At the beginning of this review it is essential to clarify the terminology that will be used to refer to the members of the Burkholderia cepacia complex and their relatives. The name B. cepacia will relate only to B. cepacia genomovar I. Strains resembling B. cepacia may belong to the B. cepacia complex, to other Burkholderia species (for instance, Burkholderia gladioli), or to species from other genera (for instance, Ralstonia pickettii) that share some phenotypic or genotypic similarities with the B. cepacia complex. B. cepacia complex bacteria and organisms that may be confused with them will be altogether referred to as B. cepacia-like organisms. Most previous reports regarding these organisms were published before the recognition of the complicated taxonomic relationships between the different members of the B. cepacia complex; it is therefore unclear to what category the presumed B. cepacia isolates described would belong. For that reason, when such literature is cited, the name “B. cepacia” will be shown in double quotes.
Chronic microbial colonization of the major airways, leading to exacerbations of pulmonary infection, is the major cause of morbidity and mortality in patients with cystic fibrosis (CF). Typical CF pathogens include Staphylococcus aureus, Pseudomonas aeruginosa, and Haemophilus influenzae (30). Other glucose nonfermenters, like Stenotrophomonas maltophilia, Alcaligenes xylosoxidans, R. pickettii, and Burkholderia gladioli, can frequently be found as well, but their role in the decline of pulmonary function is unclear (14, 19, 30). Several reports on the recovery of “B. cepacia” from CF patients appeared in the late 1970s and early 1980s (62, 63). The first detailed description of the clinical significance of “B. cepacia” colonization and infection was published in 1984 (47). In that seminal paper, Isles et al. documented the increasing prevalence of “B. cepacia” colonization and infection in the Toronto, Canada, CF treatment center and described the so-called “cepacia syndrome,” a severe progressive respiratory failure with bacteremia that occurs in about 20% of all infected CF patients. Clustering of new cases in some centers and the decrease of colonization of new patients following segregation of colonized and noncolonized patients in other centers suggested that “B. cepacia” could be transmitted between CF patients. This was confirmed by several studies (34, 64, 67, 76, 84, 94) that showed that “B. cepacia” strains can spread between CF patients via simultaneous hospital admissions or social contact outside of the hospital. As a result of these findings, new guidelines were issued to reduce the risk of “B. cepacia” acquisition. These included discontinuing sponsorship and support of CF summer camps and segregation of colonized patients. Implementation of these draconian infection control measures has a tremendous impact on the lives of CF patients, and not all patients or caregivers accept such measures (35, 36, 62, 63).
“B. cepacia” can also cause lung infections in chronic granulomatous disease patients, and infections in these patients are associated with pneumonia and septicemia and are often lethal (2, 58, 72, 96). “B. cepacia” infections in immunocompetent patients occur only sporadically, but several cases of pseudoepidemics and nosocomial infections, often caused by contaminated disinfectants and anesthetic solutions, have been reported (3, 43, 50, 107).
Despite the advances that have been made in the understanding of the epidemiology, “B. cepacia” infections still have a considerable impact on morbidity and mortality in CF patients (18, 61, 62, 63). Since “B. cepacia” is resistant to most antimicrobial agents, effective therapies are not straightforward and management efforts are therefore aimed at prevention of infection (35, 63). Several recommendations regarding infection control measures have been made, and these include that CF patients should not share hospital rooms as inpatients and should limit contact in outpatient clinics (63). However, the efficiency of infection control measures are determined by the accuracy with which “B. cepacia” is diagnosed, and poor laboratory proficiency in identification of this organism still prevails (17, 40, 75). Although several guidelines intended to enhance accurate identification of bacterial species from sputum culture have been proposed by national CF organizations and by the International Burkholderia cepacia Working Group, the degree to which these are followed varies greatly among clinical microbiology laboratories (90).
The problem is given an extra dimension by the fact that several “B. cepacia” strains have attracted attention as antagonists of soilborne plant pathogens (44, 66) and as plant-growth-promoting agents that can colonize the rhizosphere of several economic crops and thereby increase the crop yield (9, 39, 74, 83). The exceptional metabolic diversity of this organism (which allows it to use, e.g., constituents of crude oils and herbicides as carbon sources) could be put to use in the bioremediation of recalcitrant xenobiotics (8, 28, 54, 57). However, most strains used or under development for biocontrol or bioremediation purposes are taxonomically poorly characterized, and their potential hazard to the CF community is unclear (33, 37, 44, 110).
