Abstract
Background and Aims
Plant diseases caused by Pectobacterium atrosepticum are often accompanied by extensive rot symptoms. In addition, these bacteria are able to interact with host plants without causing disease for long periods, even throughout several host plant generations. There is, to date, no information on the comparative physiology/biochemistry of symptomatic and asymptomatic plant–P. atrosepticum interactions. Typical (symptomatic) P. atrosepticum infections are associated with the induction of plant responses mediated by jasmonates, which are one of the products of the lipoxygenase cascade that gives origin to many other oxylipins with physiological activities. In this study, we compared the functioning of the lipoxygenase cascade following typical and latent (asymptomatic) infections to gain better insight into the physiological basis of the asymptomatic and antagonistic coexistence of plants and pectobacteria.
Methods
Tobacco plants were mock-inoculated (control) or infected with the wild type P. atrosepticum (typical infection) or its coronafacic acid-deficient mutant (latent infection). The expression levels of the target lipoxygenase cascade-related genes were assessed by Illumina RNA sequencing. Oxylipin profiles were analysed by GC-MS. With the aim of revising the incorrect annotation of one of the target genes, its open reading frame was cloned to obtain the recombinant protein, which was further purified and characterized using biochemical approaches.
Key Results
The obtained data demonstrate that when compared to the typical infection, latent asymptomatic P. atrosepticum infection is associated with (and possibly maintained due to) decreased levels of 9-lipoxygenase branch products and jasmonic acid and increased level of cis-12-oxo-10,15-phytodienoic acid. The formation of 9-oxononanoic acid and epoxyalcohols in tobacco plants was based on the identification of the first tobacco hydroperoxide lyase (HPL) with additional epoxyalcohol synthase (EAS) activity.
Conclusions
Our results contribute to the hypothesis of the oxylipin signature, indicating that different types of plant interactions with a particular pathogen are characterized by the different oxylipin profiles of the host plant. In addition, the tobacco LOC107825278 gene was demonstrated to encode an NtHPL (CYP74C43) enzyme yielding volatile aldehydes and aldoacids (HPL products) as well as oxiranyl carbinols (EAS products).
Keywords: Latent and typical infections, lipoxygenase cascade, oxylipins, Pectobacterium, plant soft rots
INTRODUCTION
Most (if not all) phytopathogenic microorganisms are able to not only cause diseases but also colonize host plants without manifestation of symptoms (Sinclair, 1991; Takahashi et al., 2019). Moreover, the asymptomatic forms of host–pathogen interactions seem to be even more common in the environment than symptomatic interactions. The identification of physiological markers of different forms of infection (symptomatic and latent) is of particular interest, since it may provide a basis for effective disease control. However, very little information has so far been obtained on the comparative physiology and biochemistry of different types of infections.
Pectobacterium and Dickeya species belong to the soft rot Pectobacteriaceae (SRP), which cause plant tissue degradation following the production of multiple plant cell wall-degrading enzymes (PCWDEs) (Charkowski et al., 2012). Latent infections have been widely described for SRP-infected plants (Hayward, 1974; Pérombelon and Kelman, 1980; Pérombelon, 2002; Toth and Birch, 2005; Charkowski, 2007). Often, latent SRP infections can last for long periods, spanning up to or even throughout several host plant generations, until the equilibrium between host and pathogen is disturbed and the asymptomatic interaction turns to disease.
Coronafacic acid (CA) was shown in our previous study on Pectobacterium atrosepticum (Pba) and tobacco to serve as a switch that transforms latent infection into a typical symptomatic infection (Gorshkov et al., 2018). CA, which is a component of a phytotoxin coronatine described for Pseudomonas syringae (Ichihara et al., 1977; Mitchell, 1984), is a functional analogue of jasmonates. The CA-deficient Pba mutant leads to a much lower induction of the jasmonate-regulated gene LOX2 compared to the wild type, causing only latent xylem-limited infections rather than the typical infections associated with extensive colonization of the parenchyma (Gorshkov et al., 2014, 2018). This probably means that the induction of jasmonate-mediated responses is a marker of plant susceptibility to Pba and an indicator of the typical Pba infection.
Jasmonates are the products of the lipoxygenase cascade, which gives rise not only to jasmonic acid (JA) derivatives but also to a variety of oxylipin products, many of which possess physiological activities (Hughes et al., 2009; Wasternack and Feussner, 2018). The synthesis of oxylipins starts with the oxidation of polyunsaturated fatty acids (linoleic and α-linolenic) at the C9 or C13 positions by lipoxygenases (LOXs), yielding 9- and 13-hydroperoxides (HPOs). Accordingly, the LOXs are subdivided into 9- and 13-LOXs. LOX-derived HPOs are further converted via distinct metabolic branches mediated by the cytochromes P450 of the CYP74 family: allene oxide synthases (AOSs), hydroperoxide lyases (HPLs), divinyl ether synthases (DESs) and epoxyalcohol synthases (EASs). The CYP74 enzymes of each type are subdivided into 9- and 13-specific depending on the substrate utilized: 9- or 13-HPOs. AOSs transform HPOs into unstable allene oxides that are converted into ketols (α and ɣ) and cyclopentenones, including cis-12-oxo-10,15-phytodienoic acid (12-OPDA) and dinor-OPDA, which are further converted into JA via 12-OPDA reductases (OPR3 and OPR2, respectively) and β-oxidation steps (Li et al., 2005; Chini et al., 2018; Wasternack and Feussner, 2018). The HPL branch yields a bulk of short-chain volatile aldehydes (e.g. hexenals) and ω-oxo fatty acids (Noordermeer et al., 2001). DESs and EASs convert HPOs into divinyl ethers and epoxyalcohols, respectively (Galliard and Phillips, 1972; Hamberg, 1999; Lee et al., 2008; Gogolev et al., 2012; Gorina et al., 2016).
The most widely studied oxylipins are undoubtedly the JA derivatives. Jasmonates provide resistance to herbivores and participate in plant–pathogen cross-talk, abiotic stress and common physiological processes (Savchenko et al., 2014; Howe et al., 2018). HPL-synthesized hexenals are also involved in herbivore resistance. In addition, the HPL branch yields the wound phytohormone traumatin as well as compounds with bactericidal, fungicidal or antioxidant properties (Pietryczuk and Czerpak, 2011). The physiological roles of DES- and EAS-synthesized metabolites have been poorly investigated. Some divinyl ethers, epoxyalcohols and trihydroxy acids were shown to participate in the defence responses against phytopathogens (Kato et al., 1985; Williams et al., 2000; Granér et al., 2003; Prost et al., 2005; Fammartino et al., 2010; Toporkova et al., 2018a).
Oxylipin composition is considered to reflect the physiological/functional state of a plant; therefore, the term ‘oxylipin signature’ has been put forward (Weber et al., 1997). Changes in oxylipin signature have been shown to take place in the course of plant development and ageing (Hause et al., 2000; Delaplace et al., 2008), as well as during drought- and toxin-caused stress (Savchenko and Dehesh, 2014; Hanano et al., 2018). The oxylipin profile has also been demonstrated to undergo different alterations depending on the attacking herbivore: aphid and Colorado potato beetle induced the 9-LOX and 13-LOX pathways, respectively, in potato plants (Gosset et al., 2009). Variations in oxylipin signature have also been observed in potato plants when different types of infections develop, namely Phytophthora infestans symptomatic infection and Pseudomonas syringae abortive infection, associated with a hypersensitive response (Göbel et al., 2002). The enhanced synthesis of 9-LOX oxylipins, such as colnelic and colneleic acids, occurred following both types of infection; however, during Phytophthora infestans symptomatic infection the accumulation of these oxylipins took several days, compared to several hours during the Pseudomonas syringae abortive infection. In addition, the synthesis of JA was typical of abortive but not symptomatic infection (Göbel et al., 2002). The mutualistic interactions of plants with microorganisms (Cirsium arvense and Chaetomium cochlioides) have also been shown to ‘shift’ the oxylipin profile, leading to an increase in dihydrojasmonic acid, 12-OPDA and galactolipid content (Hartley et al., 2015). Latent infections, however, have not yet been compared with typical infections in terms of the oxylipin profile, as well as any different infection types caused by particular pathogen species.
Our recent study showed that the typical Pba infection in tobacco is coupled with the upregulation of LOX-, AOS- and DES-encoding genes, together with an increase in LOX enzymatic activity (Tsers et al., 2020). Whether the activities of CYP74 family enzymes also increase, and which oxylipins accumulate in infected plants remain to be determined. Additionally, latent Pba infection has not been investigated in terms of lipoxygenase cascade activity. Therefore, the aim of the present study was to compare the functioning of the lipoxygenase cascade during typical and latent Pba infections. Analyses of gene expression, the activity of corresponding enzymes and the relative abundance of different oxylipins enabled the identification of lipoxygenase cascade-related differences between typical and latent Pba infections. In addition, we carried out a re-annotation of some of the CYP74 enzymes in tobacco and identified a gene that encodes 9/13-HPL with additional EAS activity.
MATERIALS AND METHODS
Bacteria and plant growth conditions, plant inoculation
The strains of the wild type Pectobacterium atrosepticum SCRI1043 (Pba) (Bell et al., 2004) and its coronafacic acid-deficient cfa mutant (Gorshkov et al., 2018) were kindly provided by Dr Yevgeny Nikolaichik (Belarus State University, Belarus). Bacterial strains were grown overnight in lysogeny broth (LB) at 28 °C and 180 rpm. Nicotiana tabacum cv. Petit Havana SR1 plants were grown axenically in tubes in a growth chamber with a 16-h light/8-h dark cycle photoperiod. Seeds were surface-sterilized using bleach (0.8 % active chlorine) and 1 % sodium dodecyl sulphate for 30 min, washed seven times with sterile distilled water, then transferred to Murashige and Skoog medium (MS) in Petri dishes. Ten-day-old seedlings were transferred to individual sterile tubes containing MS.
Six weeks after planting, tobacco plants were infected with Pba or cfa mutant. For plant inoculation, bacteria were grown until the early stationary phase, then washed with sterile 10 mm MgSO4 and resuspended in the same solution up to a density of ~2 × 107 CFU mL−1. Sterile 10 mm MgSO4 or bacterial suspensions containing ~2 × 105 cells were placed as 10-μL drops into the bosoms of the leaves in the middle part of the stems using sterile pipette tips, and slight scratches were made simultaneously. Two days after plant inoculation, when disease symptoms were visible on Pba-infected plants, the plant material was harvested for analysis.
