Abstract
In the present study, thirty two lactic acid bacteria (LAB) were isolated from fermented Indian herbal medicine. In comparison to other strains, MNL5 had stronger bile salt hydrolase (BSH) and cholesterol-lowering properties. Furthermore, it can withstand the extreme conditions found in the GI tract, due to, e.g., pepsin, bile salts, pancreatin, and acids. Pediococcus acidilactici MNL5 was identified as a probiotic candidate after sequencing the 16S rRNA gene. The antibacterial activity of P. acidilactici MNL5 cell-free supernatants (CFS) against Escherichia coli, Staphylococcus aureus, Helicobacter pylori, Bacillus cereus, and Candida albicans was moderate. A Caenorhabditis elegans experiment was also performed to assess the effectiveness of P. acidilactici MNL5 supplementation to increase life span compared to E. coli supplementation (DAF-2 and LIU1 models) (p < 0.05). An immense reduction of the lipid droplets of C. elegans was identified through a fluorescent microscope. The drastic alteration of the expression of fat genes is related to obesity phenotypes. Hence, several paths are evolutionary for C. elegans; the results of our work highlight the nematode as an important model for obesity.
Keywords: Pediococcus acidilactici, cholesterol-lowering ability, lipid reduction, C. elegans, qPCR
1. Introduction
Probiotics are living microbial dietary supplements that impact the host by strengthening microbial gut stability [1]. Reports have documented that there is a discrepancy between information on the labels and data reported following laboratory safety assessments of various commercial probiotic products [2,3]. Lactic acid bacteria (LAB) such as Lactococcus, Pediococcus, Enterococcus, Streptococcus, Weissella, and Saccharomyces are common commercial probiotics. The primary criteria for selecting probiotic bacteria include hemolytic activity, antibiotic resistance, antimicrobial activity, pepsin and pancreatin tolerance, bile tolerance, morphologies, genotypes, and carbohydrate tolerance [4]. Different studies of LAB have noted a broad range of health-enhancing characteristics associated with microbial balance in the host gut flora [5]. In addition, probiotics have the ability to produce bile-salt hydrolase, which considerably decreases cholesterol levels [6,7]. Lactobacilli belong to the primary group known as the LAB that is Generally Recognized as Safe (GRAS) [8]. Lactobacillus and Pediococcus are able to maintain a healthy intestinal microbial ecosystem and reduce the function of cholesterol [9,10].
Furthermore, isolates showing unique features that could be beneficial in biotechnology or probiotic use could be produced by isolating and identifying new strains from un-investigated medicinal and edible plants [11,12]. Recently, C. elegans have been used in fat metabolism research. Bacterial dietary fatty acids are transformed into triglycerides, which are the main form of fat deposited on the epithelial and gut organs of C. elegans. Most of the key biosynthetic pathways of humans, such as synthesis with fatty acids, elongation, denaturation, and β-oxidation, as well as pathways for the neuropeptides [13], are well preserved in C. elegans. Fatty acid metabolism shows a vital feature of controlling stress resistance pathways, even without the activity of DAF-16, if nematode DAF-2/insulin signaling is regulated. This allows the identification of the specific aspects and pathways that act downstream of fat regulators through expression profiling and epistasis evaluation of potential metabolic genes [14]. Several studies have indicated that the consumption of LAB and other probiotics reduces coronary artery risk and the severity of symptoms of Bowel’s syndrome [15]. In our study, we identified and evaluated lactic acid bacteria based on BSH activity, cholesterol reduction capacity, probiotic capabilities, and food safety aspects. Additionally, the mechanism of action for inhibiting fat accumulation in a C. elegans gut model by P. acidilactici MNL5 was studied. This research underlines the ability of P. acidilactici MNL5 to reduce multitarget lipid lowing active substance to help in the management of obesity.
2. Results
2.1. Isolation and Identification of Cholesterol-Lowering LAB
Moola nivarana lehyam is a fermented herbal medicine, mainly applied for reducing obesity, which consist of combined dried spices such as Medhya Rasayan (Nootropic herb), Terminalia chebula: 3.10%, Emblica officinalis: 3.10%, Terminalia bellirica: 3.10%, Withania somnifera (6.85%), Zingiber officinale (3.6%), Phytophthora megasperma (root rot) (3.6%), Cyperus rotundus (7%), Tamarindus indica (7%), Aloe barbadensis miller (7%), Cissus quadrangularis (7%), Amorphophallus paeoniifolius (3%), Arenga pinnata (5%), butter (10%), where naturally fermented at 30 °C for 72 h. The lactic acid bacterial species have been isolated from fermented herbal medicine products, obtained from India. Total 32 LAB strains were examined for BSH and cholesterol assimilation (Table 1). Among them, six LAB strain expressed BSH activity and cholesterol assimilation ranged from 9.5 to 89.5%. The MNL5 had the highest cholesterol (89.5%) and BSH activity (16 mm zone of precipitation) out of 32 strains. The isolate MNL5 was compared with 16S rRNA gene sequences in GenBank and it was very similar (99% (MK928489)) with the P. acidilactici strain (Table 1, Figure 1 and Figure 2). P. acidilactici MNL5 strain content in moola nivarana lehyam 3.69 log CFU/g.
Table 1.
Assimilation of cholesterol, Haemolytic activity and BSH of isolated LAB strains from fermented herbal medicine.
| No | Strain Name | Colour Morphology | Shape | Hemolytic Activity | BSH Activity | Cholesterol Assimilation (%) |
|---|---|---|---|---|---|---|
| 1 | MNL 1 | White/Smooth | cocci | NA | ++ | 25.3 ± 0.54 g |
| 2 | MNL 2 | White | rod | NA | + | 18.5 ± 0.41 h |
| 3 | MNL 3 | White/Round | rod | NA | - | 10.2 ± 0.39 h |
| 4 | MNL 4 | White/Smooth/Round | rod | NA | - | 11.7 ± 0.24 h |
| 5 | MNL 5 | White/Smooth | cocci | NA | +++ | 89.5 ± 1.09 a |
| 6 | MNL 6 | White | rod | NA | - | 25.5 ± 0.44 g |
| 7 | MNL 8 | White | cocci | NA | - | 23.4 ± 0.38 g |
| 8 | MNL 9 | White | cocci | NA | + | 31.5 ± 0.61 f |
| 9 | MNL 10 | White/Round | rod | NA | + | 24.5 ± 0.50 g |
| 10 | MNL 11 | White/Smooth/Round | short rod | NA | ++ | 47.5 ± 0.36 e |
| 11 | GSC 5 | White/Smooth | rod | NA | + | 31.0 ± 0.37 f |
| 12 | GSC 8 | White | rod | NA | - | 12.4 ± 0.47 h |
| 13 | GSC 10 | White | short rod | NA | + | 10.5 ± 0.39 h |
| 14 | TVL 11 | White/Smooth | short rod | NA | + | 14.6 ± 0.40 h |
| 15 | TVL 12 | White | short rod | NA | - | 12.4 ± 0.55 h |
| 16 | SKML 3 | White/Round | rod | NA | + | 22.5 ± 0.51 g |
| 17 | SKML 4 | White | rod | NA | - | 9.5 ± 0.64 h |
| 18 | SKML 5 | White/Smooth | cocci | NA | +++ | 77.1 ± 0.98 b |
| 19 | SKML 6 | White/Smooth | rod | NA | ++ | 69.5 ± 0.78 c |
| 20 | SKML 8 | White/Smooth/Irregular | rod | NA | + | 53.43 ± 0.63 d |
| 21 | SKML 10 | White | cocci | NA | + | 47.56 ± 0.51 e |
| 22 | SKML 11 | White | short rod | NA | + | 45.21 ± 0.69 e |
| 23 | SKML 12 | White | cocci | NA | - | 25.80 ± 0.40 g |
| 24 | SKML 13 | White | short rod | NA | - | 22.4 ± 0.35 g |
| 25 | SK SPL 1 | White/Rough | short rod | NA | - | 21.4 ± 0.44 g |
| 26 | SK SPL 6 | White | cocci | NA | ++ | 57.4 ± 0.64 a |
| 27 | SK SPL7 | White/Smooth | cocci | NA | + | 42.5 ± 0.51 e |
| 28 | GSC 1 | White | cocci | NA | + | 38.4 ± 0.38 f |
| 29 | GSC 11 | White | short rod | NA | + | 32.5 ± 0.33 f |
| 30 | GSC 12 | White | cocci | NA | ++ | 41.5 ± 0.46 e |
| 31 | SKML 18 | White | short rod | NA | ++ | 57.8 ± 0.48 d |
| 32 | SKML 20 | White/Smooth | cocci | NA | ++ | 68.6 ± 0.64 c |
-, no precipitation; +, precipitation zone up to 10 mm; ++, precipitation zone up to 15 mm; and +++, precipitation zone up to 20 mm. NA-γ- haemolysis (no zones around colonies). Values are expressed in mean ± standard deviation (n = 3) Different superscripts (a, b, c, d, e, f, g, h) represent significantly different values (p < 0.05).
