as Saccharomyces genus is mainly composed of an inner layer with β-glucans and chitin, and outer layer with heavily glycosylated mannoproteins. Some studies suggest that the cell wall thickness depends on β-D-glucans reticular organization and that β-(1, 3)-D-glucans contents play a major role in the toxin adsorption efficacy in S. cerevisiae [72,73]. Thus, elucidating the differences between cell wall components of probiotics species might provide a potential strategy to select strains to act as the mycotoxin binder.
3.2.2. Biodegradation
In contrast to cell-binding methods, research on the biodegradation of mycotoxins by probiotics is very limited. The available literatures about mycotoxins degradation by probiotics over the last decade was summarized in Table 2. Harkai et al. (2016) successfully selected several Streptomyces strains which have shown a strong capability to degrade AFB1 and ZON mycotoxin with eliminated genotoxicity [74]. Rao et al. (2017) reported a well-known probiotic bacteria Bacillus licheniformis isolate CFR1, which can degrade aflatoxin B1 in liquid culture media with a reduction rate of 94.7%. In the same study, it was found that AFB1 could be metabolized to degradation products using a cell-free supernatant, and that treatments such as heating, proteinase K and SDS lead to the complete loss of degradation activity, indicating that the extracellular proteins or enzymes may be involved in the degradation process [75]. The application of biological detoxification in food fermentation has enormous practical significance. Juodeikiene et al. (2018) treated wheat grains with three P. pen-tosaceus strains, and the results showed a decrease in mycotoxins (DON, T-2 and HT-2 toxins) in malting wheat grains. The strains would serve as candidates for reducing mycotoxins in corresponding raw materials for beverages and certain baked-goods production. [76]. During the beer-brewing process, some stable mycotoxins in contaminated cereal-derived raw materials may survive and enter the final products, thereby it is important to screen brewers’ yeast with the ability to alleviate toxicity. In the study conducted by Nathanail et al. (2016), the lager yeast Saccharomyces pastorianus A15 may enzymatically transform fusarium trichothecenes to less toxic form in the fermentations of brewer’s wort, with reduction rates of 15% for DON, 17% for DON3Glc, 34% for HT-2 and 31% for T2 [77]. This strain might be an excellent candidate for mycotoxicosis control during food processing. It must be addressed that the biodegradation method may bring potential risks. Most of the studies did not look into the possible toxicity of by-products created during the decomposition process. For biotechnological utilization, applicable methods must be developed to monitor the potential hazardous metabolites and biological effects in mycotoxin biodegradation.
Table 2.
Degradation of mycotoxins by probiotic bacterial strains reported in the last decade.
| Mycotoxins | Microorganism | Reduction Rate (%) | Toxin Level | Degradation Condition | Reference |
|---|---|---|---|---|---|
| AFB1 | Streptomyces cacaoi subsp. Asoensis K234 | 88.34 ± 15.62 | 1 μg mL−1 | 5 days, 28 °C,170 rpm liquid LB medium |
Harkai et al. (2016) |
| Streptomyces luteogriseus K144 | 79.93 | 1 μg mL−1 | 5 days, 28 °C,170 rpm liquid LB medium |
||
| Bacillus licheniformis CFR1 | 94.73 ± 1.09 | 500 ppb | liquid nutrient broth (NB) at 37 °C, 72 h | Rao et al. (2017) | |
| T-2 | P. pentosaceus KTU05-10 | 78.0 | 12.8–19.5 μg kg−1 | malting wheat grains with bacterial suspension at 18 °C for 30 min |
Juodeikiene et al. (2018) |
| Saccharomyces pastorianus A15 | 31.0 | 5000 µg L−1 | 15 °C, 120 rpm, four days 11.5° Plato wort |
Nathanail et al. (2016) | |
| HT-2 | P. pentosaceus KTU05-10 | 79.0 | 258–819 μg L−1 | malting wheat grains with bacterial suspension at 18 °C for 30 min |
Juodeikiene et al. (2018) |
| ZEA | P. acidilactici | 38.0 | 19.5–873.7 μg L−1 | malting wheat grains with bacterial suspension at 18 °C for 30 min |
Juodeikiene et al. (2018) |
| Streptomyces rimosus (K145, K189) | 100.0 | 1 μg mL−1 | 5 days, 28 °C,170 rpm liquid LB medium |
Harkai et al. (2016) | |
| DON | P. pentosaceus KTU05-10 | 47.0 | 3370–6930 μg kg−1 | malting wheat grains with bacterial suspension at 18 °C for 30 min |
Juodeikiene et al. (2018) |
| Saccharomyces pastorianus A15 | 15.0 | 400 µg L−1 | 11.5° Plato wort, 15 °C, 120 rpm for 4 days | Nathanail et al. (2016) |
4. Biodegradation of Mycotoxins by Microbial Consortia
The biodegradation of mycotoxins that are able to transform mycotoxins into nontoxic metabolites has emerged as an alternative strategy for food and feed-safety control. However, it is difficult or even impossible to completely detoxify some complex contaminants such as aromatic compounds and mycotoxins by single-species strains. Therefore, toxin biodegradation with microbial consortia has gained increasing attentions, owing to their ability to perform more complex tasks than individual strains or species [78]. Moreover, microbial consortium is more resilient to environmental changes than single microbial species. For instance, constructed PAHs degrading microbial consortia has a higher potential for bioremediation of sites pollution due to their elevated tolerance and improved adaptation [79]. Microbial consortia have become a major strategy for soil-pollutant degradation; however, the degradation mechanism remains largely unknown. Available literatures showed that the application of microbial consortium might provide a new solution for mycotoxin biodegradation, which is a complex process involving two or more tasks that in some cases may pose insuperable challenges for one single organism.
A diversity of microbes in soil with great diversity has been reported to bio-transform complex contaminants into harmless or less toxic smaller molecules. The possible role of soil microbial communities for mycotoxins degradation was suggested by several researchers. In the early research of Beeton and Bull (1989), soil bacterial communities was found to be able to utilize T-2 toxin as a sole carbon source and energy, and the major degradation pathway involved HT-2 toxin and T-2 triol production by side chain cleavage of acetyl moieties. Compared to single bacterial function, the complete communities were more active in terms of their T-2 toxin degradation rate [80]. In their study, they isolated a bacterial consortium PGC-3 from soil by in situ enrichment and found that PGC-3 exhibited strong de-epoxidation activity on trichothecene mycotoxins under aerobic conditions. The result of the 16S rDNA sequences analysis showed that PGC-3 comprised 10 bacterial genera, among them, Desulfitobacterium increased steadily under increased DON concentrations [81].
Besides, thermophilic agricultural compost materials with abundant microorganisms were also used as the microbial source. In the work of Wang et al. (2020), a novel microbial consortium (NZDC-6) from agricultural composts with moldy corncob and cornstalk was characterized to degrade estrogenic ZEA and its cognates, α-zearalenol (α-ZAL) and β-zearalenol. This consortium was found to be thermophilic and highly effective with a degradation ratio >90% at an optimum temperature of 60 °C. The author further evaluated the potential effects of NZDC-6 on the treatment of ZEA-contaminated corncobs by semisolid fermentation. Fermentation conditions have a great influence on microbial composition and efficiency. It was found that a high solid/liquid ratio lead to the increase in dissolved oxygen content and acidity, thus inhibiting microbial ZEA degradation [82]. Zhao et al. (2019) isolated a FB1-degradingbacterial consortium SAAS79, mainly consisted of Pseudomonas, Comamonas, Delftia, Sphingobacterium, and Achromobacter genera, from spent mushroom compost. SAAS79 could transform FB1 to less toxic degradation products in 3 h with a degradation rate of 90%. Since no active single-degrader was isolated from consortium SAAS79, the authors speculated that the synergistic effect among the species in the bacterial consortium may contribute to the degradation process [83]. Similarly, TADC7, a high performance thermophilic microbial consortium constructed by Wang et al. (2017) exhibited a stable and efficacious ability to degrade high doses of AFB1 (up to 5000 μg L−1) under extreme heat condition. Isolates from the consortium were found to be able to reduce AFB1 to varying degrees, although none was comparable to the efficacy of microbial consortium TADC7, indicating that TADC7 benefits from the synergistic effects [84].