The taxonomic complexity of B. cepacia-like organisms and the lack of widespread and generally accepted identification schemes hinder sound studies that could establish the roles played by and the pathogenic significance of the different B. cepacia-like organisms. This information is crucial to propose scientifically founded policies for each of the above-mentioned problems. The purpose of this review is to present an overview both of the taxonomy of the B. cepacia complex and of the available phenotypic and genotypic methods aimed at the correct identification of these organisms.
TAXONOMY OF THE B. CEPACIA COMPLEX
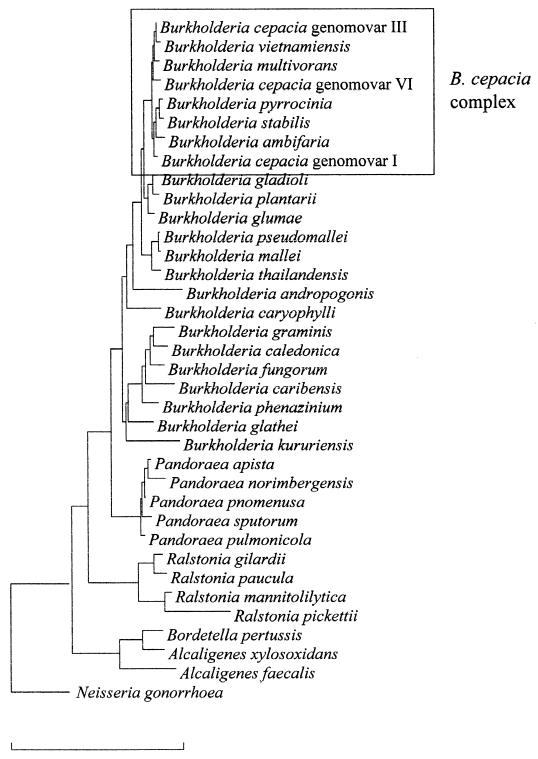
Pseudomonas cepacia was originally described by Burkholder in 1950 as the causative agent of bacterial rot of onion bulbs (13). Other names that were assigned included eugonic oxidizers group 1, Pseudomonas kingii, and Pseudomonas multivorans (49, 77, 97), but several studies clearly showed that these could be considered as synonymous names of P. cepacia and that the name P. cepacia had priority (5, 86, 92, 95). The name P. cepacia was not included in the Approved List of Bacterial Names (93) and therefore lost standing in bacterial nomenclature until 1981, when it was revived by Palleroni and Holmes (81). In 1992, P. cepacia and six other species belonging to rRNA group II of the genus Pseudomonas (Pseudomonas solanacearum, Pseudomonas pickettii, Pseudomonas gladioli, Pseudomonas mallei, Pseudomonas pseudomallei, and Pseudomonas caryophylli) (82) were transferred to the new genus Burkholderia (119). In contrast to the genus Pseudomonas, the genus Burkholderia belongs to the β-subdivision of the phylum Proteobacteria (53). Since the genus name was first assigned, the taxonomy of the genus Burkholderia has undergone considerable changes (Table 1), and the genus now includes 22 validly described species: B. cepacia (the type species), Burkholderia caryophylli, Burkholderia mallei, Burkholderia pseudomallei, Burkholderia gladioli, Burkholderia plantarii, Burkholderia glumae, Burkholderia vietnamiensis, Burkholderia andropogonis, Burkholderia multivorans, Burkholderia glathei, Burkholderia pyrrocinia, Burkholderia thailandensis, Burkholderia graminis, Burkholderia phenazinium, Burkholderia caribensis, Burkholderia kururiensis, Burkholderia ubonensis, Burkholderia caledonica, Burkholderia fungorum, Burkholderia stabilis, and Burkholderia ambifaria (1, 10, 20, 22, 23, 24, 25, 32, 102, 104, 105, 109, 117, 118, 119, 120, 122, 123). A phylogenetic tree based on 16S rRNA gene sequences, showing the positions of all the Burkholderia species and representatives of related genera, is shown in Fig. 1.