RNA extraction and cDNA library preparation
Stem sections in the asymptomatic stem zone (0.5 cm below the symptomatic, macerated area formed at the point of inoculation) were taken from Pba-infected plants for RNA extraction; the corresponding stem sections were taken from cfa mutant-infected and non-infected plants. Plant material was ground in liquid nitrogen. The obtained powder was resuspended in 1 mL ExtractRNA Reagent (Evrogen, Russia) and the subsequent procedures were performed according to the manufacturer’s instructions. Residual DNA was eliminated using a DNA-free kit (Life Technologies, USA). RNA quantity and quality were analysed using a Qubit fluorimeter (Life Technologies) and Qsep100 DNA Analyzer (Bioptic, Taiwan), respectively.
For RNA-sequencing (RNA-seq), total RNA (1 µg) was enriched by mRNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, UK). mRNA was processed using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) according to the manufacturer’s instructions. The quality and quantity of the cDNA libraries before sequencing were monitored using an Agilent 2100 Bioanalyzer (Agilent, USA) and a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA). Libraries were sequenced in three biological replicates. Sequencing was conducted on an Illumina HiSeq 2500 (Illumina, USA) at the Joint KFU–Riken Laboratory, Kazan Federal University (Russia).
Gene expression analysis
The quality of the obtained RNA-Seq reads was assessed using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads with q-score <30 and rRNA-corresponding reads were filtered out using Trimmomatic and SortMeRNA, respectively (Kopylova et al., 2012; Bolger et al., 2014). To create the optimal reference for read pseudo-alignment, the EvidentialGene package (https://sourceforge.net/projects/evidentialgene/) was used with default parameters to reduce the redundancy of tobacco coding sequence (CDS) (NCBI Assembly GCF_000715135.1). Pseudo-alignment and quantification of filtered reads were carried out using kallisto (Bray et al., 2016) with default parameters. The edgeR package (Robinson and Oshlack, 2010) was used to reveal differentially expressed genes (DEGs). Genes that had TMM-normalized read counts per million (CPM) values ≥1 in all replicates within at least one of the experimental conditions (typical/latent infection or control plants) were considered to be expressed in our study. Genes with |log2FC|>1 and false discovery rate (FDR) <0.05 were considered to be DEGs. The functional annotations were assigned to the DEGs as described earlier (Tsers et al., 2020).
Profiling of oxylipins
Two 3-g portions of plant leaves were ground in liquid nitrogen. The first portion was supplemented with 75 µg margaric acid (internal standard) and homogenized with ten volumes (w/v) of ice-cold hexane/ethyl acetate 1 : 1 (v/v). The obtained homogenates were centrifuged (8000 g, 20 min, 4 °C) and supernatants were collected. The second portion was homogenized in 6 mL of ice-cold 50 mm Tris-HCl buffer, pH 7.0, using Ultra-Turrax (IKA, Germany), filtered through cheesecloth, and incubated with 150 µg linoleic, 150 µg α-linolenic and 75 µg margaric (internal standard) acids at 23 °C for 30 min under continuous oxygen bubbling. The reaction mixture was acidified to pH 6.0 and the reaction products were extracted with a 10-mL mixture of ethyl acetate/hexane 1 : 1 (v/v).
The solvents in the obtained extracts from the first and the second portions were evaporated. The resulting total lipid extracts were dissolved in chloroform/isopropanol 2 : 1 (v/v) and passed through Supelclean LC-NH2 (3 mL) cartridges (Supelco, USA). Then, free carboxylic acids were eluted with solvent mixture ethyl acetate/acetic acid 98 : 2 (v/v). The products were methylated with ethereal diazomethane and trimethylsilylated with pyridine/hexamethyldisilazane/trimethylchlorosilane 1 : 1 : 1 (v/v/v) mixture at 23 °C for 30 min. Additionally, the products were reduced with NaBH4, then methylated and trimethylsilylated. The silylation reagents were evaporated in vacuo. Dry residues were dissolved in 100 µL of hexane and the resulting methyl esters/ Trimethylsilyl (TMS) derivatives (Me/TMS) were subjected to GC-MS analysis.
The GC-MS analyses were performed using a Shimadzu QP5050A mass spectrometer connected to a Shimadzu GC-17A gas chromatograph equipped with a Supelco MDN-5S (5 % phenyl 95 % methylpolysiloxane) fused capillary column (length, 30 m; ID 0.25 mm; film thickness, 0.25 µm). Helium at a flow rate of 30 cm s–1 was used as the carrier gas. Injections were performed in the split mode using an initial column temperature of 120 °C and injector temperature 230 °C. The column temperature was raised at 10 °C min–1 to 240 °C. Electron impact ionization (70 eV) was used.
The abundance of the target products was measured by the integration of the total ion current GC-MS chromatograms. The content of oxylipins was determined relative to the internal standard, margaric acid. Oxylipins 2–6 and 9 were quantified as Me esters or Me/TMS derivatives. Oxylipins 1, 7 and 8 were measured as Me/TMS derivatives after preliminary NaBH4 reduction (Supplementary Data Fig. S1) since the quantification of non-reduced derivatives of these compounds can yield inaccurate results. 9-Oxononanoic acid (compound 1) is a more volatile compound than its NaBH4-reduced derivative 9-hydroxynonanoic acid (compound 1a). NaBH4 reduction prevented the thermal rearrangement of hydroperoxides, yielding oxiranyl carbinols (7 and 8) during GC-MS analysis.
Molecular cloning, expression and purification of the recombinant enzyme
To clone the open reading frame (ORF) of the LOC107825278 gene, total RNA was isolated from Pba-infected tobacco plants. One microgram of RNA was used for cDNA synthesis using RevertAid reverse transcriptase (Thermo Scientific, USA) according to the manufacturer’s instructions. cDNA was used as the template for PCR with primers GACGACGACAAGATGTCTTCATTTTCCACATCTTC and GAGGAGAAGCCCGGTGTCGCTCTTTCCACTTTCTTC. Primer construction and multiple sequence alignments were performed using the Vector NTI Software V0.11.5.4 (Thermo Fisher Scientific, USA). The resulting PCR product (~1500 bp) was cloned into the expression vector pET-32 Ek/LIC (Merck KGaA, Germany) using ligation-independent cloning and sequenced to verify the presence of the LOC107825278 gene ORF (which consisted of 1452 bp). The resulting plasmid was transferred into Escherichia coli BL21(DE3)рLysS (Merck) and the recombinant enzyme was obtained as follows: 1 L of LB medium supplemented with mineral medium M9 1 : 1 (v/v) was inoculated by 10 mL of the overnight culture of E. coli BL21(DE3)рLysS carrying the target plasmid. Bacteria were grown at 37 °C, 180 rpm, to an OD600 of 0.6. The expression of the target gene was induced by the addition of 0.1 mm isopropyl-β-d-1-thiogalactopyranoside to the medium. Simultaneously, the medium was supplemented with 100 mg L–1 5-aminolevulinic acid to facilitate haem formation. To obtain the target enzyme, bacteria were harvested by centrifugation for 15 min at 4500 rpm at 4 °C and lysed with BugBuster Protein Extraction Reagent (Merck). Purification of His-tagged recombinant protein was performed using a Bio-Scale Mini Profinity IMAC cartridge in the BioLogic LP chromatographic system (Bio-Rad). The recombinant enzyme was eluted from the cartridge using 30 mm histidine. The concentration of recombinant protein was determined using the Quant-iT™ Protein HS Assay Kit (Thermo Fisher Scientific). The haemoprotein concentration was estimated using the pyridine haemochromogen assay (Schenkman and Jansson, 2006). The relative purity of the recombinant protein was estimated by SDS-PAGE and staining of the gel with Coomassie brilliant blue R-250.
Kinetics studies
The enzymatic activity of the purified recombinant enzyme was determined by monitoring the decrease of the signal at 234 nm using a РВ 2201 В spectrophotometer (SOLAR, Belarus) with substrate concentrations ranging from 5 to 150 μm. The analyses were performed in 0.6 mL of 0.1 m Na phosphate buffer (pH 6.0–9.0) at 25 °C. The initial linear regions of the kinetic curves were used to calculate the rates. The molar extinction coefficient for 9- and 13-hydroperoxides of linoleic and α-linolenic acids at 234 nm was 25 000 m−1 cm−1. Kinetic parameters were calculated by fitting the datasets to a one-site saturation model for simple ligand binding using the SigmaPlot 11 software (Systat Software Inc., USA). Five independent experiments were performed for each specified variant, and averaged values were used for calculation.
Incubations of recombinant enzyme with fatty acid hydroperoxides
(9S,10E,12Z)-9-Hydroperoxy-10,12-octadecadienoic acid (9-HPOD) and (9S,10E,12Z,15Z)-9-hydroperoxy-10,12,15-octadecatrienoic acid (9-HPOT) were prepared by incubation of linoleic and α-linolenic acids (Sigma-Aldrich, Germany), respectively, with the recombinant maize 9-lipoxygenase (GenBank: AAG61118.1) in Na-phosphate buffer (100 mm, pH 6.0) at 0 °C, under continuous oxygen bubbling. (9Z,11E,13S,15Z)-13-Hydroperoxy-9,11,15-octadecatrienoic (13-HPOT) and (9Z,11E,13S)-13-hydroperoxy-9,11-octadecadienoic (13-HPOD) acids were obtained by incubation of α-linolenic and linoleic acids, respectively, with the soybean lipoxygenase type V (Sigma-Aldrich). The extracted hydroperoxides (as free carboxylic acids) were purified twice by normal-phase HPLC on Kromasil Si (250 × 4 mm; 7 µm) under isocratic elution with solvent mixture hexane/isopropanol/acetic acid 98.4 : 1.5 : 0.1 (v/v) at a flow rate of 0.4 mL min–1. Hydroperoxides were chromatographically pure and at least 98 % optically pure, as judged by chiral-phase HPLC.
The recombinant enzyme (10 µg) was incubated with 100 µg of hydroperoxide in Na-phosphate buffer (100 mm, 10 mL, pH 7.0), at 23 °C, for 15 min. The reaction mixture was acidified to pH 6.0, and the products were extracted with a hexane/ethyl acetate 1 : 1 (v/v) mixture. Further product derivatization and GC-MS analyses were performed as described above. Additionally, the products (Me esters) were subjected to hydrogenation over PtO2, followed by trimethylsilylation.