Figure 1.
(The BSH activity of LAB isolates grown on bile salt–MRS medium as manifested by the formation of precipitation zone around the colony.
Figure 2.
The 16S rRNA sequence of Pediococcus acidilacticii MNL5 based phylogenetic tree was constructed by the neighbor-joining method. The numbers at the nodes are bootstrap values based on 0.0001 replications.
2.2. Antibiotic Resistance
All the isolates were tested with nine different antibiotics (Supplementary Table S1). Most of the bacterial isolates were susceptible with ampicillin, streptomycin, penicillin, tetracycline, kanamycin, erythromycin, gentamicin, clindamycin, and chloramphenicol. The P. acidilactici MNL5 was showed the resistance with nine antibiotics and the evaluation was according to the European Food Safety Authority. Hence, in this results supports that the consumption of P. acidilactici MNL5 increase a human health while administration with antibiotic.
2.3. Antimicrobial Activity
A broad-spectrum antimicrobial activity was observed in thirty-two isolates against gram positive and negative pathogens. Above all LAB, the MNL5 exhibits the substantial inhibitory activity against Escherichia coli, Staphylococcus aureus, Bacillus cereus and H. pylori while lesser activity observed with Candida albicans KCTC 7965 (Supplementary Table S2).
2.4. Hemolytic Activity
The tested bacteria showed γ-hemolytic (no zone effect), α-hemolysis (green zone), and none had β-hemolytic (blood lysis zone) effects (Table 1). Probiotic strain screening should consider safety issues, including specifications such as origin, identity, and lack of harmful activities [16].
2.5. Survival under In Vitro Gastrointestinal Conditions
Tolerance to Low pH, Pepsin and Bile Salts and Pancreatin
In our study, the survival of P. acidilactici MNL5 under in vitro gastrointestinal conditions like low pH (>94%), pepsin (>76%), bile salts, and pancreatin (>77%) was increased (Figure 3). According to the results, MNL5 showed significant resistance in simulated intestinal conditions.
Figure 3.
Stability of Lactic acid bacteria towards tolerance of acid, pepsin and Bile & Pancreatin. Values are expressed as the mean ± standard deviation (n = 3). Different superscripts (a, b, c, d) represent significantly different values (p < 0.05).
2.6. Chemotaxis Assays
In our experiment, the nematodes showed minimal preferences for the MNL5 pellet and strong inclinations toward broth culture (liquid) cultivation. This finding may be due to the attraction of nematodes to the diacetyl group, which is one of the secondary metabolites produced by lactic acid bacteria. Based on the results, MNL5 appears to promote nematode growth when administered as a nematode supplement. We found that significantly more animals selected the MNL5 diet supplemented with sugar than the control diet.
2.7. Worm Size Measurement
According to our results, the body size of C. elegans altered significantly throughout the first three days of the diet. Body size was increased in the E. coli OP50+Glucose diet, while MNL5+Glucose was decreased compared to the regular E. coli OP50 diet. Our findings suggest that increasing the survival rate while decreasing the size of worms employed leads to a long life for both the DAF-2 and LIU1 models. (Supplementary Figure S1).
2.8. C. elegans Gut Colonization Ability
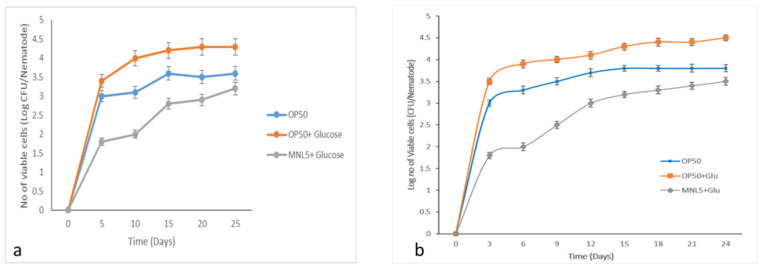
The population of gut bacteria accumulated while administered MNL5+ Glucose and E. coli OP50 significantly diminished when compared to E. coli OP50+ Glucose diet colonization (Figure 4). These results indicate that early bacterial accumulation is associated with a shorter period of life for nematodes. We also pointed out that the increased lifespan of the worm depended on the DAF-2/DAF-16 function, and on the colonization of the bacterial strains in the gut of the worms if the MNL5 strain was presented as food sources to the worms. Nevertheless, our results suggest that these newly isolated MNL5 strains might have health-promoting impacts on the host.
Figure 4.
Bacterial colonization in C. elegans. (a) DAF-2, (b) LIU1 model; the treatments such as, OP50 (Control), MNL+ GLU (P. acidilactici MNL5 with Glucose) [Treatment]), OP50+GLU (E. coli OP50 with Glucose [Positive Control]; Evaluation of MNL5 colonization (numbers of log CFU/nematode) in the nematode intestine. In each experiment, 100 worms were administered, repeated twice and plotted using OASIS II. All results are presented as the means ± standard error mean.