TABLE 1.
Overview of the genus Burkholderiaa
| Species name originally assignedb | Burkholderia species name or taxon assigned | Yr of assignment | Reference | Other name subsequently assigned | Yr of assignment | Reference |
|---|---|---|---|---|---|---|
| Pseudomonas cepacia | B. cepacia comb. nov. (B. cepacia genomovar I) | 1992 | 81, 104, 119 | |||
| Pseudomonas solanacearum | B. solanacearum comb. nov. | 1992 | 119 | Ralstonia solanacearum comb. nov. | 1995 | 120 |
| Pseudomonas pickettii | B. picketti comb. nov. | 1992 | 119 | Ralstonia pickettii comb. nov. | 1995 | 120 |
| Pseudomonas gladioli | B. gladioli comb. nov. | 1992 | 119 | |||
| Pseudomonas mallei | B. mallei comb. nov. | 1992 | 119 | |||
| Pseudomonas pseudomallei | B. pseudomallei comb. nov. | 1992 | 119 | |||
| Pseudomonas caryophylli | B. caryophylli comb. nov. | 1992 | 119 | |||
| Pseudomonas plantarii | B. plantarii comb. nov. | 1994 | 102 | |||
| Pseudomonas glumae | B. glumae comb. nov. | 1994 | 102 | |||
| B. vandii sp. nov. | 1994 | 102 | Junior synonym of B. plantarii | 1999 | 22 | |
| B. vietnamiensis sp. nov. (B. cepacia genomovar V) | 1995 | 32, 104 | ||||
| Pseudomonas cocovenenans | B. cocovenenans comb. nov. | 1995 | 123 | Junior synonym of B. gladioli | 1999 | 22 |
| Pseudomonas andropogonis | B. andropogonis comb. nov. | 1995 | 32 | |||
| B. multivorans sp. nov. (B. cepacia genomovar II) | 1997 | 104 | ||||
| Pseudomonas glathei | B. glathei comb. nov. | 1997 | 104 | |||
| Pseudomonas pyrrocinia | B. pyrrocinia comb. nov. | 1997 | 4, 104 | |||
| B. thailandensis sp. nov. | 1998 | 10 | ||||
| B. graminis sp. nov. | 1998 | 109 | ||||
| Pseudomonas phenazinium | B. phenazinium comb. nov. | 1998 | 109 | |||
| B. norimbergensis sp. nov. | 1998 | 117 | Pandoraea norimbergensis comb. nov. | 2000 | 20 | |
| B. caribensis sp. nov. | 1999 | 1 | ||||
| B. stabilis sp. nov. (B. cepacia genomovar IV) | 2000 | 104, 105 | ||||
| B. kururiensis sp. nov. | 2000 | 12 | ||||
| B. ubonensis sp. nov. | 2000 | 118 | ||||
| B. fungorum sp. nov. | 2001 | 23 | ||||
| B. caledonica sp. nov. | 2001 | 23 | ||||
| B. ambifaria sp. nov. (B. cepacia genomovar VII) | 2001 | 25 | ||||
| B. cepacia genomovar III | 1997 | 104 | ||||
| B. cepacia genomovar VI | 2001 | 24 |
Members of the B. cepacia complex are in boldface type.
FIG. 1.
Phylogenetic tree based on 16S rRNA gene sequences, showing the positions of all the Burkholderia species and of representatives of related genera. Bar, 10% sequence dissimilarity.