RESULTS
Expression of lipoxygenase cascade-related genes during typical and latent Pba infections in tobacco plants
In the tobacco genome, 12 13-LOX and 19 9-LOX genes are annotated, of which six and 12 genes, respectively, were expressed under the experimental conditions (Supplementary Data Table S1). Three 9-LOX genes were induced during both typical and latent infections; the expression levels of two of them were more than ten times greater during typical infection than during latent infection. Five genes for 13-LOX were induced during typical infection, and only one of them was also induced during latent infection. Although, during typical infection, the quantity of upregulated 13-LOX genes was greater than upregulated 9-LOX genes (five vs. three, respectively), the degree of upregulation and the absolute expression level (transcripts per million, TPM) were greater for 9-LOX genes. The highest value of the absolute expression level (TPM) for 13-LOX genes was 44, while two 9-LOX genes were expressed at TPM values of 671 and 740. The degree of upregulation (log2FC) was 1.5–5.1 for 13-LOX and 5.2–11.2 for 9-LOX (Table S1). This means that both the 9- and 13-LOX pathways of the lipoxygenase cascade were induced during the infection; the 9-LOX pathway was induced to a much greater extent in terms of gene expression than the 13-LOX pathway. The greater induction of the 9-LOX pathway compared to the 13-LOX pathway was also revealed for the latent infection.
The only HPL-encoding gene annotated in the tobacco genome was slightly induced during both types of infection to the same extent (log2FC 1.8) (Table 1). All four annotated AOS-encoding genes were expressed under the experimental conditions; one of the genes was induced during both types of infection and one additional gene was induced only during typical infection. Six of the nine annotated DES-encoding genes were expressed under the experimental conditions, and all of them were induced during typical infection. Five DES-encoding genes were also induced during latent infection, but most of them (four) were upregulated to a much lesser extent compared to the typical infection (Table 1). Both of the annotated AOC-encoding genes were induced during typical infection and one of them was upregulated during the latent infection. Six of the ten 12-OPDA-reductase encoding genes were expressed under the experimental conditions, and four of them were highly upregulated during typical infection. Two of these genes were also induced during latent infection but to a lower level than during the typical infection (Table 1).
Table 1.
Genes for CYP74 enzymes (HPLs, DESs, AOSs) as well as for AOCs and OPRs annotated (NCBI) in the tobacco genome and the pattern of their expression (red, upregulation; blue, downregulation) during latent and typical infections caused by the coronafacic acid-deficient mutant and the wild-type Pectobacterium atrosepticum, respectively
| Enzyme | Locus ID RefSeq | Expr. | Latent infection (LI) vs. control | Typical infection (TI) vs. control | LI vs. TI | ||||
|---|---|---|---|---|---|---|---|---|---|
| Up- (log2FC) | Down- (log2FC) | TPM at LI | Up- (log2FC) | Down- (log2FC) | TPM at TI | log2FC | |||
| HPL | LOC107801729 | yes | 1.8 | 3.5 | 1.8 | 2 | |||
| AOS | LOC107767065 | yes | 5.1 | 6.3 | 7.5 | 21.4 | |||
| LOC107822008 | yes | 5.6 | 8.1 | 24.5 | |||||
| LOC107832778 | yes | 39.5 | -1.7 | 5.2 | 2.2 | ||||
| LOC107783985 | yes | 22.7 | 13.5 | ||||||
| DES | LOC107799697 | yes | 7.6 | 73 | 11.5 | 753.8 | -4 | ||
| LOC107808154 | yes | 7.2 | 25.8 | 11.4 | 324.6 | -4.3 | |||
| LOC107825278 | yes | 4.1 | 6.2 | 6.9 | 27.5 | -2.8 | |||
| LOC107766501 | yes | 1.7 | 1.4 | 4.2 | 4.9 | -2.6 | |||
| LOC107806557 | yes | 1.6 | 1.5 | 2.2 | 1.3 | ||||
| LOC107819622 | yes | 19.4 | 1.2 | 39.7 | -1.7 | ||||
| LOC107766195 | no | ||||||||
| LOC107772346 | no | ||||||||
| LOC107829877 | no | ||||||||
| AOC | LOC107768393 | yes | 3.1 | 80.1 | 4.1 | 106.9 | |||
| LOC107767576 | yes | 82.2 | 1.7 | 65.7 | |||||
| OPR | LOC107766920 | yes | 2.4 | 77.5 | 4.4 | 196 | -2 | ||
| LOC107789205 | yes | 1.9 | 44.6 | 4.6 | 179.5 | -2.7 | |||
| LOC107765005 | yes | 1.9 | 11 | 18.2 | -3.9 | ||||
| LOC107793069 | yes | 0.9 | 3.1 | 3.3 | -2.5 | ||||
| LOC107773112 | yes | 75.7 | 51 | ||||||
| LOC107782790 | yes | 53.7 | 20.8 | ||||||
| LOC107766387 | no | ||||||||
| LOC107801089 | no | ||||||||
| LOC107806437 | no | ||||||||
| LOC107817070 | no | ||||||||
TPM (transcripts per million), the absolute expression level. Log2FC (log2 fold change), the relative expression level. The column ‘Expr’ shows whether a gene is expressed (yes) or not (no) under the experimental conditions. LOC107825278 (marked in bold) was cloned and characterized in the present study (see below). HPL, hydroperoxide lyase; DES, divinyl ether synthase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPR, 12-oxophytodienoic acid reductase.
We also compared the expression levels of genes encoding the components of the JA-signalling cascade: MYC2 transcription factors and TIFY/JAZ transcriptional regulators. Six of the 13 annotated MYC2 genes were induced during typical infection and none of the MYC2 genes were induced during the latent infection. TIFY/JAZ-encoding genes were found among the upregulated ones during both typical and latent infections. However, both the number of upregulated TIFY/JAZ-encoding genes and the degree of their upregulation were greater during typical infection (14 genes) than during latent infection (seven genes) (Supplementary Data Table S2). Thus, the ‘fraction’ of upregulated JA-signalling-related genes was too small during latent infection to state whether this pathway was induced during asymptomatic interaction or not. During typical infection, MYC2- and TIFY/JAZ-encoding genes were greatly induced, indicating that JA-mediated signalling events were strongly activated during symptomatic interaction.
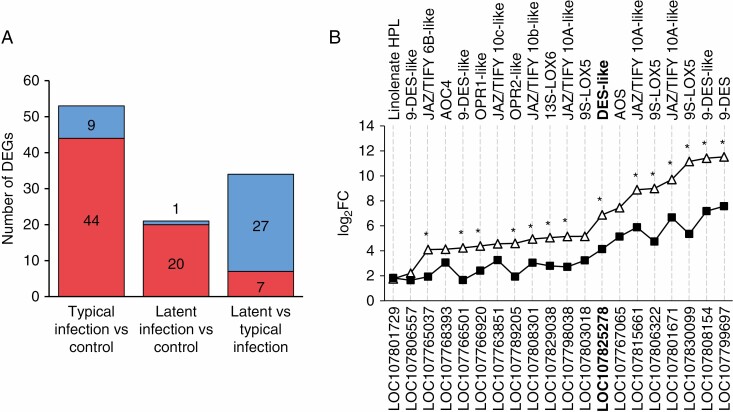
Among all target genes (encoding CYP74 enzymes, LOX, AOC, OPR, TIFY/JAZ, MYC2), 65 genes were expressed under the experimental conditions. Fifty-three of them were DEGs at typical infection vs. control, 21 were DEGs at latent infection vs. control, and 34 were DEGs at latent infection vs. typical infection (Fig. 1A, Table 1; Supplementary Data Tables S1 and S2). Forty-four and 20 DEGs were upregulated during typical and latent infection, respectively, compared to the control, and 27 DEGs were downregulated during latent infection compared to the typical infection. All genes that were upregulated during latent infection compared to the control were also upregulated during the typical infection compared to the control. Twenty-three genes were upregulated specifically during the typical infection. Twenty genes were upregulated during both typical and latent infections and 15 of these ‘infection-upregulated’ genes were induced to a much greater extent during typical infection than during the latent infection (Fig. 1B). None of the ‘infection-upregulated’ genes were induced to a greater extent during latent infection than during the typical infection. All of the genes (seven) that were upregulated during latent infection compared to the typical infection were those that were downregulated during typical infection compared to the control and simultaneously non-differentially expressed at latent infection compared to the control. In other words, the formal upregulation of these genes during latent infection compared to the typical one was not a consequence of their greater induction during latent infection; rather, this was a result of their greater downregulation during the typical infection.
Fig. 1.
The number and expression pattern of differentially expressed genes (DEGs) related to the lipoxygenase cascade and JA signalling in tobacco plants infected with the wild-type Pectobacterium atrosepticum SCRI1043 (causing the typical infection) or coronafacic acid-deficient mutant strain of P. atrosepticum (causing the latent infection). (A) Up- and downregulated DEGs are marked in red and blue, respectively. (B) Expression pattern (log2FC values) of DEGs that were upregulated during both typical (open triangles) and latent (filled squares) infections compared to control. Gene IDs are given on the lower x-axis; the product annotations (NCBI) are given on the upper x-axis. Genes that were expressed differentially during latent infection compared to typical infection are labelled with asterisks (*). HPL, hydroperoxide lyase; DES, divinyl ether synthase; OPR, 12-oxophytodienoic acid reductase; LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase. LOC107825278 (marked in bold) was cloned and characterized in the present study (see below).
Activity of lipoxygenase cascade-related enzymes during typical and latent Pba infections in tobacco plants
The activities of target enzymes – CYP74 enzymes (AOSs, HPLs, DESs and EASs) as well as 12-OPDA-reductases and AOCs – were assessed by analysing the corresponding products in plant extracts after incubation with linoleic and α-linolenic acids. These acids are converted by lipoxygenases into 9- and 13-HPOs, which further serve as substrates for the target enzymes. The products of the AOS [jasmonic acid, cis-12-oxo-10,15-phytodienoic acid (12-OPDA), 9-hydroxy-10-oxo-12-octadecenoic acid (α-ketol)], HPL (9-oxononanoic acid), DES (colneleic acid) and EAS (9,10-epoxy-11-hydroxy-12-octadecenoic acid, 9,10-epoxy-11-hydroxy-12,15-octadecadienoic acid and trihydroxy acids) branches of the lipoxygenase cascade were revealed as well as the products of 9-LOX [(9S,10E,12Z)-9-hydroperoxy-10,12-octadecadienoic acid (9-HPOD), (9S,10E,12Z,15Z)-9-hydroperoxy-10,12,15-octadecatrienoic acid (9-HPOT)] and 13-LOX activities [(9Z,11E,13S)-13-hydroperoxy-9,11-octadecadienoic acid (13-HPOD), (9Z,11E,13S,15Z)-13-hydroperoxy-9,11,15-octadecatrienoic acid (13-HPOT)] (Fig. 2). HPOs were registered in the chromatograms as hydroxy acids (9-HOD, 9-HOT, 13-HOD and 13-HOT). The mass-spectral data are presented in the Supplementary Data File S1. The structural formulae of the listed compounds are presented in Fig. 3.
