We read with interest the recent publication by Benjamin et al. (2) regarding the characterization of IS6110 insertion sites in the direct repeat (DR) region of Mycobacterium tuberculosis. This topical and relevant study described the dissection of a single molecular event leading to an altered DNA fingerprint pattern that was detected by two different strain genotyping systems, namely, IS6110 DNA fingerprinting and spoligotyping. It was shown that a single IS6110 transposition event in the DR region disrupted one of the spoligotyping primer regions, thereby resulting in simultaneous changes in the IS6110 fingerprint and spoligotype pattern. The authors rightly recommended that some caution should be applied to the interpretation of similar, but nonidentical, IS6110 DNA fingerprint and spoligotype patterns. In addition, the results corroborated the occurrence of IS6110 preferential integration loci, as previously identified by a number of investigators (3, 5, 7).
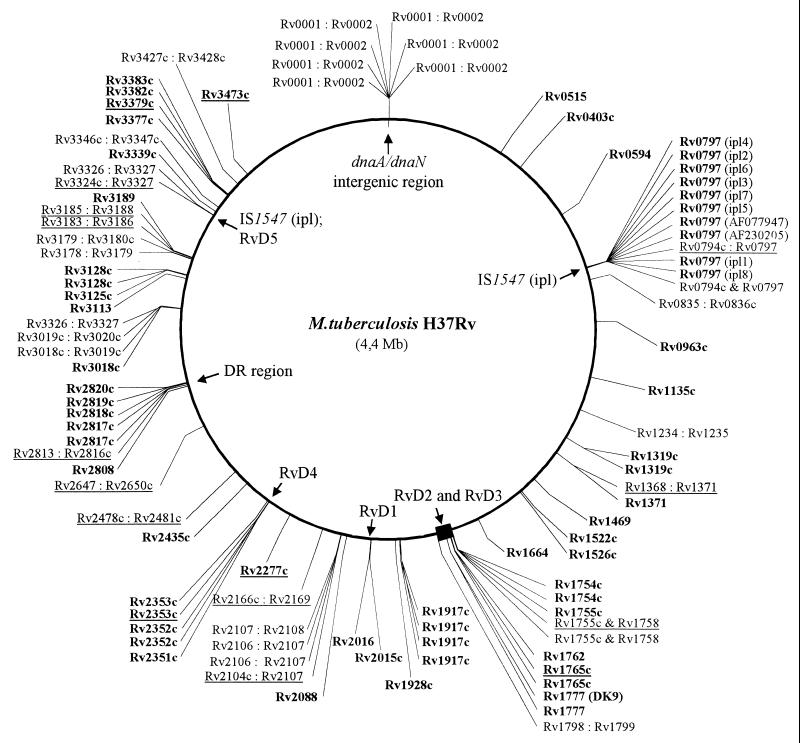
However, we would like to question the statement by the authors that “nearly all IS6110 insertions are between open reading frames…” The assertion was not referenced, but we assume that this statement was occasioned by the original reporting of IS6110 distribution in the reference strain M. tuberculosis H37Rv (4), which states that the majority of insertions occur within noncoding regions. However, this early conclusion was restricted to the analysis of a very limited number of insertion sites in M. tuberculosis H37Rv. Since this preliminary characterization, numerous investigators (ourselves included) have characterized IS6110 integration loci in clinical isolates (1, 3, 5, 7). Our analysis of insertion locus sequence data collated from various sources demonstrated that 42 of 66 discrete IS6110 insertion sites (58%) occurred within coding regions of the M. tuberculosis genome (1, 5, 7). We performed an updated analysis (inclusive of data published in the literature and DNA sequence databases since our original report), the results of which demonstrated that of 95 discrete IS6110 integration loci identified (excluding multiple insertions into the DR and ipl locus), 60% (57 of 95 sites) occur within coding regions, 33% (31 of 95 sites) occur within intergenic regions (Fig. 1), and 7% (7 of 95 sites) did not map to the M. tuberculosis H37Rv genome. This strengthens our conclusion (5) that IS6110 frequently disrupts coding regions. In addition, the revised map (Fig. 1) of the genome distribution of IS6110 insertions highlights the existence of numerous preferential integration sites in addition to those previously described (2, 3, 5, 7).
FIG. 1.
Distribution of IS6110 insertion loci relative to open reading frames (ORFs) of the Mycobacterium tuberculosis H37Rv genome. The ORF numbers of the disrupted genes are indicated in bold, while the intergenic insertions are represented by two ORF numbers separated by a colon. The M. tuberculosis H37Rv insertion sites are underlined. Multiple integration sites within a single gene or intergenic region are denoted by multiple entries with the same name. The relative positions of the DR region and the ipl locus are shown, and the positions of regions deleted from M. tuberculosis H37Rv relative to M. bovis are indicated by their RvD numbers.
In the light of the limited structural gene diversity within M. tuberculosis strains, it has been suggested that IS6110 may be capable of driving genome evolution (6). Therefore, while the biological significance of our finding remains to be elucidated, it nonetheless deserves further attention, as it clearly demonstrates the potential for the IS6110 element to impact on strain phenotype by gene disruption. Finally, our approach highlights the value of collation of data from various sources, and we advocate the establishment of a web-based genome map of IS6110 integration sites to facilitate this endeavor.
Authors' Reply
We acknowledge the extensive work by Drs. Sampson, Warren, Richardson, van der Spuy, and van Helden to characterize the IS6110 insertion sites within the M. tuberculosis genome. The statement that “nearly all IS6110 insertions are between open reading frames …” was derived from the original reporting of IS6110 distribution that focused primarily on the H37Rv strain of M. tuberculosis. We appreciate the letter correcting our error.
REFERENCES
- 1.Beggs M L, Eisenach K D, Cave M D. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:2923–2928. doi: 10.1128/jcm.38.8.2923-2928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin W H, Jr, Lok K H, Harris R, Brook N, Bond L, Mulcahy D, Robinson N, Pruitt V, Kirkpatrick D P, Kimerling M E, Dunlap N E. Identification of a contaminating Mycobacterium tuberculosis strain with a transposition of an IS6110 insertion element resulting in an altered spoligotype. J Clin Microbiol. 2001;39:1092–1096. doi: 10.1128/JCM.37.3.1092-1096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B, Jacobs W J, Cole S T. An integrated map of the genome of the tubercle bacillus Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampson S L, Warren R M, Richardson M, van Der Spuy G D, van Helden P D. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuberc Lung Dis. 1999;79:349–359. doi: 10.1054/tuld.1999.0218. [DOI] [PubMed] [Google Scholar]
- 6.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren R M, Sampson S L, Richardson M, Van Der Spuy G D, Lombard C J, Victor T C, van Helden P D. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol Microbiol. 2000;37:1405–1416. doi: 10.1046/j.1365-2958.2000.02090.x. [DOI] [PubMed] [Google Scholar]