From the mid-1990s on, several researchers noted that there was a marked heterogeneity among “B. cepacia” strains isolated from different ecological niches. These strains were tentatively classified as “B. cepacia” using a wide range of techniques (7, 15, 32, 91, 100, 101, 121). The heterogeneity among “B. cepacia” isolates made correct identification problematic, and evaluation of the techniques used showed that they were either not very sensitive, not very specific, or neither sensitive nor specific (55, 59, 60, 69, 91, 98). The remarkable diversity among presumed “B. cepacia” strains and the lack of reliable identification schemes led Vandamme et al. (104) to a polyphasic taxonomic study that demonstrated that presumed “B. cepacia” strains isolated from CF patients and other sources belonged to at least five distinct genomic species or genomovars (the term genomovar was introduced to denote phenotypically similar genomic species [103]). B. cepacia genomovar V was identified as the previously described species B. vietnamiensis (32), and the name B. multivorans was proposed for the genomic species formerly known as B. cepacia genomovar II. The remaining groups were referred to as B. cepacia genomovars I, III, and IV. This group of five genomic species was collectively referred to as the B. cepacia complex. Since B. cepacia genomovar I contains the type strain, it retains the formal binomial name B. cepacia. Following a thorough investigation of the phenotypic and genotypic characteristics of B. cepacia genomovar IV strains (105), it became obvious that this organism could be differentiated from all other members of the B. cepacia complex, and it was formally classified as B. stabilis. Subsequent polyphasic taxonomic studies identified two more members of the B. cepacia complex (24, 25). B. cepacia genomovar VI contains strains isolated from CF patients in the United States and the United Kingdom. This organism can phenotypically be differentiated from all members of the B. cepacia complex except B. multivorans. The name B. ambifaria (B. cepacia genomovar VII) was proposed for isolates from human clinical and environmental specimens, including CF patients. B. ambifaria also contains several well-characterized biocontrol strains. In addition, it was recently shown that the species B. pyrrocinia also belongs to the B. cepacia complex (4).
Within the B. cepacia complex, representatives of different species generally have DNA-DNA hybridization values between 30 and 60%, while values obtained from strains belonging to the same species are generally higher than 70%. DNA-DNA binding values obtained with other Burkholderia species are generally below 30% (22, 24, 25, 26, 32, 104). These values correspond to the three categories described in reference 106: high DNA relatedness (70% or higher) between strains of a single species, low but significant DNA relatedness below the species level, and nonsignificant DNA relatedness (30% or less). In addition, the similarities between 16S ribosomal DNA (rDNA) sequences obtained from different members of the B. cepacia complex are higher (>97.7%) than similarities between such sequences and those of other Burkholderia species (<97.0%) (Fig. 1).
IDENTIFICATION OF B. CEPACIA COMPLEX ORGANISMS
Introduction.
The identification of organisms cultured from respiratory specimens obtained from CF patients is not straightforward. Using commercial systems, members of the B. cepacia complex have been misidentified as (among others) B. gladioli, R. pickettii, Alcaligenes spp., Pseudomonas spp., S. maltophilia, Flavobacterium spp., and Chryseobacterium spp., and strains of these various species have likewise been misidentified as belonging to the B. cepacia complex (55, 75). Methods for the identification of B. cepacia-like organisms must be capable of accurately identifying such a diverse variety of gram-negative nonfermenters, both distinguishing them from the B. cepacia complex and identifying the individual members of the B. cepacia complex. In addition, these methods should be relatively quick and easy to perform, given the clinical relevance of these organisms and the relatively large number of isolates involved (for example, the Cystic Fibrosis Foundation [CFF] Burkholderia cepacia Research Laboratory and Repository receives on average 750 B. cepacia-like isolates per year [J. J. LiPuma, Int. Burkholderia cepacia Working Group Abstr. 6th Annu. Meet., 2001 {Online}]).
Phenotypical tests.
In routine clinical laboratories, the identification of putative B. cepacia complex isolates is generally performed using a combination of selective media, conventional biochemical analysis, and/or commercial systems (89, 108). Several different media have been developed for the selective isolation of B. cepacia complex isolates from sputum of CF patients. These media include P. cepacia medium (PC agar) (containing 300 U of polymyxin B per ml and 100 μg of ticarcilline per ml) (31); oxidation-fermentation agar supplemented with lactose, 300 U of polymyxin B per ml, and 0.2 U of bacitracin per ml (OFPBL agar) (113); and B. cepacia selective agar (BCSA) (containing 1% lactose and 1% sucrose in an enriched base of casein and yeast extract with 600 U of polymyxin B per ml, 10 μg of gentamicin per ml, and 2.5 μg of vancomycin per ml) (40). BCSA was reported to be superior to OFPBL and PCA in terms of rapidity (100% recovery following 72 h of incubation) and quality (70% of isolates showed good growth following 72 h of incubation) of recovery of B. cepacia complex organisms from CF respiratory specimens and inhibition of other organisms (41). Organisms not belonging to the B. cepacia complex that are capable of growth on BCSA include B. gladioli and Ralstonia spp. (41). The sensitivity and specificity of some or all of the above-mentioned media for the isolation of environmental “B. cepacia” isolates may be much lower (17), and therefore the use of other media, like PCAT medium (containing azelaic acid and tryptamine) (12) or TB-T medium (containing glucose, asparagine, trypan blue, and tetracycline) (38) may be recommended (4, 109).