During complex processing and storage, food and feed are probably co-contaminated by multiple, leading to more serious toxic effects than individual mycotoxin. Therefore, it is necessary to achieve effective co-degradation of multiple mycotoxins synchronously [85]. For instance, the co-existence of AFB1 and ZEA in agricultural products may have a higher hepatotoxicity than either of them alone [86]. TADC7, the aforementioned AFB1-degrading microbial community, was domesticated by co-culturing with AFB1 and ZEA, yielding the derived microbial consortium TMDC with a stable microbial composition and the ability to simultaneously degrade AFB1 and ZEA [87]. A promising new approach is the combination of compound probiotics (CP) with mycotoxin-degradation enzymes for the synchronous detoxification of AFB1 and ZEA. Compared to individual Bacillus subtilis, the combination with Lactobacillus casein and Candida utilis (1:1:1) significantly improved the degradability with a degradation rate of 45.49% and 44.90% for AFB1 and ZEA, respectively. The synergistic use of compound probiotics with mycotoxin-degradation enzymes (MDEs) from Aspergillus oryzae at a ratio of 3:2 could further improve degradation rates (63.95% and 73.51%) [9]. To reduce or eliminate the hazards of AFB1 and ZEA in broiler growth, CP of Bacillus subtilis, Lactobacillus casei and Candida utilis (7, 6 and 7 log CFU g−1) was added in broiler diets for a simultaneous biodegradation of the toxins. As a result, the CP addition decreased mycotoxin residues through positively regulating bacterial abundances of gut microbiota [88]. Moreover, the cell-free supernatant of compound probiotics (CFSCP) combined with MDEs has been demonstrated to be capable of alleviating the cytotoxic effects of AFB1 and ZEA on swine jejunal epithelial cells [89]. These findings shed light on the potential application of microbial consortia in the biodegradation of mycotoxins.
5. Mycotoxin Degradation by Recombinant Enzymes
Various microbiological techniques have emerged for eliminating or controlling mycotoxin over the past decades. Among them, on the investigation of enzymes that can transform mycotoxins to less toxic or nontoxic products have has been a prevalent topic [90]. A wide range of enzymes have been identified for mycotoxin degradation, including laccase, manganese peroxidase (MnP) and oxidase [91]. Although the identification, characterization and purification of mycotoxin-degrading enzymes (MDE) can sometimes be time consuming and fails to be cost-effective, the use of pure enzymes offers an attractive and unavoidable alternative to the application of whole bacterial cells, especially under conditions that are not conducive to the survival of microorganisms. Moreover, compared to the use of living microorganisms, enzymatic degradation has several superiorities such as ease-of-handling, reproducible and homogeneous performances, high efficiency and high specificity of action [23]. However, a number of MDE are naturally produced and stored in fungi intracellularly, making them particularly difficult to lysate and extract. Although various natural MDE were purified from microorganisms in recent years as reviewed in the previous literature [1,12,17,25], their application in real food and feed with complex matrices is very limited [90].
Mycotoxin degradation by a recombinant enzyme is particularly attractive because the clone and heterologous expression of enzymes via the genetic engineering technique allow for efficient production at lower economic and labor costs. For example, it is very challenging to obtain natural laccase from the fungi Streptomyces cyaneus CECT 3335 due to its slow growth rate and narrow optimum pH range. While the large-scale production of laccase was realized by the heterologous expression in Escherichia coli with the yield up to 104 mgL−1 [92]. Some studies concerning mycotoxin degradation by recombinant enzymes expressed in engineered strains are shown in Table 3. Yu et al. (2012) cloned and expressed a gene of peroxiredoxin (Prx) from Acinetobacter sp. SM04 in E. coli. The recombinant Prx was able to degrade ZEA in the presence of H2O2 efficiently. Moreover, nearly 90% of ZEA was degraded when this recombinant Prx was employed for the treatment of ZEA-contaminated corn [93]. However, the undesirable formation of inclusion bodies may limit the high overexpression of Prx. in E. coli. To eliminate the disadvantages, Tang et al. (2013) successfully expressed Prx as a secreted product in the eukaryote S. cerevisiae, which avoided the formation of inclusion bodies and complex purification step. Furthermore, S. cerevisiae conforms to the standards of GRAS by FDA in USA, which is particularly beneficial for the practical application of recombinant Prx in real matrices of foods and feeds [94]. Researchers have also tried to express the ZEA-degrading enzyme in Pichia pastoris, a robust fermentation organism with the ability to secrete protein into the medium. Xiang et al.(2016) reported a high level expressed lactone hydrolase ZHD with the specific activity of 4976.5 U mg−1 in the recombinant P. pastoris X3c derived from Gliocladium roseum by codon optimization and bio-brick method [95]. Bi et al. employed P. pastoris as a secretory expression system to express a zearalenone lactonase gene from Neurospora crassa (ZENC). The maximal enzyme activity of ZENC reached 290.6 U mL−1 in a 30-L fermenter using high-density fermentation. Moreover, ZENC was also found to be efficient in ZEN-containing materials with high degradation rate, providing a foundation for industrial application [96]. In 2018, Wang et al. expressed a novel lactonohydrolase Zhd518 in E. coli with high degrading activities against ZEA and its derivatives. The optimal temperature and pH of Zhd518 was 40 °C and 8.0, indicating that this enzyme can be used as an excellent candidate for ZEA detoxification in the food and feed industry [97]. Yang et al. (2017) reported the first successful expression of a ZEA-degrading enzyme ZHD101 in a probiotic strain Lactobacillus reuteri Pg4 without losing their probiotic properties. It is suggested that L. reuteri pNZ-zhd101 can be used as a probiotic feed additive with the function of ZEA-degradation [98].
Table 3.
Degradation of mycotoxin by recombinant enzymes.
| Enzyme | Gene Source | Target Toxin | Degrading Products | Expression System | Degrading Properties | Degradation Mechanism | Reference |
|---|---|---|---|---|---|---|---|
| Cytochrome P450 3A37 |
Turkey liver | AFB1 |
exo-AFBO aflatoxin Q1 |
E. coli |
exo-AFBO, Km: 287 ±21 µmol L−1 Vmax: 1.45 ± 0.07 nmol min−1 P450 aflatoxin Q1, Km: 302 ±51 µmol L−1 Vmax: 7.86 ±0.75 nmol min−1 |
Epoxidation | Rawal et al. (2010) |
| Peroxiredoxin (Prx) |
Acinetobacter sp. SM04 | ZEA | NM | E. coli | The optimum degradation pH and temperature was 9.0 and 70 °C in presence of H2O2 | Oxidation | Yu et al. (2012) |
| Lactonohydrolase Zhd518 |
Clonostachys rosea | ZEA | NM | E. coli | Activity of 207.0 U mg−1 with the optimal temperature and pH at 40 °C and 8.0 | NM | Wang et al. (2018) |
| Peroxiredoxin | Acinetobacter sp. SM04 | ZEA | NM | Saccharomyces cerevisiae | Optimal activity at pH 9.0, 80 °C and H2O2 concentration of 20 mmol L−1 Thermal stable, alkali resistance |
Oxidation | Tang et al. (2013) |
| Lactone hydrolase ZHD | Gliocladium roseum | ZEA | α-zearalenol and β-zearalenol | Pichia pastoris | Enzyme activity in shake flask fermentation was 22.5 U mL−1 with the specific activity of 4976.5 U mg−1 The maximum enzyme activity of the supernatant was 150.1 U mL−1 in 5-L fermenter |
Cleavage of lactone ring | Xiang et al. (2016) |
| Lactonohydrolase | Clonostachys rosea | ZEA | 1-(3,5-dihydroxy-phenyl)-10-hydroxy-1-undecen6-one | Lactobacillus reuteri Pg4 | Did not affect cell growth, acid and bile salt tolerance | Cleavage of lactone ring | Yang et al. (2017) |
| Lactonase | Neurospora crassa | ZEA | NM | Pichia pastoris | Optimal activity at pH 8.0 and 45 °C, highly stable at pH 6.0–8.0 for 1 h at 37 °C, the maximal enzyme activity reached 290.6 U mL−1 in 30-L fermenter |
NM | Bi et al. (2018) |
| Carboxylesterases, type B | Sphingopyxis sp. MTA144 | FB1 | NM | E. coli | NM | Deesterification | Heinl et al. (2010) |
| Aminotransferases, class III | HFB1 | Oxygen independence, temperature range 6–50 °C with an optimum at 35 °C, and pH adaptation 6–10 with an optimum at pH 8.5 | Deamination | ||||
| Aminotransferase FumI | Sphingopyxis sp. MTA144 | HFB1 | NM | E. coli | Optimal activity at pH 8.5 and 35 °C, low salt concentration, the kinetic parameters Km = 1.1 μmol L−1 and kcat = 104 min−1 | Eamination | Hartinger et al. (2011) |
| Putative amidase | Aspergillus niger | OTA | NM | NM | Thermostable, optimal activity at pH 6.0 and 66 °C | Hydrolysis | Dobritzsch et al. (2014) |
| N-acyl-L-amino acid amidohydrolase | Alcaligenes faecalis | OTA | β-phenylalanine | E. coli | Optimal activity at pH 6.5 and 50 °C | OTA amide bond hydrolysis | Zhang et al. (2019) |
| Dehydrogenase DepA |
Devosia mutans 17-2-E-8 | DON | 3-keto-DON | E. coli | NM | Oxidation of C3 position | Carere et al. (2017) |
| Cytochrome P450 | Sphingomonas sp. strain KSM1 | DON | 16-hydroxy-deoxynivalenol | E. coli | kcat/Km of 6.4 mmol L−1 s−1 | Hydroxylation | Ito et al. (2013) |
| Fusion ZHDCP enzyme |
Clonostachy rosea B. amyloliquefaciens ASAG |
ZEA OTA |
HZEA, DZEA, OTAα |
E. coli | 100% degradation rate at pH 7 and 30 °C in 2 h | Hydrolysis Removal of an amino acid from the end of a peptide chain |
Azam et al. (2019) |
| Manganese peroxidase |
Irpex lacteus, Phanerochaete chrysosporium, Ceriporiopsis subvermispora, Nematoloma frowardii |
AFB1 ZEA DON FB1 |
AFB1-8,9-epoxide | E. coli | In the presence of dicarboxylic acid malonate | Oxidoreduction | Wang et al. (2019) |
NM, not mentioned.