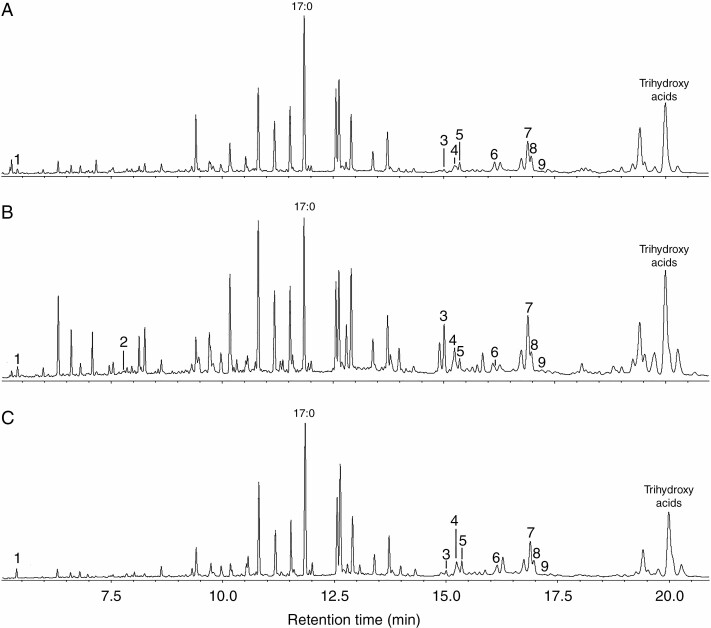
Fig. 2.
The results of total ion current GC-MS analysis of lipoxygenase cascade products (Me/TMS) in tobacco plant extracts after incubation with linoleic and α-linolenic acids: (A) control non-infected plants; (B) plants infected with the wild-type Pectobacterium atrosepticum (typical infection); (C) plants infected with the coronafacic acid-deficient P. atrosepticum mutant (latent infection). 1, 9-Oxononanoic acid (9-HPL product); 2, jasmonic acid (13-AOS branch product); 3, (8E,1′E,3′Z)-9-(1′,3′-nonadienyloxy)-8-nonenoic (colneleic) acid (9-DES product); 4, (9S,10E,12Z)-9-hydroxy-10,12-octadecadienoic acid (9-HOD, derivative of 9-LOX product); 5, (9S,10E,12Z,15Z)-9-hydroxy-10,12,15-octadecatrienoic acid (9-HOT, derivative of 9-LOX product); 6, cis-12-oxo-10,15-phytodienoic acid (13-AOS branch product); 7, 9,10-epoxy-11-hydroxy-12-octadecenoic acid (9-EAS product); 8, 9,10-epoxy-11-hydroxy-12,15-octadecadienoic acid (9-EAS product); 9, 9-hydroxy-10-oxo-12-octadecenoic acid (α-ketol, 9-AOS product); trihydroxy acids (EAS branch product); 17:0, margaric acid (internal standard). The incubation, extraction, derivatization and analysis procedures are described in the Materials and Methods. The structural formulae of the revealed oxylipins are presented in Fig. 3.
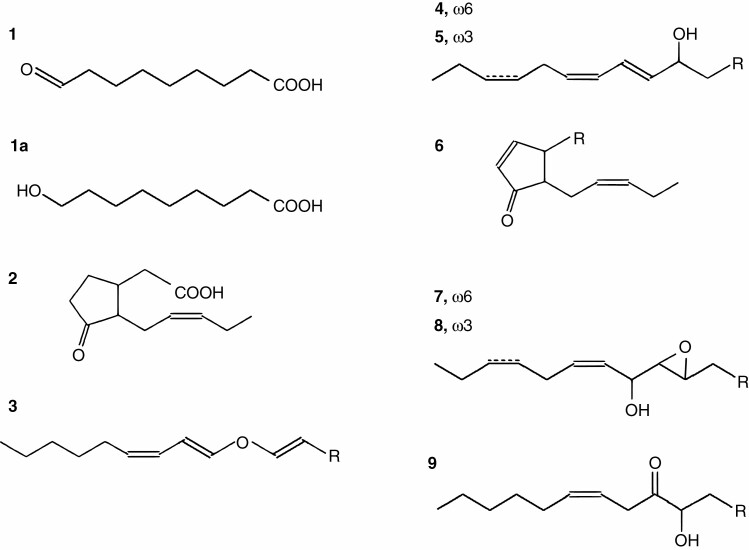
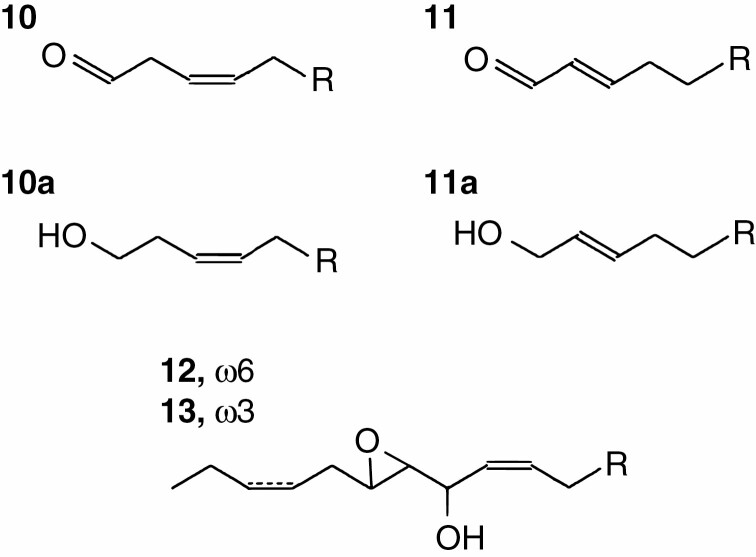
Fig. 3.
Structural formulae of lipoxygenase cascade products detected in our study. 1, 9-Oxononanoic acid; 1a, 9-hydroxynonanoic acid (NaBH4 reduced 9-oxononanoic acid); 2, jasmonic acid; 3, (8E,1′E,3′Z)-9-(1′,3′-nonadienyloxy)-8-nonenoic (colneleic) acid; 4, (9S,10E,12Z)-9-hydroxy-10,12-octadecadienoic acid (9-HOD); 5, (9S,10E,12Z,15Z)-9-hydroxy-10,12,15-octadecatrienoic acid (9-HOT); 6, cis-12-oxo-10,15-phytodienoic acid; 7, 9,10-epoxy-11-hydroxy-12-octadecenoic acid; 8, 9,10-epoxy-11-hydroxy-12,15-octadecadienoic acid; 9, 9-hydroxy-10-oxo-12-octadecenoic acid (α-ketol).
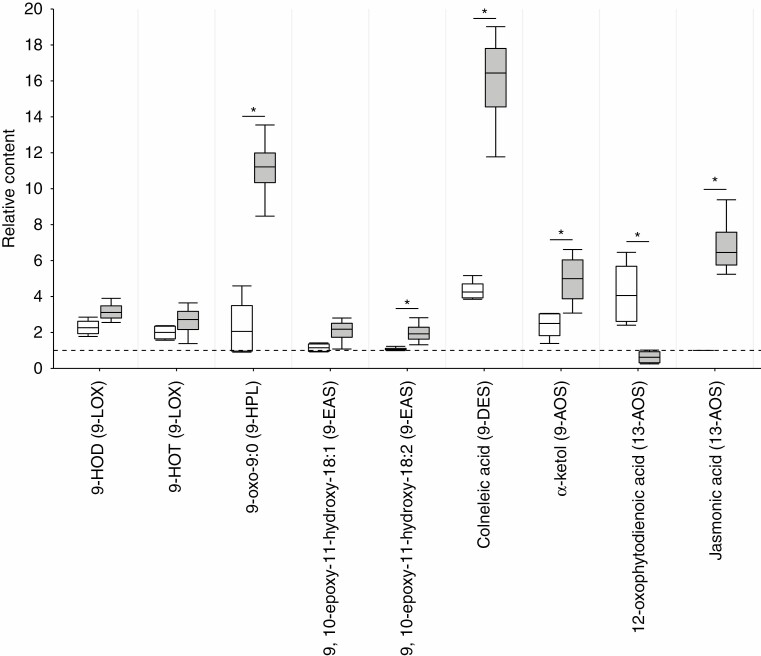
The revealed products were quantified by the integration of the total ion current GC-MS chromatograms. The relative abundance of different oxylipins in the extracts (incubated with linoleate and α-linolenate) of control plants and infected plants with typical and latent infections was considered as a measure of the target enzymatic activities. 9-LOX activity was increased in plants with typical infection compared to the control plants since, in linoleate- and α-linolenate-incubated extracts of Pba-infected plants, the levels of 9-HOD and 9-HOT (derivatives of corresponding HPOs) were 3.1- and 2.7-fold greater, respectively, than in the incubated extracts of control plants. The levels of 9-HOD and 9-HOT were also increased in the incubated extracts of plants with latent infection compared to those of control plants (Fig. 4).
Fig. 4.
Relative abundance of lipoxygenase cascade products in tobacco plant extracts after incubation with linoleic and α-linolenic acids: plants infected with the coronafacic acid-deficient Pectobacterium atrosepticum mutant (latent infection) (white boxplots); plants infected with the wild-type P. atrosepticum (typical infection) (grey boxplots). The abundance of oxylipins in the incubated extracts of control non-infected plants was taken as 1 (horizontal dashed line). The relative abundance of oxylipins was measured by integration of the total ion current GC-MS chromatograms. The content of oxylipins was normalized to the internal standard, margaric acid. Since JA (jasmonic acid) was not detected in the incubated extracts of control plants (as well as plants with latent infection), the minimal detectable peak area was assigned to JA in the extracts of control plants (and plants with latent infection) in order to calculate the relative content of JA in the extracts of plants with typical infection. The boxplots’ upper and lower whiskers extend from the fourth and second quartiles, respectively, to the extreme values no further than 1.5× interquartile range. Asterisks (*) show the significance of the difference in relative content between the experimental groups (Mann–Whitney two-sided test, n = 4, P < 0.05). 9-HOT, (9S,10E,12Z,15Z)-9-hydroxy-10,12,15-octadecatrienoic acid; 9-HOD, (9S,10E,12Z)-9-hydroxy-10,12-octadecadienoic acid; 9-oxo-9:0, 9-oxononanoic acid. The biosynthetic branch for a particular compound is given in parentheses.