There are several reports that describe the failure of most commercial test systems to identify B. cepacia complex isolates with sufficient sensitivity and specificity, with isolates commonly misidentified as B. gladioli, S. maltophilia, or Ralstonia spp. (55, 75, 89). Commercial test systems with relatively high positive predictive values (including the Vitek GNI Plus and Remel Uni-N/F Tek Plate and N/F Screen [89]) are available, but there is nevertheless a general consensus that bacterial isolates presumptively identified as belonging to the B. cepacia complex on the basis of commercial test system results should be tested for growth on BCSA, presence of lysine and ornithine decarboxylase activity, oxidation of sucrose and adonitol, presence of oxidase activity, hemolysis, pigment production, and growth at 42°C (42, 55, 75, 89, 108).
There are several phenotypic tests that allow the separation of B. gladioli, Pandoraea species, R. pickettii, A. xylosoxidans, and S. maltophilia from the B. cepacia complex (Table 2), and some of the members of the B. cepacia complex can be identified to the species or genomovar level based on phenotype. However, given the phenotypic variation that can occur within species and the frequent discrepancies between results obtained with different methodologies, the identification of B. cepacia complex based on phenotypic analysis alone should be confirmed by a reference laboratory equipped to provide more complete analyses (42). Consideration should also be given to the use of reference labs for any gram-negative nonfermenter for which species identification remains equivocal after phenotypic analysis.
TABLE 2.
Characteristics useful for the differentiation of members of the B. cepacia complex, B. gladioli, Pandoraea spp., R. pickettii, A. xylosoxidans, and S. maltophiliaa
| Result forb:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
B. cepacia
|
B. gladioli | Pandoraea species | R. pickettii | A. xylosoxidans | S. maltophilia | |||||||
| Genomovar I | Genomovar IIc | Genomovar III | Genomovar IVd | Genomovar Ve | Genomovar VI | Genomovar VIIf | ||||||
| Oxidase | + | + | + | + | + | + | + | − | v | + | + | − |
| Oxidation of: | ||||||||||||
| Sucrose | v | − | v | − | + | − | + | − | − | − | − | v |
| Adonitol | v | + | v | v | − | + | + | + | − | − | − | − |
| Lactose | v | + | v | + | + | + | + | − | − | − | − | + |
| Lysine decarboxylase | + | v | + | + | + | − | + | − | − | − | − | + |
| Ornithine decarboxylase | v | − | v | + | − | − | − | − | − | − | − | − |
| Gelatine liquefaction | v | − | v | + | − | − | + | v | − | − | − | + |
| Aesculine hydrolysis | v | − | v | − | − | − | v | v | − | v | − | + |
| β-Galactosidase activity | + | + | + | − | + | + | + | + | − | − | − | + |
| Growth at 42°C | v | + | v | − | + | + | v | − | v | v | NK | v |
| β-Hemolysis | − | − | − | − | v | − | v | − | − | − | NK | NK |
Data for members of the B. cepacia complex, B. gladioli, and R. pickettii from reference 42; data for Pandoraea spp. from reference 20; data for A. xylosoxidans from references 52 and 111; and data for S. maltophilia from references 27, 80, and 111.
+, 90% or more of the strains investigated yielded a positive reaction; −, 10% or fewer of the strains investigated yielded a positive reaction; v, between 10 and 90% of the strains investigated yielded a positive reaction; NK, not known. The numbers of strains investigated were 23 (B. cepacia genomovar I), 109 (B. multivorans), 139 (B. cepacia genomovar III), 27 (B. stabilis), 36 (B. vietnamiensis), 9 (B. cepacia genomovar VI), 18 (B. ambifaria), 27 (B. gladioli), 32 (Pandoraea spp.), and 12 (R. pickettii). The numbers of A. xylosoxidans and S. maltophilia strains investigated are not known.