2.9. C. elegans Life Span Assay
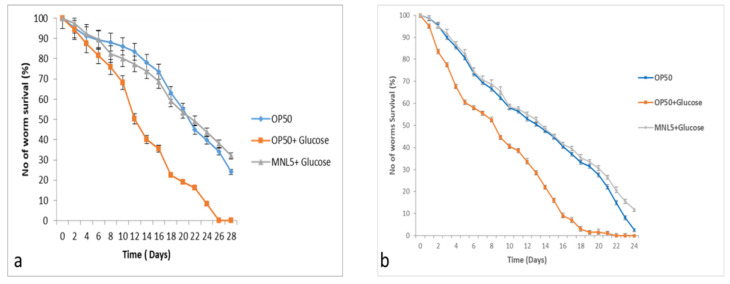
The findings revealed that P. acidilactici MNL5 has a long life span and protective nematode effects that benefit the DAF-2 and LIU1 models. The life span of C. elegans (DAF-2 and LIU1 model) increased supplemented with P. acidilactici MNL5 + Glucose (<32 ± 1 days; <27 ± 1), while compared to E. coli (29 ± 1; 24 ± 1) and E. coli OP50 + Glucose (26 ± 1; 17 ± 1) is decreased. (Figure 5). When comparing the control results observed with the test results, we found that the P. acidilactici MNL5 showed that the worms longer lifespan.
Figure 5.
Glucose supplements diet by lifespan of C. elegans. (a) DAF-2; (b) LIU1 model. The treatments such as, OP50 (Control), MNL + GLU (P. acidilactici MNL5 with Glucose) [Treatment]), OP50+GLU (E. coli OP50 with Glucose [Positive Control]; in each experiment, 100 worms were administered, repeated twice and plotted using OASIS II. All results are presented as the means ± standard error mean.
2.10. Lipid Determination by Microscope Analysis
Aging dysregulates intracellular lipid metabolism in several organisms. Experiments DAF-2 and LIU1 model showed that the Lipid droplets are decreased P. acidilactici MNL5 with Glucose supplement treatment on end day (28th and 25th day) of the experiment, similarly, the lipid droplets increased significantly after the OP50 with Glucose treatment by fluorescence microscopy analysis (Figure 6).
Figure 6.
Lipid content visualized by C. elegans body (DAF-2 and LIU1 model) by fluorescence intensity measured by ImageJ software. Microscopic images showing different treatments such as OP50 (Control), MNL+ GLU (P. acidilactici MNL5 with Glucose) [Treatment]), OP50+GLU (E.coli OP50 with Glucose [Positive Control]; (A) Nile Red staining by fluorescence microscope (20× magnification); (B) Oil Red O staining by light microscope (20× magnification). Bar diagram presents the relative lipid content of worms. The error bar represents the mean ± SEM). Different superscripts (a, b, c, d) represent significantly different values (p < 0.05).
2.11. Transcriptomic Analysis in C. elegans
In our study, we performed a demonstration of DAF-2 and LIU1 with modified fat composition, examining the relationships between particular lipids and biological functions. The qPCR experiments suggested that the three delta-9 desaturase homologs, i.e., fat-4 fat-5 and fats-6, encoding fatty acid desaturase enzyme, encoding double bond formation in a saturated fatty acid metabolism related gene expression were decreased. The obtained standard curve for each target gene with its respective regression coefficient (R2 value) and efficiency (%) is represented in Supplementary Figure S2. The developed qPCR assay was found to work efficiently over a cDNA concentration range of 1 × 102, 1 × 101, 1 × 10°, 1 × 10−1, 1 × 10−2, 1 × 10−3 ng per reaction. This experiments pathway was intended to demonstrate that the daf-2 and LIU1 model responds to the absence of lipids by either up- or down- regulating these pathways. Based on our qPCR analysis, we showed the active role of transcriptional upregulation of E. coli + glucose treated worms in increasing de novo fatty acid synthesis. However, P. acidilactici MNL5 + glucose treated worms indicated that the suppression of transcription of the fatty acid desaturase genes was downregulated. As a result, we showed that fat-4, fat-5, and fat-6 were modulated by a mechanism that suppresses lipid reduction by P. acidilactici MNL5 in a C. elegans fat gene model (Figure 7).
Figure 7.
The qPCR fold change in expression pattern of fat genes. (a) DAF-2; (b) LIU1; The bar diagram indicates the following changes in worms exposed different diets to gene expression: OP50; MNL5+ Glucose; OP50+ Glucose. Values are expressed in mean ± standard deviation (n = 3). Different superscripts (a, b, c, d) represent significantly different values (p < 0.05).
3. Discussion
In this study, lactic acid bacterial species were isolated from fermented herbal medicine. Nutritional foods are essential for the quality of life and survival of organisms. Probiotic bacteria play a significant role in digestion and survive in the gastrointestinal tract due to their acid tolerance in human gastric juices and bile in the gut. In this study, 32 LAB were identified from fermented herbal medicine products from India, and analyzed for BSH and cholesterol assimilation. Among them, P. acidilactici MNL 5 exhibited antibacterial activity with Escherichia coli ATCC 35150, Staphylococcus aureus ATCC 13,150, and Bacillus cereus ATCC 14,579. Additionally, some LAB strains were found to interact with a wide range of antimicrobial drugs. The functional activity of P. acidilactici in fermented foods leads to the production of phytase and lactic acid through homofermentation [17]. In addition, P. acidilactici was shown to enhance thep-hydroxy-benzoic, vanillic acids and crude fiber, but to reduce protein and crude fat content in fermented barley [18]. Furthermore, P. acidilactici enhanced the flavor and aroma characteristics of bioactive phenolic compounds (catechin, quercetin, hydroxybenzoic acids and myricetin) [19,20,21]. Pathogenic bacteria with elevated levels of antimicrobial activity are key effective probiotics [22]. Resistance to gastrointestinal juices throughout the intestinal tract and stomach is the main factor determining whether probiotic strains reach the gut and benefit the host [23]. Before confirming the efficiency of a probiotic bacterium, its biological impact on the host during intestinal passage must be tested [24]. The probiotic physiological activity should resist stress in digestive enzymes such as pepsin, pancreatin, bile salt, and acidic medium (pH 2.0) [6]. For example, if exposed to low pH, L. delbrueckii subsp and L. bulgaricus exhibit low survival rates [25]. In this study, the selected MNL5 strains showed good tolerance to low pH (>94%), pepsin (>76%) and bile salts and pancreatin (77%) (see Figure 3).
The nematode C. elegans is a useful in vivo host model to investigate the interactions among probiotic bacteria. Probiotics have a health-promoting effect recognitions to their ability to colonize the host gut; therefore, their adhesion and colonization abilities must be evaluated in studies of the potential benefits of their use [3,26]. The chemotaxis test was used to assess C. elegans odor reactions and their food preferences. Our experiment showed that these nematodes were disinclined to consume MNL5 pellets, instead favoring liquid culture. Additionally, the behavioral characteristics of nematodes clearly demonstrated, through a chemotaxis test, that the MNL5 strains are nontoxic. Based on these findings, it may be concluded that LAB plays a role in growth enhancement when used as a nematode food supplement. Previous studies indicated that some bacteria may colonize the intestines of worms, lengthening the lifespan of the nematode. C. elegans has been used as a model organism for over half a century, with its many similarities between mammals and microbes facilitating assessments of host–microbe interactions [27]. Coenzyme Q (CoQ), an essential coenzyme with aerobic respiration, plays a significant role in increasing C. elegans longevity. Since the lifespans of nematodes decrease following exposure to E. coli, it would appear that LAB also has a set of CoQ synthesizing enzymes that lead to lifespan extension. When LAB produces CoQ, C. elegans will have a longer lifetime [3,28]. In our study, the lifespans of DAF-2 and LIU1 decreased considerably due to lipid metabolism. The longevity of the DAF-2 and LIU1 models was significantly improved following exposure to P. acidilactici MNL5.