Recombinant enzymes have also been used for the degradation of FB1. The process involves at least two enzymatic reactions, namely, de-esterification by a carboxylesterase resulting in hydrolyzed FB1 followed by an oxidative deamination of the hydrolysis product. Based on this, Heinl et al. (2010) isolated and heterologously expressed a carboxylesterase and an aminotransferase gene from Sphingopyxis sp. MTA144 for consecutive FB1 detoxification [99]. It is worth noting that the aminotransferase (FumI) is independent of oxygen, which is especially beneficial for its applications under oxygen limited conditions such as ensilaged forage and animal intestinal tract. FumI is tolerant to a wide range of temperature (6–50 °C) and pH (6–10),with the optimum activity at 35 °C and pH 8.5 [100]. In 2014, the cloning, purification and characterization of two amidases from A. niger was reported by Dobritzsch et al. these thermo stable amidases were demonstrated to possess OTA-hydrolyzing activity with optimal activity at pH 6 and 66 °C [101]. In a recent study, Zhang et al. purified and identified a novel OTA degrading enzyme, N-acyl-L-amino acid amidohydrolase (AfOTase) from A. faecalis DSM and expressed it in E. coli. AfOTase exhibited commendable thermal and pH stability [102]. Carere et al. identified a dehydrogenase with the capability to transform DON to the less toxic 3-keto-DON [103]. Cytochrome P450s play important roles in the metabolism of endogenous and exogenous substrates. Rawal et al. (2010) described the heterologous expression and functional characterization of P4503A37 from a turkey liver with high aflatoxin B1 epoxidation activity [104]. Ito et al. (2013) reported a cytochrome P450 system with DON-catabolic activity in Sphingomonas sp. strain KSM1. This enzyme system can transform DON to 16-hydroxy-deoxynivalenol. Similar degrading abilities were also demonstrated toward nivalenol and 3-acetyl deoxynivalenol, toxins that frequently co-occur with DON [105]. These findings provide a biochemical and genetic basis for economic production of recombinant enzymes to eliminate the toxic threat.
To manage the co-contamination of different mycotoxins, the recombinant genetic engineering of two or more enzymes for the degradation of multiple mycotoxins has become a new development direction. Azam et al. (2019) reported a novel bifunctional recombinant fusion enzyme (ZHDCP) combining zearalenone hydrolase (ZHD) and carboxypeptidase (CP) by genetic engineering technology. The bifunctional enzyme was able to completely degrade ZEA and OTA in 2 h and 30 min, respectively. The final products were less harmful with reduced cell death and mitochondrial apoptosis. The result demonstrated that bifunctional purified enzyme can be more advanced and superior than the single strain or enzyme [106]. Another feasible route would be the use of a single mycotoxin-degrading enzyme with wide substrate specificity on multiple mycotoxins for the treatment of two or even more mycotoxins co-contamination. A recent study revealed that eight recombinantly produced manganese peroxidase (MnP), responsible for lignin oxidative depolymerization in fungi, could degrade four major mycotoxins including AFB1, ZEA, DON and FB1 in the presence of a dicarboxylic acid malonate [107].
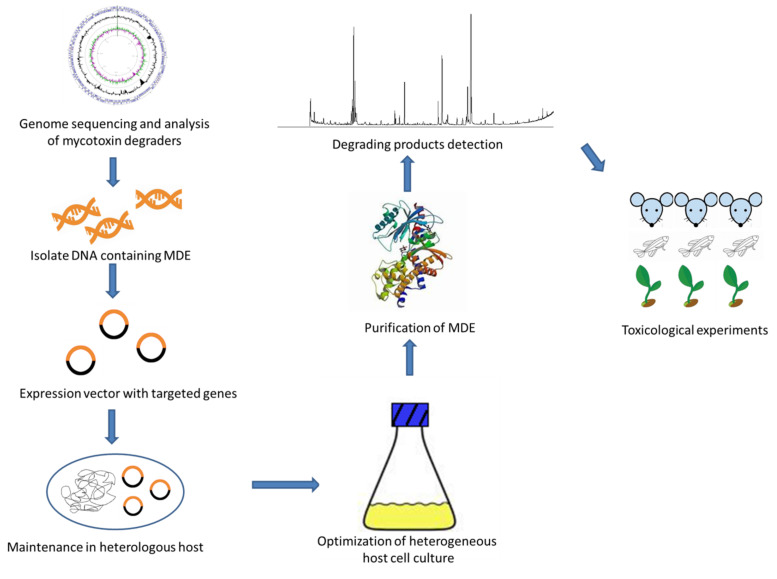
The above studies made contributions to the application of genetic engineering methods to mycotoxins degradation; however, the commercial application of recombinant MDE is still far from achievable. To provide a foundation for the industrial application of recombinant MDE, here we suggest a research route as shown in Figure 2. The screening and cloning of new degradation pathways and MDE genes are of great importance to the development of the entire industry. Novel metagenomics screening surveys may serve as the resources for the acquisition of diverse MDE from unexplored complex natural samples. To realize the efficient production of MDE, optimization of incubation and purification conditions including temperature and pH is necessary. Moreover, before the applying MDEs in real matrices, the possible toxic effects of the degrading products should be fully evaluated.
Figure 2.
The mechanism of heterogeneous expression of mycotoxin degradation enzyme (MDE).