B. multivorans.
B. stabilis.
B. vietnamiensis.
B. ambifaria.
Whole-cell protein analysis.
Data presented by Vandamme et al. (104) indicated that sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell proteins was a suitable technique for the identification of members of the B. cepacia complex. However, the comparison of the identification results obtained by this method with those obtained by other identification approaches revealed several discrepancies and a poor discrimination between B. cepacia genomovars I and III, B. stabilis, and B. ambifaria was noted. The advantages of this technique are its applicability to a wide range of organisms, the fact that little prior knowledge regarding the isolate is required, and its relative simplicity. A drawback of this method for the identification of B. cepacia-like isolates is that the whole-cell protein patterns are often characterized by a distortion of part of the banding pattern. These distortions significantly influence the correlation level between the protein patterns. Therefore, it is essential to compare the result of the numerical analysis of the protein patterns with the profiles themselves in order to delineate the clusters (21, 24, 25, 104). SDS-PAGE of whole-cell proteins remains a valuable tool for the identification of B. cepacia complex and B. cepacia-like isolates in the research setting, where experienced personnel are present for the interpretation of the protein profiles; the above-mentioned shortcomings, however, render it unsuitable for use in the clinical setting.
AFLP fingerprinting.
In the past decade, various nucleic acid sequence-based methods have been developed for the identification and typing of bacterial pathogens (73, 79). One of these methods is amplified fragment length polymorphism (AFLP) fingerprinting, a fingerprinting technique based on the selective PCR amplification of genomic restriction fragments. This method combines broad applicability with high reproducibility and discriminatory power (45, 48, 85, 87). Other data (20, 24, 25, 26) indicate that AFLP fingerprinting is a technique that can be used for the identification of members of the B. cepacia complex and other B. cepacia-like bacteria. However, the method is technically demanding and labor-intensive and radioactive formats are impractical for clinical use (79). Significant progress has been made with the fluorescent format (26, 29, 56), but the high setup costs associated with the purchase of a DNA sequencer may be prohibitive for most laboratories. The high reproducibility of the banding patterns for a given strain facilitates database construction and use of such a database for identifying new bacterial strains. However, the presence of high-intensity bands in the patterns of some strains and the intermediate taxonomic position of several strains (as revealed by DNA-DNA hybridization) may ultimately require additional testing before some strains can be conclusively identified, again making this method unsuitable for application in routine diagnostic microbiology laboratories. It is, however, a valuable tool in taxonomic studies and a welcome addition to SDS-PAGE of whole-cell proteins for the identification of organisms easily misidentified by the latter method.
Whole-cell fatty acid analysis.
The high degree of automation, the relative simplicity, and the fairly low costs associated with whole-cell fatty acid analysis make it a valuable technique for rapid identification of isolates in clinical laboratories (112). However, Vandamme et al. (104) reported the failure of whole-cell fatty acid analysis to distinguish between the first five known species of the B. cepacia complex, and more-recent data (105) confirmed this conclusion. It was also shown that fatty acid analysis cannot differentiate members of the B. cepacia complex from B. gladioli (116; Clode, F. E., A. Louise, L. Metherel, and T. L. Pitt, Letter, Am. J. Respir. Crit. Care Med. 160:374–375, 1999; M. Wilsher, J. Kolbe, A. J. Morris, D. F. Welch, and P. A. R. Vandamme, Authors' Reply to Letter, Am. J. Resp. Crit. Care Med. 160:374–375, 1999). From the comparison of published data, it is obvious that there are qualitative and quantitative differences in the fatty acid composition of members of the B. cepacia complex and other B. cepacia-like species, like Pandoraea spp. (20) and Ralstonia spp. (21), but considering standard deviations, it seems questionable whether these differences will suffice to identify all new isolates to the species level. Therefore, all organisms identified by whole-cell fatty acid analysis as belonging to the B. cepacia complex, B. gladioli, the genus Pandoraea, or the genus Ralstonia should be further investigated with methods more suitable for identification of B. cepacia-like isolates to the species level. A main advantage of this technique is the existence of a commercial database (Microbial ID) for identification of isolates that allows the rapid separation of B. cepacia complex organisms and related organisms both from other gram-negative nonfermenters (like P. aeruginosa and S. maltophilia) and from Enterobacteriaceae. The technique can also be used to assign isolates that cannot be classified with other screening methods to a major phylogenetic lineage.