C. elegans extends the lifespan, and requires the activity, of DAF-16, FOXO longevity controlled transcription factors, known as IGF-I Signaling Pathways (IIS), in response to insulin/IGF. As several studies have shown, elevated glucose levels in C. elegans reduce their fertility and lifespan; indeed, the efficiency of regulating oxidation increases substantially with increasing exposure to glucose [29]. C. elegans has been used in studies of metabolism related diseases, such as the regulation of fat storage and obesity [30]. The transparency of C. elegans makes it easy to view the fluorescent Nile Red, a commonly used dye for staining and measurement of lipid stores within worm intestine [31]. In our experiment, on the 15th day of P. acidilactici MNL5 and glucose supplement treatment, we observed a significant reduction in lipid droplets after OP50 with glucose lipid droplets is increased (Figure 6). Drosophila leads, for example to, delayed growth and mutations in the insulin/IGF receptor, as well as and somatotropic signaling disruptions in young worms, leading to lower body size in mammals and reduced IGF-1 circulation [32]. Accordingly, fatty acid oxidation related to fat metabolism in mice is upregulated by L. curvatus HY7601, L. plantarum KY1032, and B. breve B-3 [22,33,34,35]. Thus, our results support the hypothesis that the modulation of lipid metabolism is a regulatory mechanism for probiotics. Through qPCR, the active role of de novo fatty acid synthesis in the transcriptional upregulation of E. coli + glucose treated worms was shown. However, P. acidilactici MNL5 + glucose-treated worms showed that transcription repression in the fatty acid desaturase genes is decreased. Therefore, we have shown that P. acidilactici MNL5 fat 4, fat 5, and fat 6 are regulated via a mechanism that suppress lipid accumulation through the C. elegans (DAF-2 and LIU1) model.
4. Materials and Methods
4.1. Isolation and Characterization of Cholesterol Reducing LAB
LAB isolated from fermented herbal medicine [MNL–Moola nivarana lehyam, SKML—Solaimalai Kandan Kathri Lehiyam, SKSPL–Sarkaraikolli special Lehiyam, TVL—Thuthuvalai Lehyam, GSC—Gastone Lehiyam] collected from Vadalur Arutjothi Vaithiyasalai [P] Limited, Tiruchirappalli, Tamil Nadu, India. Moola nivarana lehyam: the primary herbal plant is Withania somnifera, which belongs to the Solanaceae family and is known as Indian Ginseng or Ashwagandha and is known in Ayurvedic medicine as MedhyaRasayan (Nootropic herb), Terminalia chebula: 3.10%, Emblica officinalis: 3.10%, Terminalia Bellirica: 3.10%, Withania somnifera: 6.85%, Zingiber officinale (3.6%), Phytophthora megasperma (root rot) (3.6%), Cyperus rotundus (7%), Tamarindus indica (7%), Aloe barbadensis miller (7%), Cissus quadrangularis (7%), Amorphophallus paeoniifolius (3%), Arenga pinnata (5%), butter (10%) mixed with Honey, further naturally fermented at 30 °C for 72 h. Colony morphology, staining, motility, and other biochemical tests were conducted on the isolates. Screening of LAB strains from fermented herbal medicines for cholesterol removal was carried out by determination of BSH activity and cholesterol assimilation.
4.2. BSH Activityand Cholesterol Assimilation
For BSH activity measurement, 10 mL of culture grown in MRS broth was spotted onto BSH screening medium, which consisted of de Man Rogosa and Sharpe (MRS; Difco, Sparks, MD, USA) agar supplemented with 0.5% (w/v) sodium salt taurodeoxycholic acid (TDCA, Sigma Aldrich, St. Louis, MO, USA) and 0.37 g/L of CaCl2 [36]. Plates were incubated anaerobically at 37 °C using GasPak EZ anaerobe container systems (Becton, Dickinson and Company, Sparks, MD, USA), after which BSH activity was determined by measuring the diameters of precipitation zones. BSH activity was expressed based on the diameters of precipitation zones on BSH screening medium:-no precipitation; +, precipitation zone of up to 10 mm; ++, precipitation zone of up to 15 mm; and +++, precipitation zone of up to 20 mm.
Cholesterol assimilation was determined using the method of Rudel and Morris [37]. LAB cells grown overnight were inoculated (1%) and cultivated anaerobically using GasPak EZ anaerobe container systems (Becton, Dickinson and Company) at 37 °C in MRS broth supplemented with 0.5% (w/v) oxgall (Sigma Aldrich, St. Louis, MO, USA) and 0.1 g/L of water-soluble cholesterol (Sigma Aldrich). Following incubation, cells were harvested (9950× g, 5 min, 4 °C), after which 1 mL of supernatant was added to 2 mL of 33% (w/v) potassium hydroxide and 3 mL of 95% (v/v) ethanol. The mixture was shaken well for 1 min and then heated for 10 min in a 60 °C water bath. After cooling in cold water, 5 mL of hexane was added, followed by mixing and addition of 1 mL of distilled water. The tube was allowed to stand for 10 min at room temperature for phase separation, after which 3 mL of the hexane phase was transferred to a clean tube. The hexane phase was then evaporated under a nitrogen stream. The concentrated phase was added to 4 mL of freshly prepared O-phthalaldehyde (Sigma-Aldrich, 0.5 mg of O-phthalaldehyde/mL of acetic acid), mixed, and permitted to stand at room temperature for 10 min. Following the addition of 2 mL of concentrated sulfuric acid and incubation for 10 min, the absorbance at 550 nm was read using a spectrophotometer (Amersham Biosciences, Uppsala, Sweden). Absorbance values were compared to those obtained using cholesterol standard.
4.3. Molecular Identification of LAB
Using universal primer, 16S rRNA (27F-5′ AGA GTT TGA TCM TGG CTC AG 3′ and 1492R-5′ GGT TAC CTT GTT ACG ACT 3′) for bacteria, the genotype and species level identification was performed at Macrogen, Seoul, Korea. Entirely attained gnomic sequences were analyzed using the Basic Local Alignment Search Tool (BLAST) and further the sequences were entered in Gen Bank, NCBI (Bankit). All cultures were incubated at 37 °C for 24 h on MRS agar.
4.4. Safety Aspect of LAB
4.4.1. Antibiotic Susceptibility
Isolated strains were tested against 9 antibiotics (30 µg L−1) with different modes of actions [38] were tested against Gentamicin (Gen), Imipenem (Imp); Erythromycin (Ery), Novobiocin (Nov), Tetracycline (Tet), Clindamycin (Cli), Meropenem (Mer), Ampicillin (Amp), and Penicillin (Pen) based on disc diffusion method. The nutrient agar was scrutinized for the diameter of the inhibition zone was measured with calipers at 37 °C for 24 h. [38]. All antibiotics were obtained (Sigma Aldrich, St. Louis, MO, USA) and tested against LAB using a disc diffusion method. After 24 h incubation, agar plates have been evaluated for the presence or absence of an inhibition zone at 37 °C.