6. Application, Prospect and Challenges
6.1. Unexplored Mechanisms of Mycotoxins Degradation
Over the last decade, knowledge about new organisms and metabolic pathways on the biological mycotoxin degradation has grown rapidly. Tracking the fate of converted products from mycotoxins created through the metabolism of living organisms or via enzyme reaction is necessary for commercial application. However, most studies report on preliminary investigation of degradation ability without characterizing the intermediate metabolites and final products that are sometimes hardly analyzable under laboratory conditions. A full understanding of the structure and stability of degradation products could provide useful information for evaluating their potential side-effects and health impact. Traditional analysis methods, based on instruments such as LC-MS/MS, GC-MS/MS, and nuclear magnetic resonance (NMR) were not able to detect all the metabolites involved in the conversion of mycotoxins. New strategies for elucidating a pathway of biodegradation must be developed. Wang et al. (2020) reported a metabolomics-guided analysis to reveal metabolism during DON degradation catalyzed by bacterial consortium IFSN-C1 [108]. In the study of Pinedo et al. (2018) isotopic labeling experiments with 13C-labeled substrates helped to reveal the detoxification pathway of patulin to desoxypatulinic acid (DPA) by Rhodotorula kratochvilovae LS11 via the hydrolysis of the γ-lactone ring and, subsequently, enzymatic modifications [109]. With the development of genome sequencing technology and gene bank database, advanced high-throughput approaches, such as functional metagenomic and comparative transcriptomic, provide robust tools for the identification of novel genes responsible involved in biodegradation. In the study of Yu et al. (2019), community-level BPA degradation pathway was constructed based on the integrated data of metabolites and metagenomics [110]. Moreover, Zhang et al. (2019) performed a transcriptome analysis to investigate mechanisms involved in OTA degradation by Yarrowia lipolytica Y-2. Notably, the gene ECM14 encoding putative metallocarboxypeptidase ECM14 was up-regulated in Y. lipolytica Y-2 when treated with OTA [111]. As supplementary of experimental research, computational study with bioinformatic analysis could help to improve the understanding of the enzyme functions. Tomin et al. (2019) performed an extensive computational study to investigate the affinity of an aflatoxin oxidase (AFO) from Armillariella tabescens toward AFB1, and elucidated the details of the enzyme structure. The results of this study strongly support that AFO is a typical member of the dipeptidyl peptidase III family [112].
6.2. Biosafety Evaluation of Degradation Products
Over recent years, mycotoxin degradation by biological methods has been studied extensively. However, the possible toxicity of residual by-products formed in the degradation process has been overlooked. It is noticeable that toxins are not necessarily degraded into water and carbon dioxide [113]. It is therefore important to evaluate the biosafety of the degradation products to explain the mechanism and of biological mycotoxin degradation and lay a foundation for its commercial application. So far, only a limited number of reports have elucidated the structure of some converted products and performed short-term preliminary tests for the estimation of their toxicity. Zhu et al. (2017) summarized the models and methodologies for toxicity determination of mycotoxins and by-products in a review paper [11]. Hosts that are used in toxicity tests vary from animals that are ranked highly within the evolutionary tree to bacteria. Specific cell lines, such as MCF-7 cells, may be more suitable for testing certain toxins including estrogenic activity and cytotoxicity. Recently, the zebrafish has emerged as an important toxicological model, based on which easier routes have been developed for the evaluation of teratogenicity [114]. Haq et al. (2016) employed the zebrafish embryo toxicity assay (ZETA) to evaluate the teratogenicity of OTA and its degradation product OTα. Results of this study suggested that ZETA could be regarded as a useful and sensitive tool for evaluating the safety of detoxification strategies for contaminants including microbial toxins [115]. Since rigid parameters for the assessment of the safety of detoxification products have not been yet sufficiently established, it is necessary to have a full understanding of any associated risks before industrial applications of biological degradation agents.
6.3. Limited Practical Application in Real Matrices
Despite the extensive research on the biological degradation of mycotoxins by microorganisms and their enzymes, the practical application in food and feeds matrices is limited. Since the degradation process under conditions of commercial-scale production is much more complex, laboratory experiments might not always reflect practices in industrial processing. The possible reasons are that the degradation activities can be easily affected by multiple factors, such as co-factor requirement, pH, ionic-strength, temperature, etc. Given the above situation, novel non-pathogenic strains with high toxin-degradation performance and good stability are in considerable demand for practical use. Besides, the survival and adaptability of microorganisms under simulated conditions should be fully evaluated for mycotoxin detoxification in animal guts. Moreover, from an economical point of view, the cost of a lengthy incubation time and the fermentation materials should be lowered. For example, studies reported in the literature have found fermentation with LAB was an effective approach for mycotoxin degradation as summarized in an excellent review of Adebiyi et al. (2019) [116]. This food-processing technique not only provides a cheap, readily available and sustainable strategy for mycotoxin control, but also ensure the beneficial sensorial, nutritional and health composition of food. LAB strains with antifungal activity are also be reported to contribute to the safety of malt modification. In a pilot-scale malting trial with barley grains infected with F. culmorum, the application of Lactobacillus reuteri R29 cfs produced in 3 °P substrate successfully reduced mycotoxin DON by 83%, which represents a practical and inexpensive alternative to mycotoxin control in the malt industry [117].
7. Conclusions
The widespread contamination of mycotoxins has attracted worldwide attention. Many strategies have been developed for mycotoxin removal, and biological degradation is considered as the safest strategy. Probiotics, which possess plenty of beneficial properties, are reported to be useful for mycotoxins removal. The application of a microbial consortium provides a promising solution for the effective co-degradation of multiple mycotoxins. Besides, with the development of genetic engineering technology, recombinant degrading enzymes are increasingly favored in biological detoxification. It is necessary to develop recombinant degrading enzymes capable of degrading multi-toxins. Currently, however, most biological detoxification technologies have not yet been developed for the commercial scale, since a thorough understanding of the degradation pathways and the potential health effects of the degradation products does not yet exist. To facilitate the commercial application of these approaches, future studies should focus on explanining of the detailed degradation mechanisms, conducting-toxicology studies of metabolites, and developing novel analytical methods. Given the great diversity of complex matrices, further research is recommended to highlight the field-applicable degradation technologies for the control of mycotoxins. In the future, more efforts should be dedicated to exploring strategies for mycotoxins biodegradation that are cost-effective, highly efficient, instrumently simple and easy to operate for large-scale application in industry.