PCR-based identification.
Several candidate PCR assays aimed at the identification of “B. cepacia” have been described previously (16, 51, 78, 101) but most of these assays were developed before the recognition that the B. cepacia complex consists of several species. In addition, most relied on published DNA sequence data derived from analyses of culture collection strains that, in retrospect, are poorly representative of the total diversity within the B. cepacia complex. Most of the studies regarding PCR-based identification of members of the B. cepacia complex that have been carried out so far have been based on the diversity within the nucleotide sequences of the 16S and/or 23S rDNAs and were either aimed at the development of species- and/or genomovar-specific primers or RFLP analysis of the PCR-amplified 16S rRNA gene (6, 11, 24, 25, 65, 70, 88, 114, 115). The results from these studies clearly indicate that B. multivorans, B. vietnamiensis, and B. cepacia genomovar VI each can be separated from all other members of the B. cepacia complex. B. cepacia genomovars I and III, B. stabilis, B. ambifaria, and B. pyrrocinia can be identified as a group, but the variation within the rRNA operon is obviously too small to separate all members of the B. cepacia complex, and because of this discriminatory limitation, Mahenthiralingam et al. (70) developed a novel PCR-based identification assay based on the recA gene. The recA gene shows 94 to 95% similarity between the different genomovars, and typically 98 to 99% similarity can be found within the genomovars. However, B. cepacia genomovar I and III each contain two subpopulations with a different recA allele. At the moment of this writing, recA gene-derived primer pairs are available for the identification of B. cepacia genomovar I, B. cepacia genomovar III, B. multivorans, B. stabilis, B. vietnamiensis, and B. ambifaria (no primers are available yet for B. pyrrocinia or B. cepacia genomovar VI) (25, 70). In addition to recA gene-derived species-specific primers, a recA gene-based RFLP approach, enabling the recognition of multiple types within each genomovar, was developed (70).
The development of these novel molecular tools has provided the scientific community with quick, easy, and scientifically sound ways of identifying individual strains belonging to this taxonomically complex group of organisms. The disadvantages of the PCR-based methods include the need for appropriate measures to avoid cross-contamination (including the use of negative controls and the use of different areas for PCR manipulations) and the fact that PCR primers are not available for all B. cepacia-like organisms (e.g., no published primers are available yet for the identification of Pandoraea or Ralstonia species). In addition, care should be taken in the interpretation of negative PCR results (i.e., in distinguishing between true- and false-negative results), and in general it can be stated that laboratories engaging in PCR-based identification of B. cepacia complex organisms should be appropriately equipped at the technical level and should comply with stringent quality control requirements to exclude misidentifications (46).
B. cepacia experimental strain panel.
Recently, a panel of 30 well-characterized strains representative of B. cepacia genomovars I and III, B. stabilis, B. multivorans, and B. vietnamiensis was assembled (71). The main reason for the assembly of this panel was that identification, epidemiological, and virulence studies all would benefit from the use of a defined set of representative strains. Since the assembly of the panel, several new taxa belonging to the B. cepacia complex have been described, and representative strains of these new taxa will have to be included in an updated version of the experimental strain panel.
CONCLUSIONS
It can be concluded that most of the methods necessary to identify B. cepacia-like organisms are available. The choice of what identification tools to use depends on their availability and the mission of the laboratory involved. In the research laboratory, a polyphasic approach (aimed at the integration of different kinds of data and information) (106) seems appropriate. Firstly, isolates should be assigned to a major phylogenetic group (such as the B. cepacia complex or the genus Pandoraea) using SDS-PAGE, whole-cell fatty acid analysis, or 16S rDNA sequence analysis. In addition, members of the B. cepacia complex that cannot unequivocally be identified to the genomovar level should be included in complementary screening methods like RFLP fingerprinting of the recA gene and/or AFLP fingerprinting. The identity of strains can then be confirmed using recA gene-based PCR assays or 16S rDNA RFLP fingerprinting. The mission and therefore the challenge posed by the identification of B. cepacia-like organisms for routine clinical microbiology laboratories is different. Strains isolated on selective media and tentatively identified as belonging to the B. cepacia complex using commercial systems should be confirmed with the classical biochemical tests described. The present state of the art indicates that isolates that are considered to be putative members of the B. cepacia complex after additional testing should be further examined by the genotypic methods discussed above. Laboratories equipped to augment routine evaluation with genotypic analyses have been established (e.g., the CFF Burkholderia cepacia Research Laboratory and Repository, as well as the Canadian Burkholderia cepacia Research and Referral Repository [for more information, please see the website http://go.to/cepacia]). The development of additional PCR-based identification systems and their wider use will have an important impact on studies that seek to elucidate the epidemiology and natural history of human infections due to B. cepacia-like organisms.