4.4.2. Antibacterial Activity
The antibacterial activity was evaluated using the LAB cell-free supernatants by the agar well diffusion method. All isolates screened were cultivated in MRS Broth and incubated 24 h at 37 °C. To extract the bacterial supernatant, a culture centrifugation with isolates was spin for 25 min at 4 °C, using 6000× g [3,39]. These human pathogens, Escherichia coli ATCC 35150, Staphylococcus aureus ATCC 13,150, H. pylori ATCC 43,504, Bacillus cereus ATCC 14,579 and Candida albicans KCTC 7965 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and Korean Collection for Type Cultures (KCTC, Seoul, Korea).
4.4.3. Hemolytic Activity
The hemolytic activity was evaluated using the method described earlier [40]. Briefly, the isolate was streaked on nutrient agar plate supplemented with 5% goat blood and incubated for 24 h–48 h at 37 °C to detect patterns of hemolytic activity.
4.5. Gastrointestinal Survival under In Vitro Conditions
4.5.1. Tolerance to Low pH
The tolerance to low pH of selected lactic acid bacterial strains was determined using the method described by Mokoena [41], with some modifications. Briefly, the active strains grown in MRS broth were inoculated (1.5%) in 10 mL of fresh MRS broth, incubated at 37 °C for 24 h and then centrifuged at 6000× g for 15 min. The pellets were suspended in sterile phosphate-buffered saline (PBS; Gibco™, Thermo Fisher Scientific, Waltham, MA, USA) containing 9 g/L NaCl (Sigma Aldrich, St. Louis, MO, USA), 9 g/L Na2HPO4⋅2H2O and 1.5 g/L KH2PO4 adjusted to a pH of 2.5. The mixture was incubated at 37 °C for 4 h, and samples were taken at time 0 and after 4 h. These samples were serially diluted in sterile saline (0.85% NaCl) solution and plated on MRS agar; the viable cells were determined after incubation at 37 °C for 24 h, and the percentage of survival of the bacteria was calculated as follows: % survival = [CFU of viable cells survived/CFU of initial viable cells inoculated] × 100.
4.5.2. Tolerance to Pepsin
To test the bacterial tolerance to pepsin, a simulated gastric juice was prepared by adding 3 mg/mL pepsin (Sigma Aldrich, St. Louis, MO, USA) to sterile saline solution (0.85% NaCl, w/v) and adjusting the pH to 2.5. The fluid was inoculated with active cultures at an inoculum size of 1% (v/v) and incubated at 37 °C for 4 h. The viable cells were determined at an initial (T1) and final incubation time (T2) by the spread plate method by Oh et al. [42]. The percentage survival of the bacteria was calculated as follows: % survival = [CFU of viable cells survived/CFU of initial viable cells inoculated × 100] percentage of survival [3,42].
4.5.3. Resistance of Pancreatin
The resistance of pancreatin was tested as described by Oh et al. [42], with some modifications [3]. Briefly, 0.3% (w/v) bile salts (Sigma Aldrich, St. Louis, MO, USA) and 1 mg/mL pancreatin (Sigma Aldrich, St. Louis, MO, USA) were dissolved in sterile saline solution (0.85% NaCl, w/v) adjusted to pH 8.0. The fluid was inoculated with 1% (v/v) LAB cultures and incubated at 37 °C for 6 h. The viable cells were determined before and after incubation by the spread plate method. The percentage survival of the bacteria was calculated as follows: % survival = [CFU of survivors/CFU of inoculums × 100].
4.6. In Vivo Studies
4.6.1. Maintain the C. elegans
In the Caenorhabditis elegans, the gene daf-2 encodes the insulin-like growth factor 1 receptor (IGF-1). The C. elegans (DAF-2(e1370) and LIU1 (ldrIs1) model) are obtained from the Caenorhabditis Genetics Center (CGC), Minnesota. Then E. coli OP50 was placed in the NGM agar plates to supplement the arrested harmonized L1 worm population.
4.6.2. C. elegans Synchronizing
The young adult worms hatched from eggs were treated with sterile water containing 0.5 M NaOH and 0.5% bleaching solution (sodium hypochlorite) for synchronization. After vortex mixing at 2 min intervals, worms were washed three times in M9 buffer (3.0 g KH2PO4, 6.0 g Na2HPO4, 0.5 g NaCl, 1.0 g NH4Cl, H2O/1 L, sterilized at 121 °C) via centrifugation (1200× g for 200 s). To prepare worms for all assays, approximately 3000 synchronized eggs were hatched in M9 buffer at 20 °C overnight. The L1 larvae were subsequently transferred to nematode growth medium (NGM) agar with a lawn of E. coli OP50 and incubated at 25 °C for 48 h to reach the L4 stage. The test was performed by Wong et al. [43] method.
4.6.3. Chemotaxis Assay
The chemotaxis test method was followed by Margie et al. [44]. Thirty L4 stage C. elegans worms were administered MNL5 and OP50 diet include Petri plates. To further examine the response C. elegans has to a glucose diet; we have established an experiment to determine whether animals distinguish between a spot of food that is the standard OP50 E. coli diet and a spot of MNL5 mixed with glucose. At 1 h following the assays, the attractive and counter-attracting paths have been identified as the number of animals. For each condition, triplicate chemotaxis analyses were performed. For each experiment, a chemotaxis index (CI) was calculated using the following formula: Chemotaxis index (CI) = [number of worms in the LAB bacteria strain–number of worms in OP50 (control)/total number of worms applied]. A + 1.0 score indicates maximal attraction towards the target and represents 100% of the worms arriving in the quadrants containing the chemical target. An index of −1.0 is evidence of maximal repulsion.
4.6.4. Establishment of Glucose Diet with LAB
C. elegans are used for a synchronized age test, and the experiment was developed in fluid M9 buffer, and eggs are treated with sodium hypochlorite. Our study aimed at achieving glucose focus in a concentration of 10–15 mmol/L C. elegans in the entire body. The 50 μL of glucose (18 mg) diet with LAB, which is seeded at 37 °C at 4 h with NGM plates. L4 larvae are moved to the nematode growth medium after four hours.
4.6.5. Bacterial Colonization Assay
A bacterial colonization analysis was conducted for the C. elegans gut colonization of P. acidilactici MNL5, led by Antunes et al. [45]. On the first day, lactic acid bacteria were seeded on NGM plates containing 100 worms. On the 3rd, 5th, 7th, 9th and 2th day of incubation, the total number of colonies present in the gut of the young adult (L4) stage worm was determined using the following procedure: Five worms were washed in 5 μL drops of M9 buffer to inhibit pharyngeal pumping and expulsion. The washed nematodes were placed in a 1.5 mL centrifugation tube containing 50 μL of PBS buffer with 1% Triton X-100 and mechanically disrupted using a mortar and pestle. Worm lysate was diluted into PBS and plated on MRS agar at a temperature of 37 °C. LAB colonies have been enumerated for the calculation of nematode-bacteria. Compared to E. coli with lactic acid bacteria, the procedure was followed Bito et al. [46].