Author Contributions
L.L. and M.X. wrote the original draft. D.W. supervised and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the GDAS’ Project of Science and Technology Development (2020GDASYL-20200103026) and the National Natural Science Foundation of China (Grant Number 32001812). This work is partly supported by Guangdong Basic and Applied Basic Research Foundation (2019B1515120002).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Taheur F.B., Kouidhi B., al Qurashi Y.M.A., Salah-Abbes J.B., Chaieb K. Review: Biotechnology of mycotoxins detoxification using microorganisms and enzymes. Toxicon. 2019;160:12–22. doi: 10.1016/j.toxicon.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Savi G.D., Piacentini K.C., Scussel V.M. Ozone treatment efficiency in Aspergillus and Penicillium growth inhibition and mycotoxin degradation of stored wheat grains (Triticum aestivum L.) J. Food Process. Preserv. 2015;39:940–948. doi: 10.1111/jfpp.12307. [DOI] [Google Scholar]
- 3.Temba B.A., Sultanbawa Y., Kriticos D.J., Fox G.P., Harvey J.J., Fletcher M.T. Tools for defusing a major global food and feed safety risk: Nonbiological postharvest procedures to decontaminate mycotoxins in foods and feeds. J. Agric. Food Chem. 2016;64:8959–8972. doi: 10.1021/acs.jafc.6b03777. [DOI] [PubMed] [Google Scholar]
- 4.Klaric M.S., Cvetnic Z., Pepeljnjak S., Kosalec I. Co-occurrence of aflatoxins, ochratoxin A, fumonisins, and zearalenone in cereals and feed, determined by competitive direct enzyme-linked immunosorbent assay and thin-layer chromatography. Arh. Hig. Rada Toksikol. 2009;60:427–434. doi: 10.2478/10004-1254-60-2009-1975. [DOI] [PubMed] [Google Scholar]
- 5.Schatzmayr G., Zehner F., Taubel M., Schatzmayr D., Klimitsch A., Loibner A.P., Binder E.M. Microbiologicals for deactivating mycotoxins. Mol. Nutr. Food Res. 2006;50:543–551. doi: 10.1002/mnfr.200500181. [DOI] [PubMed] [Google Scholar]
- 6.Bocarov-Stancic A., Adamovic M., Salma N., Bodroza-Solarov M., Vuckovic J., Pantic V. In vitro efficacy of mycotoxins adsorption by natural mineral adsorbents. Biotechnol. Anim. Husb. 2011;27:1241–1251. doi: 10.2298/BAH1103241B. [DOI] [Google Scholar]
- 7.He J., Zhou T., Young J.C., Boland G.J., Scott P.M. Chemical and biological transformations for detoxification of trichothecene mycotoxins in human and animal food chains: A review. Trends Food Sci. Technol. 2010;21:67–76. doi: 10.1016/j.tifs.2009.08.002. [DOI] [Google Scholar]
- 8.Li P., Su R., Yin R., Lai D., Wang M., Liu Y., Zhou L. Detoxification of mycotoxins through biotransformation. Toxins. 2020;12:121. doi: 10.3390/toxins12020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W., Chang J., Wan P., Liu C., Yin Q., Lu F., Gao T. Effect of the combined compound probiotics with mycotoxin–degradation enzyme on detoxifying aflatoxin B1, and zearalenone. J. Toxicol. Sci. 2018;43:377–385. doi: 10.2131/jts.43.377. [DOI] [PubMed] [Google Scholar]
- 10.Afsah-Hejri L., Hajeb P., Ehsani R.J. Application of ozone for degradation of mycotoxins in food: A review. Compr. Rev. Food Sci. Food Saf. 2020;19:1777–1808. doi: 10.1111/1541-4337.12594. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y., Hassan Y.I., Lepp D., Shao S., Zhou T. Strategies and methodologies for developing microbial detoxification systems to mitigate mycotoxins. Toxins. 2017;9:130. doi: 10.3390/toxins9040130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyagin I., Efremenko E. Enzymes for detoxification of various mycotoxins: Origins and mechanisms of catalytic action. Molecules. 2019;24:2362. doi: 10.3390/molecules24132362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato I., Ito M., Ishizaka M., Ikunaga Y., Sato Y., Yoshida S., Koitabashi M., Tsushima S. Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. FEMS Microbiol. Lett. 2012;327:110–117. doi: 10.1111/j.1574-6968.2011.02461.x. [DOI] [PubMed] [Google Scholar]
- 14.Liuzzi V.C., Fanelli F., Tristezza M., Haidukowski M., Picardi E., Manzari C., Lionetti C., Grieco F., Logrieco A.F., Thon M.R., et al. Transcriptional analysis of Acinetobacter sp. neg1, capable of degrading ochratoxin a. Front. Microbiol. 2016;7:2162. doi: 10.3389/fmicb.2016.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monachese M., Burton J.P., Reid G. Bioremediation and tolerance of humans to heavy metals through microbial processes: A potential role for probiotics? Appl. Environ. Microbiol. 2012;78:6397–6404. doi: 10.1128/AEM.01665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajasekharan S.K., Lee J.H., Zhao Y., Lee J. The mycotoxin zearalenone hinders Candida albicans biofilm formation and hyphal morphogenesis. Indian J. Microbiol. 2018;58:19–27. doi: 10.1007/s12088-017-0690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Xie Y. Review on microbial degradation of zearalenone and aflatoxins. Grain Oil Sci. Technol. 2020;3:117–125. doi: 10.1016/j.gaost.2020.05.002. [DOI] [Google Scholar]
- 18.Liu B., Peng X., Meng X. Effective biodegradation of mycotoxin patulin by porcine pancreatic lipase. Front. Microbiol. 2018;9:615. doi: 10.3389/fmicb.2018.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Nussairawi M., Risa A., Garai E., Varga E., Szabo I., Csenki-Bakos Z., Kriszt B., Cserhati M. Mycotoxin biodegradation ability of the Cupriavidus Genus. Curr. Microbiol. 2020;77:2430–2440. doi: 10.1007/s00284-020-02063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva L.J.G., Pereira A., Pena A., Lino C.M. Citrinin in foods and supplements: A review of occurrence and analytical methodologies. Foods. 2020;10:14. doi: 10.3390/foods10010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marin S., Ramos A.J., Cano-Sancho G., Sanchis V. Mycotoxins: Occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. 2013;60:218–237. doi: 10.1016/j.fct.2013.07.047. [DOI] [PubMed] [Google Scholar]
- 22.Khan G.I. Eco-friendly bacterial biodegradation of mycotoxins. Pure Appl. Biol. 2020;9:1708–1717. doi: 10.19045/bspab.2020.90181. [DOI] [Google Scholar]
- 23.Loi M., Fanelli F., Liuzzi V.C., Logrieco A.F., Mule G. Mycotoxin biotransformation by native and commercial enzymes: Present and future perspectives. Toxins. 2017;9:111. doi: 10.3390/toxins9040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gil-Serna J., Vázquez C., Patiño B. Mycotoxins in functional beverages: A Review. Beverages. 2020;6:52. doi: 10.3390/beverages6030052. [DOI] [Google Scholar]
- 25.Vanhoutte I., Audenaert K., de Gelder L. Biodegradation of mycotoxins: Tales from known and unexplored worlds. Front. Microbiol. 2016;7:561. doi: 10.3389/fmicb.2016.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter G., Pereg L. A review on the relation between soil and mycotoxins: Effect of aflatoxin on field, food and finance. Eur. J. Soil Sci. 2019;70:882–897. doi: 10.1111/ejss.12813. [DOI] [Google Scholar]
- 27.Adebo O.A., Njobeh P.B., Sidu S., Adebiyi J.A., Mavumengwana V. Aflatoxin B1, degradation by culture and lysate of a Pontibacter specie. Food Control. 2017;80:99–103. doi: 10.1016/j.foodcont.2017.04.042. [DOI] [Google Scholar]
- 28.Afshar P., Shokrzadeh M., Raeisi S.N., Ghorbani-HasanSaraei A., Nasiraii L.R. Aflatoxins biodetoxification strategies based on probiotic bacteria. Toxicon. 2020;178:50–58. doi: 10.1016/j.toxicon.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Nazhand A., Durazzo A., Lucarini M., Souto E.B., Santini A. Characteristics, occurrence, detection and detoxification of aflatoxins in foods and feeds. Foods. 2020;9:644. doi: 10.3390/foods9050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polak-Sliwinska M., Paszczyk B. Trichothecenes in food and feed, relevance to human and animal health and methods of detection: A systematic review. Molecules. 2021;26:454. doi: 10.3390/molecules26020454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bezuidenhout C.C., Prinsloo M., van der Walt A.M. Multiplex PCR-based detection of potential fumonisin-producing Fusarium in traditional African vegetables. Environ. Toxicol. 2006;21:360–366. doi: 10.1002/tox.20193. [DOI] [PubMed] [Google Scholar]