The activity of 9-EAS was slightly increased in the extracts of plants with typical, but not with the latent, infection compared to that in the extracts of control plants. The greatest increase in activity during typical infection was observed for 9-HPL and 9-DES: the levels of 9-oxononanoic and colneleic acids were 11.5- and 17.0-fold greater in the incubated extracts of Pba-infected plants than in the extracts of control plants. In the incubated extracts of plants with latent infection, the levels of 9-oxononanoic and colneleic acids were lower than in the extracts of plants with typical infection, but 2.0- and 4.3-fold, respectively, greater than in the extracts of control plants. The content of α-ketol (the product of 9-AOS) was increased 5.0- and 2.5-fold in the incubated extracts of plants with typical and latent infections, respectively, compared to the extracts of control plants. JA (the product of the 13-AOS branch) was detected only in the incubated extracts of plants with typical infection. The level of 12-OPDA, the precursor of JA, did not differ in the extracts of control plants and plants with typical infection, but it was 4.0-fold increased in the extracts of plants with latent infection (Fig. 4).
Together, the results indicate that, during typical infection, the 9-LOX pathway of the cascade was induced entirely (9-LOX, 9-AOS, 9-HPL, 9-DES, 9-EAS) compared to control plants, while, within the 13-LOX pathway, only the 13-AOS branch was induced. During latent infection, the 9-LOX pathway was also mostly induced (except for 9-EAS); however, the induction level during latent infection was lower than during the typical infection. The 13-AOS branch was also the only branch of the 13-LOX pathway that was induced during the latent infection (as well as during typical infection). Different products of the 13-AOS branch were accumulated in the linoleate- and α-linolenate-incubated extracts of plants with typical and latent infections: JA at typical infection and 12-OPDA at latent infection.
Content of oxylipins in tobacco plants during typical and latent Pba infections
The endogenous oxylipins were detected only in trace amounts in control plants and in plants with latent infection (Fig. 5). In plants with typical infection, the products of the 9-LOX (9-HPOs detected as 9-HOD and 9-HOT), 9-HPL (9-oxononanoic acid detected as NaBH4-reduced derivative 9-hydroxynonanoic acid), DES (colneleic acid) and EAS (9,10-epoxy-11-hydroxy-12-octadecenoic and 9,10-epoxy-11-hydroxy-12,15-octadecadienoic acids) branches of the lipoxygenase cascade were revealed. These data are mostly consistent with the results of the determination of enzymatic activities, with one exception. The products of the 9- and 13-AOS branches (JA, 12-OPDA and α-ketol) were not detected among the endogenous oxylipins despite the upregulation of the corresponding enzymes and genes during typical infection (Figs 2 and 4, Table 1).
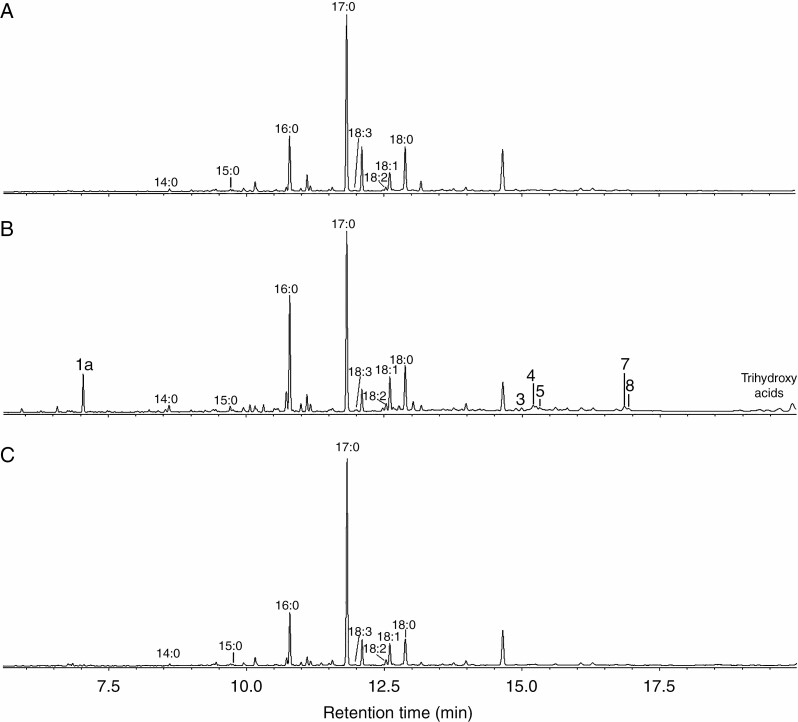
Fig. 5.
The total ion current GC-MS chromatograms of endogenous oxylipins (Me/TMS with preliminary NaBH4 reduction) of intact plants (A) and plants infected with the wild-type Pectobacterium atrosepticum (typical infection) (B) or coronafacic acid-deficient P. atrosepticum mutant (latent infection) (C). 1a, 9-Hydroxynonanoic acid (NaBH4-reduced 9-oxononanoic acid, 9-HPL product); 3, colneleic acid (9-DES product); 4, 9-HOD (derivative of 9-LOX product); 5, 9-HOT (derivative of 9-LOX product); 7, 9,10-epoxy-11-hydroxy-12-octadecenoic acid (9-EAS product); 8, 9,10-epoxy-11-hydroxy-12,15-octadecadienoic acid (9-EAS product). 14:0, myristic acid; 15:0, pentadecanoic acid; 16:0, palmitic acid; 17:0, margaric acid (internal standard); 18:0, octadecanoic acid; 18:1, octadecenoic acid; 18:2, linoleic acid; 18:3, α-linolenic acid. Trihydroxy acids are products of epoxyalcohol hydrolysis. Extraction, derivatization and analysis procedures are described in the Materials and Methods. The structural formulae of the revealed oxylipins are presented in Fig. 3.
Identification of tobacco gene encoding the enzyme with HPL and EAS activities
The results obtained from the gene expression analysis, determination of enzymatic activities in plant extracts and measurement of the endogenous content of oxylipins were mostly consistent with each other, with a couple of exceptions. We have shown that 9-HPL and 9-EAS products are accumulated in infected plants and infected plant extracts after incubation with linoleic and α-linolenic acids. The only gene annotated as HPL-encoding (13-HPL) was only slightly upregulated during infection, while EAS-encoding genes were not annotated in the tobacco genome. We presumed that such a contradiction was a consequence of the incorrect reference annotation of one or more of the target genes.
CYP74 enzymes have several catalytically essential domains with different amino acid sequences depending on the type of catalysis. Therefore, the particular enzymatic activity can be predicted based on the sequence of these domains, namely the ‘F/L toggle’ and I-helix groove region (previously ‘hydroperoxide-binding domain’, Toporkova et al., 2018b).
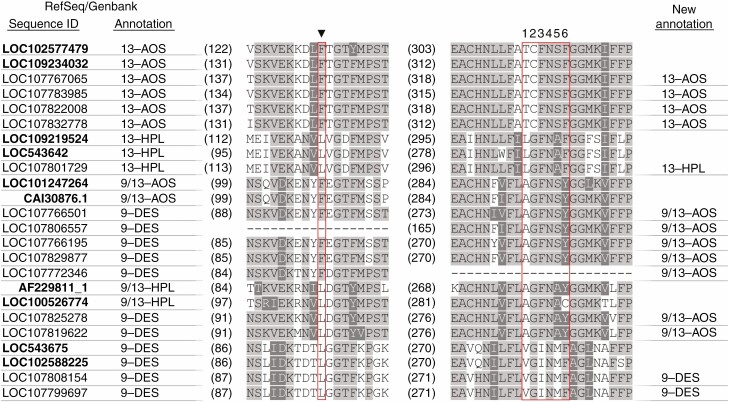
These domains were analysed in all tobacco gene products corresponding to CYP74 enzymes (Fig. 6). The analysis showed that for seven genes (all annotated as 9-DES-encoding), the annotation is likely to be incorrect. Five of these genes presumably encode 9/13-AOSs. Three of these genes were not expressed under our experimental conditions and two of them were upregulated during both typical and latent infections. The upregulation of these genes may contribute to the accumulation of 9- and 13-AOS branch products; however, it cannot explain the increased levels of 9-HPL and 9-EAS products during infection. In turn, two genes (LOC107819622 and LOC107825278) annotated as 9-DES-encoding presumably encode 9/13-HPLs (Fig. 6). One of these two genes (LOC107819622) was not induced during latent infection and was induced only slightly during the typical infection (Table 1), while the other one (LOC107825278) was highly upregulated during both latent (Log2FC 4.1) and typical (Log2FC 6.9) infections compared to non-infected plants. Consequently, if LOC107825278 indeed encodes 9-HPL, this could explain the accumulation of HPL products in the infected plant. Moreover, at least six CYP74C enzymes previously described as 9/13-HPLs possess dual HPL/EAS activities (Toporkova et al., 2018b). Thus, the LOC107825278-encoded enzyme is an obvious candidate that is presumably responsible for the increase in not only HPL products but also EAS products in plants during Pba infection. The name CYP74C43 has been assigned to the LOC107825278-encoded protein (Dr D. Nelson, The University of Tennessee Health Science Center, USA, personal communication).
Fig. 6.
Multiple alignment of partial amino acid sequences of the representatives of canonical CYP74 enzymes (marked in bold) of different plant species as well as tobacco CYP74 enzymes and their reannotation based on the sequence of ‘F/L toggle’ (marked with arrowhead) and I-helix groove region (numbered 1–6). The listed canonical CYP74 enzymes belong to Solanum tuberosum (StAOS2, LOC102577479; StAOS3, CAI30876.1; StDES, LOC102588225), Nicotiana attenuata (NaAOS, LOC109234032; NaHPL, LOC109219524), Solanum lycopersicum (LeHPL, LOC543642; LeAOS3, LOC101247264; LeDES, LOC543675), Vitis vinifera (VvHPL2, LOC100526774) and Cucumis sativus (CsHPL, AAF64041.1).
To verify this hypothesis, the ORF of LOC107825278 was cloned into the pET-32 Ek/LIC vector (Novagen, USA) to yield the recombinant CYP74C43 protein. The catalytic properties of CYP74C43, including the substrate and product specificities, were studied.