4.6.6. Lifespan Assay
C. elegans longevity was measured using the Smolentsevae et al. [47] method already explained. In the test, 50 μL of the TS and MRS broth contain bacteria (OP50-Control and MNL5- test) that were seeded over a 35 mm NGM plate (5-fluorodeoxyuridine FUdR 40 mM for prevents the reproduction) in 24 h at 37 °C. The 100 L4 stage C. elegans have been incubated at 20 °C and counted every 24 h as dead worms. Every three days, living worms were moved to fresh NGM plates with bacterial target strain. To compare the effects of glucose against MNL5, first worms with OP50 were cultivated and, the mean lifespan were determined by Chelliah et al. [3]. The three experiments for each strain were performed.
4.6.7. Body Size Measurements
The Olympus SZ 61 zoom stereomicroscope connected to HK3.1 CMOS Camera was used to photograph individual animals after the 0 and 15th day. The size of a worm body measured through a central nematode axis has been assessed by the use of ToupViewTM 3.7 software after cultivating. On at least three independent experiments, at least five nematodes were pictured.
4.6.8. Nile Red and Oil Red O (ORO) Staining Methods
The lipid reduction studies using Nile red and Oil Red O staining methods. The L4 worms used in this study were washed by liquid M9 medium and treated by sodium hypochlorite treatment. Then drop of Nile Red (0.05 μg/mL) solution were added to the worms, which were then incubated for 30 min, washed with 25% ethanol twice, and photographed in a Fluorescence microscope (Olympus CKX53, Tokyo, Japan) analysis. The fresh ORO solution was prepared by diluting stock (0.5% ORO in isopropanol) to a 60% solution with water, filtering (0.45 µM filter), stirring at room temperature overnight, and filtering again just before use. Afterwards, L4 worms were collected, washed, and fixed in 60% isopropanol for 5 min. Then, the fixed worms were incubated in the ORO solution for 6 h in a wet chamber with gentle shaking in the dark, washed with PBS, and mounted on a 2% agarose pad for microscopy visualization [48]. All of the worms were photographed with same setups and same exposure times.
4.6.9. Fluorescence Quantification
The fluorescence intensity was quantified by Image J software. Nile red and Oil Red O stained lipids were predominantly found in the gut, and the anterior region was significantly brighter than the posterior. The region for integrating fluorescence density measurements was chosen from the intestine’s anterior front to vulva. Six L4 worms were randomLy chosen for analysis. For Image J analysis, pictures were captured at a magnification of 20×.
4.6.10. RNA Extraction and Primer Deign
The gene expression of C. elegans (DAF-2& LIU1) has been analyzed in E. coli OP50, E. coli OP50 + glucose and P. acidilactici MNL5+glucose, fed worm populations. Synchronized populations were obtained under specific feed conditions from embryos isolated with gravy adults. After feeding time (12 days of 50% of population), worms were obtained with an M9 buffer and washed three times in Eppendorf tubes for disturbed by sonication (3 pulses at 10 W, 20 s/pulse) [49]. Following the manufacturer’s instructions with certain modifications, Total RNA has been extracted from nematodes using RNAiso Plus (Takara Co., Ltd., Kusatsu, Japan). RNA quantification was performed with the Eppendorf AG22331 Biospectrophotometer (Eppendorf AG, Hamburg, Germany). 260/280 and 260/230 absorption ratios assessed RNA purity.
Primer design Specific primers for the differentiation of lipids targeted on selective genes metabolism is involved in the regulation of DAF-2/insulin signaling. The respective primers (fat-4, fat-5, fat-6) were designed using Primer Express® Software v3.0.1, (Applied Biosystems; Foster City, CA, USA) and were prepared as described by Bioneer Corporation (Daejeon, Korea). The primers used in the study along with their primer sequence and product size are shown in Supplementary Table S1. In addition, to enhance the detection range of the primers, the International Union of Pure and Applied Chemistry (IUPAC, Research Triangle, NC, USA) was used for confirmation. The primers were designed to keep the melting temperature (Tm) of the PCR amplicons between 70 and 85 °C and each amplicon Tm was separated by approximately 2 °C (Supplementary Figure S2, Supplementary Table S3). All amplicons were predicted using the Bio-Edit Software [50] and by applying Primer-BLAST tool and Tm, specificity of the designed primers was determined [14].
4.6.11. Quantitative PCR Analyses
All qPCR assays were performed using StepOne™ real-time PCR System (Applied Biosystems, Foster City, CA, USA). For each reaction, 10 μL of MeltDoctor™ HRM Master Mix, 2 μL of genomic DNA (10 ng μL−1), 0.5 μL of each primer (10 pg μL−1) and 7 μL of double distilled water were added. Gene expression differences between treated and untreated worms were quantified using the relative quantification 2−∆∆Ct method [51].
4.7. Statistical Analysis
Reproducibility of the qPCR was determined with three independent experiments conducted in duplicates. Data collected from quantitative tests (acid tolerance, pepsin & pancreatin, antibacterial activity, antibiotic susceptibility, and cholesterol assimilation), further the results were The data obtained from each experiment was expressed as mean and standard deviation by Microsoft Office Excel 2010 (Microsoft Corporation, Redmond, Washington, DC, USA).
5. Conclusions
The present work reveals that fermented medicines contain probiotic bacteria. As such, their use may lead decreased cholesterol concentrations. Additionally, it was found that P. acidilactici MNL5 bacteria have better survival ability in the GI tract. Our results showed longer lifespans for P. acidilactici MNL5 and shorter colonization for C. elegans. The transcription of genes encoding fatty acid desaturase was significantly suppressed in P. acidilactici MNL5 treated worms, according to our qPCR results. These results confirm that P. acidilactici MNL5 is a viable approach to lipid reduction, supplementing dietary efforts for lipid reduction in the DAF-2 and LIU1 model for obesity management. In addition, the information presented in this paper will support future studies on mice obesity models.
Acknowledgments
The author would like to thank the Agricultural and Life Science Research Institute, Kangwon National University for their scientific support.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23031276/s1.
Author Contributions
The manuscript was written in detail and sectioned for specialized discussion with the respective authors in research. Writing—original draft; K.B., R.C., Writing—review & editing (Isolation of Probiotics, characterization, analysis and Conceptualization), Methodology—K.B., R.C., M.V.A., D.-H.O., Data curation, Formal analysis, Validation–K.B., R.C., F.E., A.T., V.S., P.A., Funding acquisition, Project administration, Resources, Supervision—D.-H.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by National Research Foundation of Korea (NRF)] [Grant number: 2018007551], Brain Korea (B.K.) 21 Plus Project [Grant number: 4299990913942] and Brain Pool Fellowship program [Grant number: 2020H1D3A1A02081423] through the Ministry of Science and ICT, Republic of Korea.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated for this study are available from the corresponding authors upon reasonable request.