- 32.Zhu M., Cen Y., Ye W., Li S., Zhang W. Recent advances on macrocyclic trichothecenes, their bioactivities and biosynthetic pathway. Toxins. 2020;12:417. doi: 10.3390/toxins12060417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khaneghah A.M., Farhadi A., Nematollahi A., Vasseghian Y., Fakhri Y. A systematic review and meta-analysis to investigate the concentration and prevalence of trichothecenes in the cereal-based food. Trends Food Sci. Technol. 2020;102:193–202. doi: 10.1016/j.tifs.2020.05.026. [DOI] [Google Scholar]
- 34.Van der Fels-Klerx H., Stratakou I. T-2, toxin and HT-2, toxin in grain and grain-based commodities in Europe: Occurrence, factors affecting occurrence, co-occurrence and toxicological effects. World Mycotoxin J. 2010;3:349–367. doi: 10.3920/WMJ2010.1237. [DOI] [Google Scholar]
- 35.McCormick S.P., Stanley A.M., Stover N.A., Alexander N.J. Trichothecenes: From simple to complex mycotoxins. Toxins. 2011;3:802–814. doi: 10.3390/toxins3070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocha O., Ansari K., Doohan F.M. Effects of trichothecene mycotoxins on eukaryotic cells: A review. Food Addit. Contam. 2005;22:369–378. doi: 10.1080/02652030500058403. [DOI] [PubMed] [Google Scholar]
- 37.Zhou T., He J., Gong J. Microbial transformation of trichothecene mycotoxins. World Mycotoxin J. 2008;1:23–30. doi: 10.3920/WMJ2008.x003. [DOI] [Google Scholar]
- 38.Stanciu O., Juan C., Miere D., Berrada H., Loghin F., Manes J. First study on trichothecene and zearalenone exposure of the Romanian population through wheat-based products consumption. Food Chem. Toxicol. 2018;121:336–342. doi: 10.1016/j.fct.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Kumar P., Mahato D.K., Sharma B., Borah R., Haque S., Mahmud M.M.C., Shah A.K., Rawal D., Bora H., Bui S. Ochratoxins in food and feed: Occurrence and its impact on human health and management strategies. Toxicon. 2020;187:151–162. doi: 10.1016/j.toxicon.2020.08.031. [DOI] [PubMed] [Google Scholar]
- 40.Bayman P., Baker J.L. Ochratoxins: A global perspective. Mycopathologia. 2006;162:215–223. doi: 10.1007/s11046-006-0055-4. [DOI] [PubMed] [Google Scholar]
- 41.Reddy L., Bhoola K. Ochratoxins-food contaminants: Impact on human health. Toxins. 2010;2:771–779. doi: 10.3390/toxins2040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rai A., Dixit S., Singh S.P., Gautam N.K., Das M., Tripathi A. Presence of zearalenone in cereal grains and its exposure risk assessment in indian population. J. Food Sci. 2018;83:3126–3133. doi: 10.1111/1750-3841.14404. [DOI] [PubMed] [Google Scholar]
- 43.Al-Jaal B.A., Jaganjac M., Barcaru A., Horvatovich P., Latiff A. Aflatoxin, fumonisin, ochratoxin, zearalenone and deoxynivalenol biomarkers in human biological fluids: A systematic literature review, 2001–2018. Food Chem. Toxicol. 2019;129:211–228. doi: 10.1016/j.fct.2019.04.047. [DOI] [PubMed] [Google Scholar]
- 44.Kowalska K., Habrowska-Gorczynska D.E., Piastowska-Ciesielska A.W. Zearalenone as an endocrine disruptor in humans. Environ. Toxicol. Pharmacol. 2016;48:141–149. doi: 10.1016/j.etap.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Popiel D., Koczyk G., Dawidziuk A., Gromadzka K., Blaszczyk L., Chelkowski J. Zearalenone lactonohydrolase activity in Hypocreales and its evolutionary relationships within the epoxide hydrolase subset of a/b-hydrolases. BMC Microbiol. 2014;14:82. doi: 10.1186/1471-2180-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y.-S., Zhou Y., Lu S.-Y., Guo D.-J., Ren H.-L., Meng X.-M., Zhi B.-H., Lin C., Wang Z., Li X.-B., et al. Development of a one-step test strip for rapid screening of fumonisins B1, B2, and B3, in maize. Food Control. 2012;24:72–77. doi: 10.1016/j.foodcont.2011.09.005. [DOI] [Google Scholar]
- 47.Mirasoli M., Buragina A., Dolci L.S., Simoni P., Anfossi L., Giraudi G., Roda A. Chemiluminescence-based biosensor for fumonisins quantitative detection in maize samples. Biosens. Bioelectron. 2012;32:283–287. doi: 10.1016/j.bios.2011.11.039. [DOI] [PubMed] [Google Scholar]
- 48.Silva L.J., Pena A., Lino C.M., Fernandez M.F., Manes J. Fumonisins determination in urine by LC-MS-MS. Anal. Bioanal. Chem. 2010;396:809–816. doi: 10.1007/s00216-009-3231-9. [DOI] [PubMed] [Google Scholar]
- 49.Voss K.A., Smith G.W., Haschek W.M. Fumonisins: Toxicokinetics, mechanism of action and toxicity. Animal Feed Sci. Technol. 2007;137:299–325. doi: 10.1016/j.anifeedsci.2007.06.007. [DOI] [Google Scholar]
- 50.Morgavi D.P., Riley R.T. Fusarium and their toxins: Mycology, occurrence, toxicity, control and economic impact. Animal Feed Sci. Technol. 2007;137:199–200. doi: 10.1016/j.anifeedsci.2007.06.001. [DOI] [Google Scholar]
- 51.Diao E., Hou H., Hu W., Dong H., Li X. Removing and detoxifying methods of patulin: A review. Trends Food Sci. Technol. 2018;81:139–145. doi: 10.1016/j.tifs.2018.09.016. [DOI] [Google Scholar]
- 52.Rodriguez A., Luque M.I., Andrade M.J., Rodriguez M., Asensio M.A., Cordoba J.J. Development of real-time PCR methods to quantify patulin-producing molds in food products. Food Microbiol. 2011;28:1190–1199. doi: 10.1016/j.fm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 53.Tannous J., el Khoury R., Snini S.P., Lippi Y., el Khoury A., Atoui A., Lteif R., Oswald I.P., Puel O. Sequencing, physical organization and kinetic expression of the patulin biosynthetic gene cluster from Penicillium expansum. Int. J. Food Microbiol. 2014;189:51–60. doi: 10.1016/j.ijfoodmicro.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H., Ahima J., Yang Q., Zhao L., Zhang X., Zheng X. A review on citrinin: Its occurrence, risk implications, analytical techniques, biosynthesis, physiochemical properties and control. Food Res. Int. 2021;141:110075. doi: 10.1016/j.foodres.2020.110075. [DOI] [PubMed] [Google Scholar]
- 55.Wan M.L.Y., Forsythe S.J., El-Nezami H. Probiotics interaction with foodborne pathogens: A potential alternative to antibiotics and future challenges. Crit. Rev. Food Sci. Nutr. 2019;59:3320–3333. doi: 10.1080/10408398.2018.1490885. [DOI] [PubMed] [Google Scholar]
- 56.Parvez S., Malik K.A., Kang S.A., Kim H.Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006;100:1171–1185. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 57.Dalié D.K.D., Deschamps A.M., Richard-Forget F. Lactic acid bacteria—Potential for control of mould growth and mycotoxins: A review. Food Control. 2010;21:370–380. doi: 10.1016/j.foodcont.2009.07.011. [DOI] [Google Scholar]
- 58.Gaggia F., Mattarelli P., Biavati B. Probiotics and prebiotics in animal feeding for safe food production. Int. J. Food Microbiol. 2010;141:S15–S28. doi: 10.1016/j.ijfoodmicro.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 59.Kesarcodi-Watson A., Kaspar H., Lategan M.J., Gibson L. Probiotics in aquaculture: The need, principles and mechanisms of action and screening processes. Aquaculture. 2008;274:1–14. doi: 10.1016/j.aquaculture.2007.11.019. [DOI] [Google Scholar]
- 60.Zoghia A., Khosravi-Darani K., Sohrabvandib S. Surface binding of toxins and heavy metals by probiotics. Mini Rev. Med. Chem. 2014;14:84–98. doi: 10.2174/1389557513666131211105554. [DOI] [PubMed] [Google Scholar]