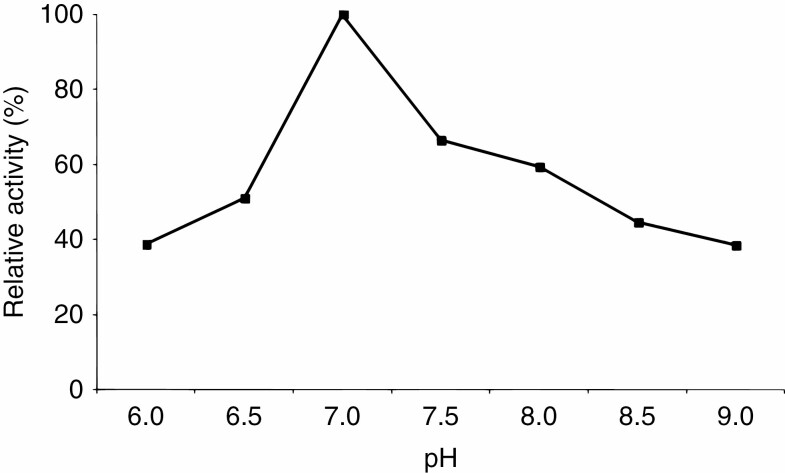
The optimum pH for the catalytic activity of CYP74C43 was 7.0 (Fig. 7). Na-phosphate buffer (100 mm) with the corresponding pH was used for all studies of the catalytic activities of CYP74C43. The affinity and catalytic activity of CYP74C43 for 9-HPOD and 9-HPOT were much higher than for 13-hydroperoxides, as indicated by the Michaelis constant (Km) and Catalytic constant (kcat) values, respectively (Table 2). Thus, according to the kcat/Km ratio values, 9-HPOs of α-linolenic and linoleic acids are the preferred substrates for CYP74C43.
Fig. 7.
The dependence of the CYP74C43 catalytic activity on the pH value of the reaction mixture.
Table 2.
Kinetic parameters and substrate specificities of CYP74C43
| Substrate | kcat (s–1) | KM (μm) | k cat/KM (μm–1 s–1) | Substrate specificity (% 9(S)-HPOD) |
|---|---|---|---|---|
| 9-HPOD | 1590 | 73.9 | 21.5 | 100 |
| 9-HPOT | 1324.6 | 62.5 | 21.2 | 98.6 |
| 13-HPOD | 704.3 | 89.3 | 7.9 | 36.7 |
| 13-HPOT | 610.9 | 136.6 | 4.5 | 20.9 |
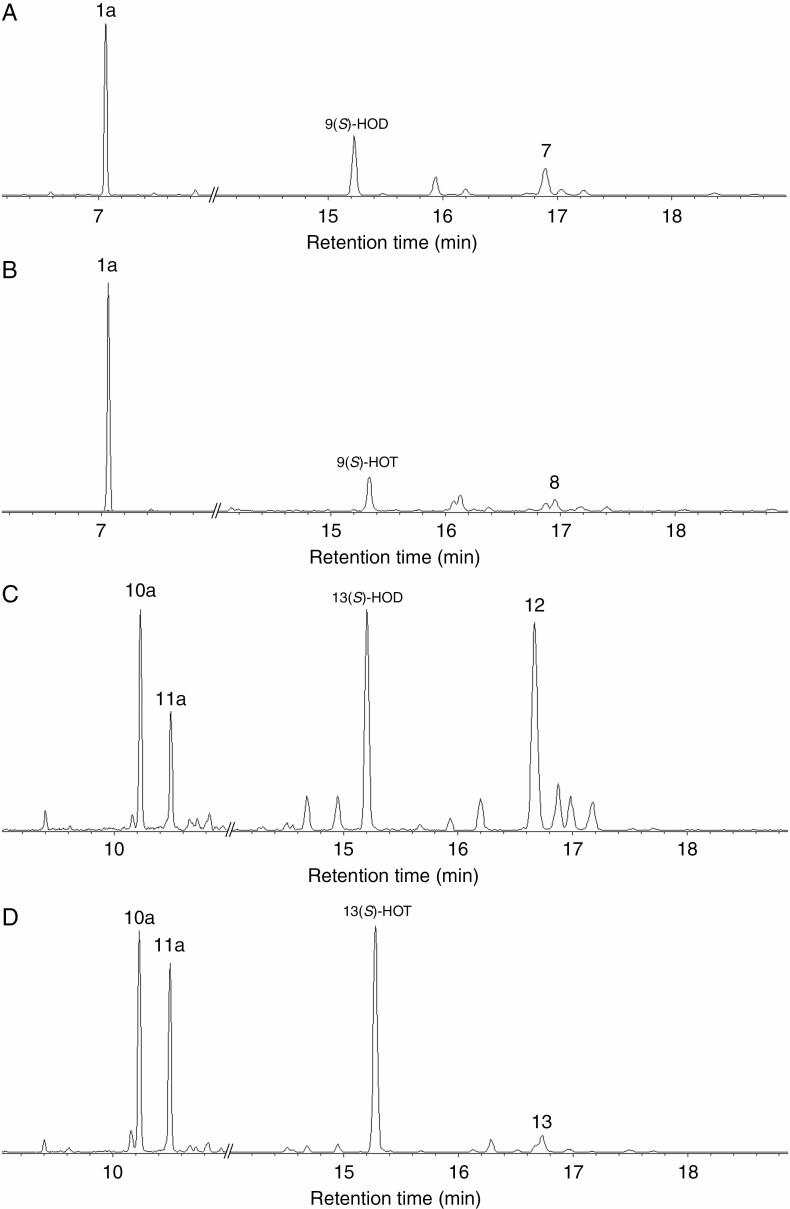
The NaBH4-reduced products (Me/TMS) of 9-HPOD, 9-HPOT, 13-HPOD and 13-HPOT conversion by CYP74C43 were subjected to GC-MS analyses. The 9-HPOD conversion by CYP74C43 afforded the main product 1a, 9-hydroxynonanoic acid (NaBH4-reduced 9-oxononanoic acid, 9-HPL product), and the minor product 7, 9,10-epoxy-11-hydroxy-12-octadecenoic acid (EAS product) (Fig. 8). The 9-HPOT conversion resulted in the formation of the main product 1a and the minor product 8, 9,10-epoxy-11-hydroxy-12,15-octadecadienoic acid (EAS product). The incubation of 13-HPOD and 13-HPOT with CYP74C43 resulted in the appearance of two major peaks, 10a and 11a, corresponding to (9Z)-12-hydroxy-9-dodecenoic and (10E)-12-hydroxy-10-dodecenoic acids, respectively (NaBH4-reduced corresponding aldoacids, 13-HPL products) (Figs 8 and 9). Additionally, oxiranyl carbinols (EAS products) 11-hydroxy-12,13-epoxy-9-octadecenoic (compound 12) and 11-hydroxy-12,13-epoxy-9,15-octadecadienoic (compound 13) acids were detected after the incubation of CYP74C43 with 13-HPOD and 13-HPOT, respectively. The mass spectral data of the discussed compounds are presented in the Supplementary Data. The structural formulae of the listed compounds are presented in Fig. 9.
Fig. 8.
The total ion current GC-MS chromatograms of products (Me/TMS with preliminary NaBH4 reduction) of incubation of 9-HPOD (A), 9-HPOT (B), 13-HPOD (C), and 13-HPOT (D) with CYP74C43. 1a, 9-Hydroxynonanoic acid; 7, 9,10-epoxy-11-hydroxy-12-octadecenoic acid; 8, 9,10-epoxy-11-hydroxy-12,15-octadecadienoic acid; 10a, (9Z)-12-hydroxy-9-dodecenoic acid; 11a, (10E)-12-hydroxy-9-dodecenoic acid; 12, 11-hydroxy-12,13-epoxy-9-octadecenoic acid; 13, 11-hydroxy-12,13-epoxy-9,15-octadecadienoic acid. Incubation, extraction, derivatization and analysis procedures are described in the Materials and Methods. The structural formulae of the products are presented in Figs 3 and 9. 13(S)-HOT, (9Z,11E,13S,15Z)-13-hydroxy-9,11,15-octadecatrienoic acid; 13(S)-HOD, (9Z,11E,13S)-13-hydroxy-9,11-octadecadienoic acid; 9(S)-HOT, (9S,10E,12Z,15Z)-9-hydroxy-10,12,15-octadecatrienoic acid; 9(S)-HOD, (9S,10E,12Z)-9-hydroxy-10,12-octadecadienoic acid.
Fig. 9.
Structural formulae of the products of 13-HPO conversions by the CYP74C43. 10, (9Z)-12-oxo-9-dodecenoic acid (13-HPL product); 10a, (9Z)-12-hydroxy-9-dodecenoic acid (NaBH4-reduced compound 10); 11, (10E)-12-oxo-10-dodecenoic acid (13-HPL product); 11a, (10E)-12-hydroxy-10-dodecenoic acid (NaBH4-reduced compound 11); 12, 11-hydroxy-12,13-epoxy-9-octadecenoic acid (13-EAS product); 13, 11-hydroxy-12,13-epoxy-9,15-octadecadienoic acid (13-EAS product).
Together, our results show that LOC107825278 indeed encodes HPL (ascribed name NtHPL) with additional EAS activity. Therefore, its upregulation during infection explains the accumulation of HPL and EAS products in the infected plants.
DISCUSSION
In the present study, we performed a comprehensive comparative analysis of the functioning of the lipoxygenase cascade following typical and latent Pba infections in tobacco plants. Typical infection has been shown in our previous study to be coupled with the activation of lipoxygenase cascade-related gene expression (Tsers et al., 2020); however, the oxylipin profiles and CYP74 enzyme activities were not compared in intact and infected plants. The latent Pba infection had not previously been characterized in terms of lipoxygenase cascade functioning. However, it was shown that the cfa mutant of Pba, deficient in coronafacic acid (functional analogue of JA), was unable to trigger host plant JA-mediated responses and cause typical soft rot symptoms; instead, it caused a xylem-limited latent infection (Gorshkov et al., 2018). Therefore, it is reasonable to presume that an interconnection between the functioning of the lipoxygenase cascade and infection type may exist.
To verify this hypothesis, we compared the expression profiles of lipoxygenase cascade-related genes, the activities of corresponding enzymes, and the content of endogenous oxylipins during tobacco–Pba (typical infection) and tobacco–cfa mutant (latent infection) interactions. Most of the obtained results were consistent with each other. During typical infection, the 9-LOX pathway was predominantly activated. During latent infection, the 9-LOX pathway was also activated compared to non-infected plants but to a much lesser extent than during the typical infection. Within the 13-LOX pathway, the activities of enzymes related to only the 13-AOS-branch were activated during both typical and latent infections. The induced activity of 13-AOS-branch enzymes led to the accumulation of different products in plant extracts depending on the infection type: JA was accumulated in the extracts of plants with typical infection, while 12-OPDA was accumulated in the extracts of plants with latent infection.