Conflicts of Interest
The authors declare conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Food and Agriculture Organization . Food and Agriculture Organization; Quebec City, QC, Canada: 2001. The State of Food and Agriculture 2001. No. 33. [Google Scholar]
- 2.Fasoli S., Marzotto M., Rizzotti L., Rossi F., Dellaglio F., Torriani S. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int. J. Food Microbiol. 2003;82:59–70. doi: 10.1016/S0168-1605(02)00259-3. [DOI] [PubMed] [Google Scholar]
- 3.Chelliah R., Choi J.G., Hwang S.B., Park B.J., Daliri E.B.M., Kim S.H., Wei S., Ramakrishnan S.R., Oh D.H. In vitro and in vivo defensive effect of probiotic LAB against Pseudomonas aeruginosa using Caenorhabditis elegans model. Virulence. 2018;9:1489–1507. doi: 10.1080/21505594.2018.1518088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khusro A., Aarti C., Dusthackeer A., Agastian P. Anti-tubercular and probiotic properties of coagulase-negative staphylococci isolated from Koozh, a traditional fermented food of South India. Microb. Pathog. 2018;114:239–250. doi: 10.1016/j.micpath.2017.11.054. [DOI] [PubMed] [Google Scholar]
- 5.Rajoka M.S.R., Shi J., Zhu J., Shao D., Huang Q., Yang H., Jin M. Capacity of lactic acid bacteria in immunity enhancement and cancer prevention. Appl. Microbiol. Biotechnol. 2017;101:35–45. doi: 10.1007/s00253-016-8005-7. [DOI] [PubMed] [Google Scholar]
- 6.Choi E.A., Chang H.C. Cholesterol-lowering effects of a putative probiotic strain Lactobacillus plantarum EM isolated from kimchi. LWT—Food Sci. Technol. 2015;62:210–217. doi: 10.1016/j.lwt.2015.01.019. [DOI] [Google Scholar]
- 7.Miremadi F., Ayyash M., Sherkat F., Stojanovska L. Cholesterol reduction mechanisms and fatty acid composition of cellular membranes of probiotic Lactobacilli and Bifidobacteria. J. Funct. Foods. 2014;9:295–305. doi: 10.1016/j.jff.2014.05.002. [DOI] [Google Scholar]
- 8.Pinto M.G.V., Franz C.M., Schillinger U., Holzapfel W.H. Lactobacillus spp. within vitro probiotic properties from human faeces and traditional fermented products. Int. J. Food Microbiol. 2006;109:205–214. doi: 10.1016/j.ijfoodmicro.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Heo W., Lee E.S., Cho H.T., Kim J.H., Lee J.H., Yoon S.M., Kim Y.J. Lactobacillus plantarum LRCC 5273 isolated from Kimchi ameliorates diet-induced hypercholesterolemia in C57BL/6 mice. Biosci. Biotechnol. Biochem. 2018;82:1964–1972. doi: 10.1080/09168451.2018.1497939. [DOI] [PubMed] [Google Scholar]
- 10.Huang T.C., Chan H.Y., Wann S.Y., Lin F.M., Lee F.L., Liao C.C. Isolated Pediococcus Acidilactici 05b0111 and Method of Producing Exopolysaccharide. US9873899B2. U.S. Patent. 2015 November 19;
- 11.Pingitore E., Jimenez M.E., Dallagnol A., Belfiore C., Fontana C., Fontana P., Plumed-Ferrer C. Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT—Food Sci. Technol. 2016;71:288–294. doi: 10.1016/j.lwt.2016.03.046. [DOI] [Google Scholar]
- 12.Ortu S., Felis G.E., Marzotto M., Deriu A., Molicotti P., Sechi L.A., Zanetti S. Identification and functional characterization of Lactobacillus strains isolated from milk and Gioddu, a traditional Sardinian fermented milk. Int. Dairy J. 2017;35:1312–1320. doi: 10.1016/j.idairyj.2007.02.008. [DOI] [Google Scholar]
- 13.Ashrafi K. Obesity and the regulation of fat metabolism. WormBook Online Rev. C Elegans Biol. 2007:1–20. doi: 10.1895/wormbook.1.130.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Tan X.R., Li S.J., Zhang X.X. LncRNA NEAT1 promotes hepatic lipid accumulation via regulating miR-146a-5p/ROCK1 in nonalcoholic fatty liver disease. Life Sci. 2019;235:116829. doi: 10.1016/j.lfs.2019.116829. [DOI] [PubMed] [Google Scholar]
- 15.Belguesmia Y., Alard J., Mendil R., Ravallec R., Grangette C., Drider D., Cudennec B. In vitro probiotic properties of selected lactobacilli and multi-strain consortium on immune function gut barrier strengthening and gut hormone secretion. J. Funct. Foods. 2019;57:382–391. doi: 10.1016/j.jff.2019.04.028. [DOI] [Google Scholar]
- 16.World Health Organization . Report of a Joint FAO/OIE/WHO Expert Consultation on Antimicrobial Use in Aquaculture and Antimicrobial Resistance, Seoul, Korea, 13–16 June.2006. World Health Organization; Geneva, Switzerland: 2006. [Google Scholar]
- 17.Bhagat D., Raina N., Kumar A., Katoch M., Khajuria Y., Slathia P.S., Sharma P. Probiotic properties of a phytase producing Pediococcus acidilactici strain SMVDUDB2 isolated from traditional fermented cheese product, Kalarei. Sci. Rep. 2020;10:1926. doi: 10.1038/s41598-020-58676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartkiene E., Mozuriene E., Lele V., Zokaityte E., Gruzauskas R., Jakobsone I., Bartkevics V. Changes of bioactive compounds in barley industry by-products during submerged and solid state fermentation with antimicrobial Pediococcus acidilactici strain LUHS29. Food Sci. Nutr. 2020;8:340–350. doi: 10.1002/fsn3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A., Noda M., Sugiyama M., Ahmad A., Kaur B. Production of Functional Buttermilk and Soymilk Using Pediococcus acidilactici BD16 (alaD+) Molecules. 2021;26:4671. doi: 10.3390/molecules26154671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albano H., Todorov S.D., van Reenen C.A., Hogg T., Dicks L.M., Teixeira P. Characterization of two bacteriocins produced by Pediococcus acidilactici isolated from “Alheira”, a fermented sausage traditionally produced in Portugal. Int. J. Food Microbiol. 2007;116:239–247. doi: 10.1016/j.ijfoodmicro.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Moyano S., Martín A., Benito M.J., Hernández A., Casquete R., de Guia Córdoba M. Application of Lactobacillus fermentum HL57 and Pediococcus acidilactici SP979 as potential probiotics in the manufacture of traditional Iberian dry-fermented sausages. Food Microbiol. 2011;28:839–847. doi: 10.1016/j.fm.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Rahmdel S., Shekarforoush S.S., Hosseinzadeh S., Torriani S., Gatto V. Antimicrobial spectrum activity of bacteriocinogenic Staphylococcus strains isolated from goat and sheep milk. J. Dairy Sci. 2019;102:2928–2940. doi: 10.3168/jds.2018-15414. [DOI] [PubMed] [Google Scholar]
- 23.Barathikannan K., Chelliah R., Rubab M., Daliri E.B.-M., Elahi F., Kim D.-H., Agastian P., Oh S.-Y., Oh D.H. Gut Microbiome Modulation Based on Probiotic Application for Anti-Obesity: A Review on Efficacy and Validation. Microorganisms. 2019;7:456. doi: 10.3390/microorganisms7100456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalesi S., Bellissimo N., Vandelanotte C., Williams S., Stanley D., Irwin C. A review of probiotic supplementation in healthy adults: Helpful or hype? Eur. J. Clin. Nutr. 2019;73:24. doi: 10.1038/s41430-018-0135-9. [DOI] [PubMed] [Google Scholar]