- 61.Shetty P.H., Jespersen L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Technol. 2006;17:48–55. doi: 10.1016/j.tifs.2005.10.004. [DOI] [Google Scholar]
- 62.Chiocchetti G.M., Jadan-Piedra C., Monedero V., Zuniga M., Velez D., Devesa V. Use of lactic acid bacteria and yeasts to reduce exposure to chemical food contaminants and toxicity. Crit. Rev. Food Sci. Nutr. 2019;59:1534–1545. doi: 10.1080/10408398.2017.1421521. [DOI] [PubMed] [Google Scholar]
- 63.Hatab S., Yue T., Mohamad O. Removal of patulin from apple juice using inactivated lactic acid bacteria. J. Appl. Microbiol. 2012;112:892–899. doi: 10.1111/j.1365-2672.2012.05279.x. [DOI] [PubMed] [Google Scholar]
- 64.Peltonen K., El-Nezami H., Haskard C., Ahokas J., Salminen S. Aflatoxin B1, binding by dairy strains of lactic acid bacteria and bifidobacteria. J. Dairy Sci. 2001;84:2152–2156. doi: 10.3168/jds.S0022-0302(01)74660-7. [DOI] [PubMed] [Google Scholar]
- 65.Prete V.D., Rodriguez H., Carrascosa A.V. In vitro removal of ochratoxin a by wine lactic acid bacteria. J. Food Prot. 2007;70:2155–2160. doi: 10.4315/0362-028X-70.9.2155. [DOI] [PubMed] [Google Scholar]
- 66.Sangsila A., Faucet-Marquis V., Pfohl-Leszkowicz A., Itsaranuwat P. Detoxification of zearalenone by Lactobacillus pentosus strains. Food Control. 2016;62:187–192. doi: 10.1016/j.foodcont.2015.10.031. [DOI] [Google Scholar]
- 67.Vinderola G., Ritieni A. Role of probiotics against mycotoxins and their deleterious effects. J. Food Res. 2014;4:10–21. doi: 10.5539/jfr.v4n1p10. [DOI] [Google Scholar]
- 68.Chapot-Chartier M.P., Kulakauskas S. Cell wall structure and function in lactic acid bacteria. Microb. Cell Factories. 2014;13:S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niderkorn V., Morgavi D.P., Aboab B., Lemaire M., Boudra H. Cell wall component and mycotoxin moieties involved in the binding of fumonisin B1, and B2, by lactic acid bacteria. J. Appl. Microbiol. 2009;106:977–985. doi: 10.1111/j.1365-2672.2008.04065.x. [DOI] [PubMed] [Google Scholar]
- 70.Haskard C., Binnion C., Ahokas J. Factors affecting the sequestration of aflatoxin by Lactobacillus rhamnosus strain GG. Chem. Biol. Interact. 2000;128:39–49. doi: 10.1016/S0009-2797(00)00186-1. [DOI] [PubMed] [Google Scholar]
- 71.Hernandez-Mendoza A., Guzman-de-Pena D., Garcia H.S. Key role of teichoic acids on aflatoxin B binding by probiotic bacteria. J. Appl. Microbiol. 2009;107:395–403. doi: 10.1111/j.1365-2672.2009.04217.x. [DOI] [PubMed] [Google Scholar]
- 72.Jouany J.P., Yiannikouris A., Bertin G. The chemical bonds between mycotoxins and cell wall components of Saccharomyces cerevisiae have been identified. Arch. Zootech. 2005;8:26–50. [Google Scholar]
- 73.Yiannikouris A., Poughon L., Cameleyre X., Dussap C.G., François J., Bertin G., Jouany J.P. A novel technique to evaluate interactions between Saccharomyces cerevisiae cell wall and mycotoxins: Application to zearalenone. Biotechnol. Lett. 2003;25:783–789. doi: 10.1023/A:1023576520932. [DOI] [PubMed] [Google Scholar]
- 74.Harkai P., Szabó I., Cserháti M., Krifaton C., Risa A., Radó J., Balázs A., Berta K., Kriszt B. Biodegradation of aflatoxin-B1, and zearalenone by Streptomyces sp. collection. Int. Biodeterior. Biodegrad. 2016;108:48–56. doi: 10.1016/j.ibiod.2015.12.007. [DOI] [Google Scholar]
- 75.Rao K.R., Vipin A.V., Hariprasad P., Appaiah K.A.A., Venkateswaran G. Biological detoxification of Aflatoxin B1, by Bacillus licheniformis CFR1. Food Control. 2017;71:234–241. [Google Scholar]
- 76.Juodeikiene G., Bartkiene E., Cernauskas D., Cizeikiene D., Zadeike D., Lele V., Bartkevics V. Antifungal activity of lactic acid bacteria and their application for Fusarium mycotoxin reduction in malting wheat grains. LWT Food Sci. Technol. 2018;89:307–314. doi: 10.1016/j.lwt.2017.10.061. [DOI] [Google Scholar]
- 77.Nathanail A.V., Gibson B., Han L., Peltonen K., Ollilainen V., Jestoi M., Laitila A. The lager yeast Saccharomyces pastorianus removes and transforms Fusarium trichothecene mycotoxins during fermentation of brewer’s wort. Food Chem. 2016;203:448–455. doi: 10.1016/j.foodchem.2016.02.070. [DOI] [PubMed] [Google Scholar]
- 78.Festa S., Coppotelli B.M., Morelli I.S. Bacterial diversity and functional interactions between bacterial strains from a phenanthrene-degrading consortium obtained from a chronically contaminated-soil. Int. Biodeterior. Biodegrad. 2013;85:42–51. doi: 10.1016/j.ibiod.2013.06.006. [DOI] [Google Scholar]
- 79.Zafra G., Absalon A.E., Anducho-Reyes M.A., Fernandez F.J., Cortes-Espinosa D.V. Construction of PAH-degrading mixed microbial consortia by induced selection in soil. Chemosphere. 2017;172:120–126. doi: 10.1016/j.chemosphere.2016.12.038. [DOI] [PubMed] [Google Scholar]
- 80.Beeton S., Bull A.T. Biotransformation and detoxification of T-2, toxin by soil and freshwater bacteria. Appl. Environ. Microbiol. 1989;55:190–197. doi: 10.1128/aem.55.1.190-197.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He W.J., Yuan Q.S., Zhang Y.B., Guo M.W., Gong A.D., Zhang J.B., Wu A.B., Huang T., Qu B., Li H.P., et al. Aerobic de-epoxydation of trichothecene mycotoxins by a soil bacterial consortium isolated using in situ soil enrichment. Toxins. 2016;8:277. doi: 10.3390/toxins8100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y., Zhao C., Zhang D., Zhao M., Peng M., Guo P., Cui Z. Microbial degradation of zearalenone by a novel microbial consortium, NZDC-6, and its application on contaminated corncob by semisolid fermentation. J. Agric. Food Chem. 2020;68:1634–1644. doi: 10.1021/acs.jafc.9b05343. [DOI] [PubMed] [Google Scholar]
- 83.Zhao Z., Zhang Y., Gong A., Liu N., Chen S., Zhao X., Li X., Chen L., Zhou C., Wang J. Biodegradation of mycotoxin fumonisin B1, by a novel bacterial consortium SAAS79. Appl. Microbiol. Biotechnol. 2019;103:7129–7140. doi: 10.1007/s00253-019-09979-6. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y., Zhao C., Zhang D., Zhao M., Zheng D., Lyu Y., Cheng W., Guo P., Cui Z. Effective degradation of aflatoxin B1, using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 2017;224:166–173. doi: 10.1016/j.biortech.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 85.Kosicki R., Błajet-Kosicka A., Grajewski J., Twarużek M. Multiannual mycotoxin survey in feed materials and feedingstuffs. Animal Feed Sci. Technol. 2016;215:165–180. doi: 10.1016/j.anifeedsci.2016.03.012. [DOI] [Google Scholar]
- 86.Sun L.H., Lei M.Y., Zhang N.Y., Gao X., Li C., Krumm C.S., Qi D.S. Individual and combined cytotoxic effects of aflatoxin B1, zearalenone, deoxynivalenol and fumonisin B1, on BRL 3A rat liver cells. Toxicon. 2015;95:6–12. doi: 10.1016/j.toxicon.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y., Zhao C., Zhang D., Zhao M., Zheng D., Peng M., Cheng W., Guo P., Cui Z. Simultaneous degradation of aflatoxin B1, and zearalenone by a microbial consortium. Toxicon. 2018;146:69–76. doi: 10.1016/j.toxicon.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 88.Chang J., Wang T., Wang P., Yin Q., Liu C., Zhu Q., Lu F., Gao T. Compound probiotics alleviating aflatoxin B1, and zearalenone toxic effects on broiler production performance and gut microbiota. Ecotoxicol. Environ. Saf. 2020;194:110420. doi: 10.1016/j.ecoenv.2020.110420. [DOI] [PubMed] [Google Scholar]
- 89.Huang W., Chang J., Wang P., Liu C., Yin Q., Song A., Gao T., Dang X., Lu F. Effect of compound probiotics and mycotoxin degradation enzymes on alleviating cytotoxicity of swine jejunal epithelial cells induced by aflatoxin B(1) and zearalenone. Toxins. 2019;11:12. doi: 10.3390/toxins11010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hartinger D., Moll W. Fumonisin elimination and prospects for detoxification by enzymatic transformation. World Mycotoxin J. 2011;4:271–283. doi: 10.3920/WMJ2011.1285. [DOI] [Google Scholar]