Some of the results for gene expression, enzymatic activities and oxylipin content displayed contradictions. The first was that no products of the 9- and 13-AOS branches among endogenous oxylipins were revealed, despite the fact that the expression of the corresponding genes as well as the levels of JA, 12-OPDA and α-ketol in the linoleate- and α-linolenate-incubated extracts of the infected plants were greater compared to the controls, indicating that the activities of corresponding enzymes were increased in infected plants. In other words, we found enhanced gene expression and activities of the AOS-branch-related enzymes in the infected plants but we did not detect corresponding oxylipins in planta. This contradiction is probably related to the fact that the content of AOS-branch-related oxylipins in planta was below the detectable level, even if their concentration was increased during the infection. JA and its precursor 12-OPDA might be further converted into downstream products, such as volatile methyl-jasmonates or conjugates with amino acids (e.g. JA-Ile). However, the low levels of these oxylipins (at least JA) did not hamper their physiological activity. The physiological activity of JA was evidently manifested during typical (but not latent) infection in the form of a prominent upregulation of JA-responsive genes, the products of which are involved in JA-mediated signalling (MYC2 and TIFY/JAZ). Thus, the relatively low content of JA (and possibly other AOS-branch-related oxylipins) was sufficient to provide its physiological activity during typical infection.
The second contradiction was that the large increase in 9-HPL products and the moderate increase in 9-EAS products in infected plants, as well as the enhanced activity of the corresponding enzymes, were not in accordance with the gene expression data: the only HPL-encoding gene (13-HPL) annotated in the tobacco genome was only slightly induced during the infections, while the EAS-encoding genes, as well as the 9-HPL-encoding genes, were not annotated in the tobacco genome. We found that this contradiction was due to the incorrect annotation of one of the genes, namely LOC107825278, which was highly upregulated during both types of infection. This gene is annotated as a 9-DES-encoding one; however, the amino acid sequences of the ‘F/L toggle’ and I-helix groove region of the LOC107825278-encoded enzyme correspond to 9/13-HPLs, many of which possess dual HPL/EAS activities (Toporkova et al., 2018b). The dual activity is rather often revealed in the CYP74s, including not only HPLs but also AOSs and DESs (Song et al., 1993; Hughes et al., 2008; Toporkova et al., 2020). To verify whether LOC107825278 indeed encodes 9/13-HPL, its ORF was cloned and the corresponding recombinant enzyme NtHPL was shown to be 9/13-HPL with additional EAS activity. Thus, the use of a combination of approaches and methods in our study allowed us to neutralize some ‘blind spots’ in the given method and to obtain a comprehensive view of the lipoxygenase cascade’s functioning in different types of infection.
Our study revealed three major oxylipin-related distinctions for the typical and latent Pba infections (Fig. 10). First, the 9-LOX pathway is differentially activated depending on the infection type: during latent infection, the degree of activation of this pathway was lower than during the typical infection. Presumably, a large increase in the products of the 9-LOX pathway exacerbates the development of disease symptoms and thus represents a susceptibility-related hallmark of the host plant to Pba. Second, the induction of JA-biosynthetic enzyme activity and JA-mediated signalling is a characteristic feature of the typical (but not latent) infection. These data support our hypothesis that JA is a switch that triggers the transition of a latent asymptomatic infection to a typical symptomatic one (this study; Gorshkov et al., 2018; Tsers et al., 2020).
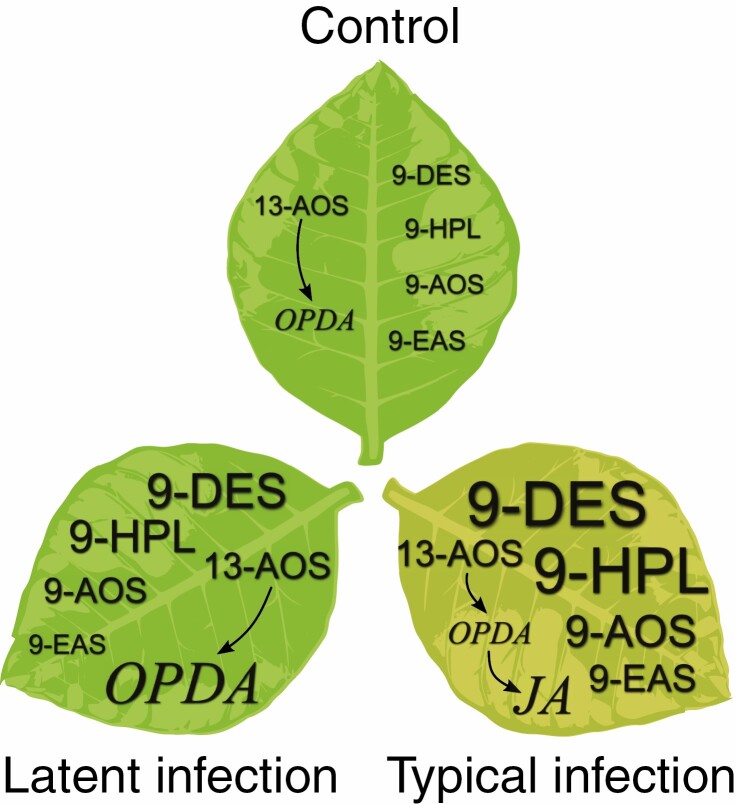
Fig. 10.
Schematic representation of the modulation of the lipoxygenase cascade during typical and latent Pectobacterium atrosepticum infections compared to control non-infected plants. HPL, hydroperoxide lyase; DES, divinyl ether synthase; AOS, allene oxide synthase; EAS, epoxyalcohol synthase; OPDA, cis-12-oxo-10,15-phytodienoic acid; JA, jasmonic acid.
Third, the only oxylipin, the content of which was increased in the linoleate- and α-linolenate-incubated extracts of plants with latent infection compared to the incubated extracts of plants with typical infection (as well as control plants), was 12-OPDA. The relationship between plant resistance to pathogens (Pseudomonas syringae) and the increased level of 12-OPDA as well as the decreased level of JA has been previously demonstrated by using tomato plants with the silenced OPR gene (Scalschi et al., 2020). The accumulation of different 13-AOS-branch products during typical and latent infections (JA and 12-OPDA, respectively) is evidently related to the differential activity of the particular enzymes of this branch. The higher the ratio of AOS/12-OPDA-reductase activities, the higher the ratio of 12-OPDA/JA levels. The results of our gene expression analysis imply a higher ratio of AOS/12-OPDA-reductase activities in plants with latent infection compared to plants with typical infection: the sums of the absolute expression values (TPM) of all AOS genes (including those that were reannotated in our study) were almost similar for the two infection types (77 and 71 for latent and typical infections, respectively), while the sum of TPM values of all 12-OPDA-reductase genes was lower for the latent infection (254) compared to the typical one (469). Such a difference in the ratios of AOS/OPDA-reductase activities may presumably lead to the accumulation of different products during different types of infection. Since JA is likely to contribute to plant susceptibility to Pba, the final steps of JA biosynthesis can be repressed during latent infection, resulting in the accumulation of the JA-precursor – 12-OPDA. Whether the increased level of 12-OPDA contributes to the restriction of disease progression, leading to plant tolerance and the development of latent infection, is unknown. However, the role of 12-OPDA in plant tolerance can easily be presumed. 12-OPDA is not only a precursor of JA but has also been demonstrated to exert its own physiological activities. 12-OPDA and JA were shown to act via different signalling pathways and to have different target genes (Brader et al., 2001; Sasaki et al., 2001; Turner et al., 2002; Sasaki-Sekimoto et al., 2005; Chini et al., 2009; Savchenko et al., 2014; Monte et al., 2020). Moreover, 12-OPDA was considered to act as the antagonist of JA in regulating singlet oxygen-induced programmed cell death (Danon et al., 2005). In addition, 12-OPDA displays a growth inhibitory effect towards some phytopathogens, including Pectobacterium carotovorum (Prost et al., 2005). Therefore, the role of 12-OPDA in the possible maintenance of latent Pba infection and prevention of the typical infection deserves further in-depth investigation.
Thus, our results demonstrate that when compared to the typical infection, latent asymptomatic Pba infection is associated with (and possibly maintained due to) decreased levels of JA and 9-LOX pathway (9-DES, 9-HPL, 9-AOS, 9-EAS branches) products and an increased level of 12-OPDA (Fig. 10). Our results contribute to the hypothesis of the oxylipin signature, indicating that different types of plant interaction with a particular pathogen are characterized by the different oxylipin profiles of the host plant.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. File S1: Details for the analysis of the spectral data. Fig. S1: The results of TIC GC-MS analysis of lipoxygenase cascade products in tobacco plant extracts after incubation with linoleic and α-linolenic acids. Tables S1 and S2 (as one.xls file): details on the expression of lipoxygenase- and JA signalling proteins-encoding genes in tobacco during typical and latent infection caused by Pba or Cfa, respectively.
FUNDING
This work was supported by the Russian Science Foundation: gene expression and oxylipin analyses (grant number 19-14-00194), and obtaining and characterizing the recombinant enzyme (grant number 20-14-00338). Bioinformatics procedures were performed within the frameworks of the government assignment for the Federal Research Center “Kazan scientific center of Russian Academy of Sciences”.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
The raw RNA-Seq data generated in this study are available from the corresponding author upon request.