- 25.Park D.Y., Ahn Y.T., Park S.H., Huh C.S., Yoo S.R., Yu R., Sung M.K., McGregor R.A., Choi M.S. Supplementation of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS ONE. 2013;8:e59470. doi: 10.1371/journal.pone.0059470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irazoqui J.E., Urbach J.M., Ausubel F.M. Evolution of host innate defence: Insights from C. elegans and primitive invertebrates. Nat. Rev. Immunol. 2010;10:47. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skora S., Mende F., Zimmer M. Energy scarcity promotes a brain-wide sleep state modulated by insulin signaling in C. elegans. Cell Rep. 2018;22:953–966. doi: 10.1016/j.celrep.2017.12.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revtovich A.V., Lee R., Kirienko N.V. Interplay between mitochondria and diet mediates pathogen and stress resistance in Caenorhabditis elegans. PLoS Genet. 2019;15:e1008011. doi: 10.1371/journal.pgen.1008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Zhang L., Zhang L., Wang W., Wei S., Wang J., Che H., Zhang Y. Effects of sugars and lipids on the growth and development of Caenorhabditis elegans. BioRxiv. 2019:615807. doi: 10.1101/615807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martorell P., Llopis S., Gonzalez N., Chenoll E., Lopez-Carreras N., Aleixandre A., Genovés S. Probiotic strain Bifidobacterium animalis subsp. lactis CECT 8145 reduces fat content and modulates lipid metabolism and antioxidant response in Caenorhabditis elegans. J. Agric. Food Chem. 2016;64:3462–3472. doi: 10.1021/acs.jafc.5b05934. [DOI] [PubMed] [Google Scholar]
- 31.Jones K.T., Ashrafi K. Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Dis. Models Mech. 2009;2:224–229. doi: 10.1242/dmm.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers D.B., Androschuk A., Rosenfelt C., Langer S., Harding M., Bolduc F.V. Insulin signaling is acutely required for long-term memory in Drosophila. Front. Neural Circuits. 2015;9:8. doi: 10.3389/fncir.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen A.L., Lum K.M., Lara-Gonzalez P., Ogasawara D., Cognetta A.B., To A., Parsons W.H., Simon G.M., Desai A., Petrascheck M., et al. Pharmacological convergence reveals a lipid pathway that regulates C. elegans lifespan. Nat. Chem. Biol. 2019;15:453–462. doi: 10.1038/s41589-019-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aranaz P., Navarro-Herrera D., Zabala M., Romo-Hualde A., López-Yoldi M., Vizmanos J.L., Milagro F.I., González-Navarro C.J. Phenolic compounds reduce the fat content in Caenorhabditis elegans by affecting lipogenesis, lipolysis, and different stress responses. Pharmaceuticals. 2020;13:355. doi: 10.3390/ph13110355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahlby C., Lee Conery A., Bray M.A., Kamentsky L., Larkins-Ford J., Sokolnicki K.L., Veneskey M., Michaels K., Carpenter A.E., O’Rourke E.J. High- and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods. 2014;68:492–499. doi: 10.1016/j.ymeth.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannock G.W., Dashkevicz M.P., Feighner S.D. Lactobacilli and bile salt hydrolase in the murine intestinal tract. Appl. Environ. Microbiol. 1989;55:1848–1851. doi: 10.1128/aem.55.7.1848-1851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudel L.L., Morris M.D. Determination of cholesterol using o-phthalaldehyde. J. Lipid Res. 1973;14:364–366. doi: 10.1016/S0022-2275(20)36896-6. [DOI] [PubMed] [Google Scholar]
- 38.Das P., Mukherjee S., Sen R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J. Appl. Microbiol. 2008;104:1675–1684. doi: 10.1111/j.1365-2672.2007.03701.x. [DOI] [PubMed] [Google Scholar]
- 39.Aslam S.N., Stevenson P.C., Kokubun T., Hall D.R. Antibacterial and antifungal activity of cicerfuran and related 2-arylbenzofurans and stilbenes. Microbiol. Res. 2009;164:191–195. doi: 10.1016/j.micres.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Manhar A.K., Bashir Y., Saikia D., Nath D., Gupta K., Konwar B.K., Kumar R., Namsa N.D., Mandal M. Cellulolytic potential of probiotic Bacillus Subtilis AMS6 isolated from traditional fermented soybean (Churpi): An in-vitro study with regards to application as an animal feed additive. Microbiol. Res. 2016;186:62–70. doi: 10.1016/j.micres.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Mokoena M.P., Mutanda T., Olaniran A.O. Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages. Food Nutr. Res. 2016;60:29630. doi: 10.3402/fnr.v60.29630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh A., Daliri E.B.M., Oh D.H. Screening for potential probiotic bacteria from Korean fermented soybean paste: In vitro and Caenorhabditis elegans model testing. LWT—Food Sci. Technol. 2018;88:132–138. doi: 10.1016/j.lwt.2017.10.007. [DOI] [Google Scholar]
- 43.Wong A., Boutis P., Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margie O., Palmer C., Chin-Sang I.C. C. elegans chemotaxis assay. J. Vis. Exp. 2013;74:50069. doi: 10.3791/50069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antunes C.A., Clark L., Wanuske M.T., Hacker E., Ott L., Simpson-Louredo L., de Luna M.D.G., Hirata R., Jr., Mattos-Guaraldi A.L., Hodgkin J., et al. Caenorhabditis elegans star formation and negative chemotaxis induced by infection with corynebacteria. Microbiology. 2016;162:84–93. doi: 10.1099/mic.0.000201. [DOI] [PubMed] [Google Scholar]
- 46.Bito T., Matsunaga Y., Yabuta Y., Kawano T., Watanabe F. Vitamin B12 deficiency in Caenorhabditis elegans results in loss of fertility, extended life cycle, and reduced lifespan. FEBS Open Bio. 2013;3:112–117. doi: 10.1016/j.fob.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smolentseva O., Gusarov I., Gautier L., Shamovsky I., DeFrancesco A.S., Losick R., Nudler E. Mechanism of biofilm-mediated stress resistance and lifespan extension in C. elegans. Sci. Rep. 2017;7:7137. doi: 10.1038/s41598-017-07222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barathikannan K., Venkatadri B., Khusro A., Al-Dhabi N.A., Agastian P., Arasu M.V., Kim Y.O. Chemical analysis of Punica granatum fruit peel and it’s in vitro and in vivo biological properties. BMC Complementary Altern. Med. 2016;16:264. doi: 10.1186/s12906-016-1237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chelliah R., Wei S., Park B.J., Kim S.H., Park D.S., Kim S.H., Hwan K.S., Oh D.H. Novel motB as a potential predictive tool for identification of B. cereus, B. thuringiensis and differentiation from other Bacillus species by triplex real-time PCR. Microb. Pathog. 2017;111:22–27. doi: 10.1016/j.micpath.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 50.Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Information Retrieval Ltd.; London, UK: 1999. pp. 95–98. (Nucleic Acids Symposium Series). [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated for this study are available from the corresponding authors upon reasonable request.