- 91.Alberts J.F., Gelderblom W.C., Botha A., van Zyl W.H. Degradation of aflatoxin B(1) by fungal laccase enzymes. Int. J. Food Microbiol. 2009;135:47–52. doi: 10.1016/j.ijfoodmicro.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 92.Ece S., Lambertz C., Fischer R., Commandeur U. Heterologous expression of a Streptomyces cyaneus laccase for biomass modification applications. AMB Express. 2017;7:86. doi: 10.1186/s13568-017-0387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu Y., Wu H., Tang Y., Qiu L. Cloning, expression of a peroxiredoxin gene from Acinetobacter sp. SM04, and characterization of its recombinant protein for zearalenone detoxification. Microbiol. Res. 2012;167:121–126. doi: 10.1016/j.micres.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 94.Tang Y., Xiao J., Chen Y., Yu Y., Xiao X., Yu Y., Wu H. Secretory expression and characterization of a novel peroxiredoxin for zearalenone detoxification in Saccharomyces cerevisiae. Microbiol. Res. 2013;168:6–11. doi: 10.1016/j.micres.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 95.Xiang L., Wang Q., Zhou Y., Yin L., Zhang G., Ma Y. High-level expression of a ZEN-detoxifying gene by codon optimization and biobrick in Pichia pastoris. Microbiol. Res. 2016;193:48–56. doi: 10.1016/j.micres.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 96.Bi K., Zhang W., Xiao Z., Zhang D. Characterization, expression and application of a zearalenone degrading enzyme from Neurospora crassa. AMB Express. 2018;8:194. doi: 10.1186/s13568-018-0723-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang M., Yin L., Hu H., Selvaraj J.N., Zhou Y., Zhang G. Expression, functional analysis and mutation of a novel neutral zearalenone-degrading enzyme. Int. J. Biol. Macromol. 2018;118:1284–1292. doi: 10.1016/j.ijbiomac.2018.06.111. [DOI] [PubMed] [Google Scholar]
- 98.Yang W.C., Hsu T.C., Cheng K.C., Liu J.R. Expression of the Clonostachys rosea lactonohydrolase gene by Lactobacillus reuteri to increase its zearalenone-removing ability. Microb Cell Fact. 2017;16:69. doi: 10.1186/s12934-017-0687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heinl S., Hartinger D., Thamhesl M., Vekiru E., Krska R., Schatzmayr G., Moll W.D., Grabherr R. Degradation of fumonisin B1, by the consecutive action of two bacterial enzymes. J. Biotechnol. 2010;145:120–129. doi: 10.1016/j.jbiotec.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 100.Hartinger D., Schwartz H., Hametner C., Schatzmayr G., Haltrich D., Moll W.D. Enzyme characteristics of aminotransferase FumI of Sphingopyxis sp. MTA144, for deamination of hydrolyzed fumonisin B(1) Appl. Microbiol. Biotechnol. 2011;91:757–768. doi: 10.1007/s00253-011-3248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dobritzsch D., Wang H., Schneider G., Yu S. Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochem. J. 2014;462:441–452. doi: 10.1042/BJ20140382. [DOI] [PubMed] [Google Scholar]
- 102.Zhang H., Zhang Y., Yin T., Wang J., Zhang X. Heterologous expression and characterization of a novel ochratoxin a degrading enzyme, N-acyl-L-amino acid amidohydrolase, from Alcaligenes faecalis. Toxins. 2019;11:518. doi: 10.3390/toxins11090518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carere J., Hassan Y.I., Lepp D., Zhou T. The enzymatic detoxification of the mycotoxin deoxynivalenol: Identification of DepA from the DON epimerization pathway. Microb. Biotechnol. 2018;11:1106–1111. doi: 10.1111/1751-7915.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rawal S., Yip S.S.M., Coulombe R.A. Cloning, expression and functional characterization of cytochrome P450, 3A37, from turkey liver with high aflatoxin B1, epoxidation activity. Chem. Res. Toxicol. 2010;23:1322–1329. doi: 10.1021/tx1000267. [DOI] [PubMed] [Google Scholar]
- 105.Ito M., Sato I., Ishizaka M., Yoshida S., Koitabashi M., Yoshida S., Tsushima S. Bacterial cytochrome P450, system catabolizing the Fusarium toxin deoxynivalenol. Appl. Environ. Microbiol. 2013;79:1619–1628. doi: 10.1128/AEM.03227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Azam M.S., Yu D., Liu N., Wu A. Degrading ochratoxin A and zearalenone mycotoxins using a multifunctional recombinant enzyme. Toxins. 2019;11:301. doi: 10.3390/toxins11050301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang X., Qin X., Hao Z., Luo H., Yao B., Su X. Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins. 2019;11:566. doi: 10.3390/toxins11100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang G., Wang Y., Man H., Lee Y.W., Shi J., Xu J. Metabolomics-guided analysis reveals a two-step epimerization of deoxynivalenol catalyzed by the bacterial consortium IFSN-C1. Appl. Microbiol. Biotechnol. 2020;104:6045–6056. doi: 10.1007/s00253-020-10673-1. [DOI] [PubMed] [Google Scholar]
- 109.Pinedo C., Wright S.A.I., Collado I.G., Goss R.J.M., Castoria R., Hrelia P., Maffei F., Duran-Patron R. Isotopic labeling studies reveal the patulin detoxification pathway by the biocontrol yeast Rhodotorula kratochvilovae LS11. J. Nat. Prod. 2018;81:2692–2699. doi: 10.1021/acs.jnatprod.8b00539. [DOI] [PubMed] [Google Scholar]
- 110.Yu K., Yi S., Li B., Guo F., Peng X., Wang Z., Wu Y., Alvarez-Cohen L., Zhang T. An integrated meta-omics approach reveals substrates involved in synergistic interactions in a bisphenol A (BPA)-degrading microbial community. Microbiome. 2019;7:16. doi: 10.1186/s40168-019-0634-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang X., Yang H., Zheng X., Zhao L., Gu X., Wang K., Apaliya M.T., Ahima J., Zhang H. Protein and transcript profiling analysis of the response of Yarrowia lipolytica Y-2, in the degradation of ochratoxin A. Ann. Appl. Biol. 2019;175:98–110. doi: 10.1111/aab.12515. [DOI] [Google Scholar]
- 112.Tomin M., Tomic S. Oxidase or peptidase? A computational insight into a putative aflatoxin oxidase from Armillariella tabescens. Proteins. 2019;87:390–400. doi: 10.1002/prot.25661. [DOI] [PubMed] [Google Scholar]
- 113.Ianiri G., Idnurm A., Wright S.A., Duran-Patron R., Mannina L., Ferracane R., Ritieni A., Castoria R. Searching for genes responsible for patulin degradation in a biocontrol yeast provides insight into the basis for resistance to this mycotoxin. Appl. Environ. Microbiol. 2013;79:3101–3115. doi: 10.1128/AEM.03851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Berry J.P., Gantar M., Gibbs P.D., Schmale M.C. The zebrafish (Danio rerio) embryo as a model system for identification and characterization of developmental toxins from marine and freshwater microalgae. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007;145:61–72. doi: 10.1016/j.cbpc.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haq M., Gonzalez N., Mintz K., Jaja-Chimedza A., de Jesus C.L., Lydon C., Welch A., Berry J.P. Teratogenicity of ochratoxin A and the degradation product, ochratoxin alpha, in the zebrafish (Danio rerio) embryo model of vertebrate development. Toxins. 2016;8:40. doi: 10.3390/toxins8020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Adebiyi J.A., Kayitesi E., Adebo O.A., Changwa R., Njobeh P.B. Food fermentation and mycotoxin detoxification: An African perspective. Food Control. 2019;106:106731. doi: 10.1016/j.foodcont.2019.106731. [DOI] [Google Scholar]
- 117.Oliveira P., Brosnan B., Jacob F., Furey A., Coffey A., Zannini E., Arendt E.K. Lactic acid bacteria bioprotection applied to the malting process. Part II: Substrate impact and mycotoxin reduction. Food Control. 2015;51:444–452. doi: 10.1016/j.foodcont.2014.11.011. [DOI] [Google Scholar]