LITERATURE CITED
- Akitami I, Shiraishi K, Sato H, et al. 1977. The structure of coronatine. Journal of the American Chemical Society 99: 636–637. [Google Scholar]
- Bell KS, Sebaihia M, Pritchard L, et al. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proceedings of the National Academy of Sciences of the United States of America 101: 11105–11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G, Tas E, Palva ET. 2001. Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiology 126: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Charkowski AO. 2007. The soft rot erwinia. In: Gnanamanickam S.S., eds. Plant-associated bacteria. Berlin: Springer, 423–505. [Google Scholar]
- Charkowski A, Blanco C, Condemine G, et al. 2012. The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Annual Review of Phytopathology 50: 425–449. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. 2009. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. The Plant Journal: for Cell and Molecular Biology 59: 77–87. [DOI] [PubMed] [Google Scholar]
- Chini A, Monte I, Zamarreño AM, et al. 2018. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nature Chemical Biology 14: 171–178. [DOI] [PubMed] [Google Scholar]
- Danon A, Miersch O, Felix G, Camp RG, Apel K. 2005. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. The Plant Journal: for Cell and Molecular Biology 41: 68–80. [DOI] [PubMed] [Google Scholar]
- Delaplace P, Rojas-Beltran J, Frettinger P, du Jardin P, Fauconnier M-L. 2008. Oxylipin profile and antioxidant status of potato tubers during extended storage at room temperature. Plant Physiology and Biochemistry 46: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Fammartino A, Verdaguer B, Fournier J, et al. 2010. Coordinated transcriptional regulation of the divinyl ether biosynthetic genes in tobacco by signal molecules related to defense. Plant Physiology and Biochemistry: PPB 48: 225–231. [DOI] [PubMed] [Google Scholar]
- Galliard T, Phillips DR. 1972. The enzymic conversion of linoleic acid into 9-(nona-1′,3′-dienoxy)non-8-enoic acid, a novel unsaturated ether derivative isolated from homogenates of Solanum tuberosum tubers. Biochemical Journal 129: 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel C, Feussner I, Hamberg M, Rosahl S. 2002. Oxylipin profiling in pathogen-infected potato leaves. Biochimica et Biophysica Acta 1584: 55–64. [DOI] [PubMed] [Google Scholar]
- Gogolev YV, Gorina SS, Gogoleva NE, Toporkova YY, Chechetkin IR, Grechkin AN. 2012. Green leaf divinyl ether synthase: gene detection, molecular cloning and identification of a unique CYP74B subfamily member. Biochimica et Biophysica Acta 1821: 287–294. [DOI] [PubMed] [Google Scholar]
- Gorina SS, Toporkova YY, Mukhtarova LS, et al. 2016. Oxylipin biosynthesis in spikemoss Selaginella moellendorffii: molecular cloning and identification of divinyl ether synthases CYP74M1 and CYP74M3. Biochimica et Biophysica Acta 1861: 301–309. [DOI] [PubMed] [Google Scholar]
- Gorshkov V, Daminova A, Ageeva M, et al. 2014. Dissociation of a population of Pectobacterium atrosepticum SCRI1043 in tobacco plants: formation of bacterial emboli and dormant cells. Protoplasma 251: 499–510. [DOI] [PubMed] [Google Scholar]
- Gorshkov V, Gubaev R, Petrova O, et al. 2018. Transcriptome profiling helps to identify potential and true molecular switches of stealth to brute force behavior in Pectobacterium atrosepticum during systemic colonization of tobacco plants. European Journal of Plant Pathology 152: 957–976. [Google Scholar]
- Gosset V, Harmel N, Göbel C, et al. 2009. Attacks by a piercing-sucking insect (Myzus persicae Sultzer) or a chewing insect (Leptinotarsa decemlineata Say) on potato plants (Solanum tuberosum L.) induce differential changes in volatile compound release and oxylipin synthesis. Journal of Experimental Botany 60: 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granér G, Hamberg M, Meijer J. 2003. Screening of oxylipins for control of oilseed rape (Brassica napus) fungal pathogens. Phytochemistry 63: 89–95. [DOI] [PubMed] [Google Scholar]
- Hamberg M. 1999. An epoxy alcohol synthase pathway in higher plants: biosynthesis of antifungal trihydroxy oxylipins in leaves of potato. Lipids 34: 1131–1142. [DOI] [PubMed] [Google Scholar]
- Hanano A, Shaban M, Almousally I, Murphy DJ. 2018. Identification of a dioxin-responsive oxylipin signature in roots of date palm: involvement of a 9-hydroperoxide fatty acid reductase, caleosin/peroxygenase PdPXG2. Scientific Reports 8: 13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SE, Eschen R, Horwood JM, Gange AC, Hill EM. 2015. Infection by a foliar endophyte elicits novel arabidopside-based plant defence reactions in its host, Cirsium arvense. The New Phytologist 205: 816–827. [DOI] [PubMed] [Google Scholar]
- Hause B, Stenzel I, Miersch O, et al. 2000. Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. The Plant Journal: for Cell and Molecular Biology 24: 113–126. [DOI] [PubMed] [Google Scholar]
- Hayward AC. 1974. Latent infections by bacteria. Annual Review of Phytopathology 12: 87–97. [Google Scholar]
- Howe GA, Major IT, Koo AJ. 2018. Modularity in jasmonate signaling for multistress resilience. Annual Review of Plant Biology 69: 387–415. [DOI] [PubMed] [Google Scholar]
- Hughes RK, De Domenico S, Santino A. 2009. Plant cytochrome CYP74 family: biochemical features, endocellular localisation, activation mechanism in plant defence and improvements for industrial applications. Chembiochem: A European Journal of Chemical Biology 10: 1122–1133. [DOI] [PubMed] [Google Scholar]
- Hughes RK, Yousafzai FK, Ashton R, et al. 2008. Evidence for communality in the primary determinants of CYP74 catalysis and of structural similarities between CYP74 and classical mammalian P450 enzymes. Proteins 72: 1199–1211. [DOI] [PubMed] [Google Scholar]
- Ichihara A, Kunio S, Hiroji S, et al. 1977. The structure of coronatine. Journal of the American Chemical Society 99: 636–637. [Google Scholar]
- Kato T, Yamaguchi Y, Abe N, et al. 1985. Structure and synthesis of unsaturated trihydroxy C18 fatty: acids in rice plants suffering from rice blast disease. Tetrahedron Letters 26: 2357–2360. [Google Scholar]
- Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics (Oxford, England) 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- Lee DS, Nioche P, Hamberg M, Raman CS. 2008. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455: 363–368. [DOI] [PubMed] [Google Scholar]
- Li C, Schilmiller AL, Liu G, et al. 2005. Role of beta-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. The Plant Cell 17: 971–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RE. 1984. A naturally occurring structural analogue of the phytotoxin coronatine. Phytochemistry 23: 791–793. [Google Scholar]
- Monte I, Kneeshaw S, Franco-Zorrilla JM, et al. 2020. An ancient COI1-independent function for reactive electrophilic oxylipins in thermotolerance. Current Biology 30: 962–971.e3. [DOI] [PubMed] [Google Scholar]
- Noordermeer MA, Veldink GA, Vliegenthart JF. 2001. Fatty acid hydroperoxide lyase: a plant cytochrome p450 enzyme involved in wound healing and pest resistance. Chembiochem: A European Journal of Chemical Biology 2: 494–504. [DOI] [PubMed] [Google Scholar]
- Pérombelon MCM. 2002. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathology 51: 1–12. [Google Scholar]
- Pérombelon MCM, Kelman A. 1980. Ecology of the soft rot erwinias. Annual Review of Phytopathology 18: 361–387. [Google Scholar]
- Pietryczuk A, Czerpak R. 2011. Effect of traumatic acid on antioxidant activity in Chlorella vulgaris (Chlorophyceae). Plant Growth Regulation 65: 279–286. [Google Scholar]
- Prost I, Dhondt S, Rothe G, et al. 2005. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiology 139: 1902–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Asamizu E, Shibata D, et al. 2001. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Research: An International Journal for Rapid Publication of Reports on Genes and Genomes 8: 153–161. [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, et al. 2005. Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. The Plant Journal: for Cell and Molecular Biology 44: 653–668. [DOI] [PubMed] [Google Scholar]
- Savchenko T, Dehesh K. 2014. Drought stress modulates oxylipin signature by eliciting 12-OPDA as a potent regulator of stomatal aperture. Plant Signaling & Behavior 9: e28304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko TV, Zastrijnaja OM, Klimov VV. 2014. Oxylipins and plant abiotic stress resistance. Biochemistry. Biokhimiia 79: 362–375. [DOI] [PubMed] [Google Scholar]
- Scalschi L, Llorens E, García-Agustín P, Vicedo B. 2020. Role of jasmonic acid pathway in tomato plant-Pseudomonas syringae interaction. Plants, 9: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkman JB, Jansson I. 2006. Spectral analyses of cytochromes P450. Methods in Molecular Biology (Clifton, N.J.) 320: 11–18. [DOI] [PubMed] [Google Scholar]
- Sinclair JB. 1991. Latent infection of soybean plants and seeds by fungi. Plant Disease 75: 220. [Google Scholar]
- Song WC, Baertschi SW, Boeglin WE, Harris TM, Brash AR. 1993. Formation of epoxyalcohols by a purified allene oxide synthase. Implications for the mechanism of allene oxide synthesis. The Journal of Biological Chemistry 268: 6293–6298. [PubMed] [Google Scholar]
- Takahashi I, Hosomi K, Nagatake T, et al. 2019. Persistent colonization of non-lymphoid tissue-resident macrophages by Stenotrophomonas maltophilia. International Immunology 32: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toporkova YY, Bessolitsyna EK, Smirnova EO, et al. 2018. a. Antimicrobial activity of geometric isomers of etherolenic acid—the products of plant lipoxygenase cascade. Doklady Biochemistry and Biophysics 480: 139–142. [DOI] [PubMed] [Google Scholar]
- Toporkova YY, Gorina SS, Bessolitsyna EK, et al. 2018. b. Double function hydroperoxide lyases/epoxyalcohol synthases (CYP74C) of higher plants: identification and conversion into allene oxide synthases by site-directed mutagenesis. Biochimica et Biophysica Acta. Molecular and Cell Biology of Lipids 1863: 369–378. [DOI] [PubMed] [Google Scholar]
- Toporkova YY, Smirnova EO, Iljina TM, Mukhtarova LS, Gorina SS, Grechkin AN. 2020. The CYP74B and CYP74D divinyl ether synthases possess a side hydroperoxide lyase and epoxyalcohol synthase activities that are enhanced by the site-directed mutagenesis. Phytochemistry 179: 112512. [DOI] [PubMed] [Google Scholar]
- Toth IK, Birch PR. 2005. Rotting softly and stealthily. Current Opinion in Plant Biology 8: 424–429. [DOI] [PubMed] [Google Scholar]
- Tsers I, Gorshkov V, Gogoleva N, Parfirova O, Petrova O, Gogolev Y. 2020. Plant soft rot development and regulation from the viewpoint of transcriptomic profiling. Plants 9: 1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A. 2002. The jasmonate signal pathway. The Plant Cell 14 (Suppl.): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Feussner I. 2018. The oxylipin pathways: biochemistry and function. Annual Review of Plant Biology 69: 363–386. [DOI] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE. 1997. Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proceedings of the National Academy of Sciences of the United States of America 94: 10473–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M, Salas JJ, Sanchez J, Harwood JL. 2000. Lipoxygenase pathway in olive callus cultures (Olea europaea). Phytochemistry 53: 13–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-Seq data generated in this study are available from the corresponding author upon request.