The early detection of B. cepacia complex and B. cepacia-like bacteria is extremely important both for the CF patient as well as for the CF community. However, a recent study (90) indicated that less than half of U.S. centers surveyed employ “B. cepacia”-specific selective media or incubate cultures for extended periods, both of which improve the yield of this organism. The use of these up-to-date culture techniques is technically not demanding and should be the expected standard of care in every CF center worldwide. Continuing education with regard to this issue is crucial. Apart from detection, correct identification of B. cepacia-like bacteria is extremely important. Therefore, priority should be given to the continuous evaluation of existing PCR-based methods (and other methods used for identification) with a view to keeping them up-to-date with respect to the increasing biodiversity found within the B. cepacia complex. It will also be useful to develop alternative PCR-based identification assays and expand the existing assays to related taxa like R. pickettii and Pandoraea spp. The use of up-to-date laboratory techniques for the proper detection and identification of B. cepacia complex organisms in respiratory cultures of CF patients will be beneficial to patients and CF centers, enhance the accuracy of national CF registries, and provide the basis for further studies. The improved diagnosis of infections caused by members of the B. cepacia complex and other B. cepacia-like organisms will help with the interpretation of the results from clinical outcome studies, and by doing so, will provide crucial information regarding the pathogenicity and/or transmissibility of specific strains involved.
ACKNOWLEDGMENTS
We thank the U.S. Cystic Fibrosis Foundation and the United Kingdom Cystic Fibrosis Trust for their continuing support. T.C. gratefully acknowledges support received from the Caroll Haas Research Fund in Cystic Fibrosis.
REFERENCES
- 1.Achouak W, Christen R, Barakat M, Martel M-H, Heulin T. Burkholderia caribensis sp. nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int J Syst Bacteriol. 1999;49:787–794. doi: 10.1099/00207713-49-2-787. [DOI] [PubMed] [Google Scholar]
- 2.Ahlin A, De Boer M, Roos D, Leusen J, Smith C I, Sundin U, Rabbani H, Palmblad J, Elinder G. Prevalence, genetics and clinical presentation of chronic granulomatous disease in Sweden. Acta Paediatr. 1995;84:1386–1394. doi: 10.1111/j.1651-2227.1995.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 3.Aoun M, Van der Auwera P, Devleeshouwer C, Daneau D, Seraj N, Meunier F, Gerain J. Bacteraemia caused by non-aeruginosa Pseudomonas species in a cancer centre. J Hosp Infect. 1992;22:307–316. doi: 10.1016/0195-6701(92)90016-f. [DOI] [PubMed] [Google Scholar]
- 4.Balandreau J, Viallard V, Cournoyer B, Coenye T, Laevens S, Vandamme P. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl Environ Microbiol. 2001;67:982–985. doi: 10.1128/AEM.67.2.982-985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballard R W, Palleroni N J, Doudoroff M, Stanier R Y. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970;60:199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- 6.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J Clin Microbiol. 1999;37:1335–1339. doi: 10.1128/jcm.37.5.1335-1339.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bevivino A, Tabacchioni S, Chiarini L, Carusi M V, Del Gallo M, Visca P. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology. 1994;140:1069–1077. doi: 10.1099/13500872-140-5-1069. [DOI] [PubMed] [Google Scholar]
- 8.Bhat M A, Tsuda M, Horiike K, Nozaki M, Vaidyanathan C S, Nakazawa T. Identification and characterization of a new plasmid carrying genes for degradation of 2:4-dichlorophenoxyacetate from Pseudomonas cepacia CSV90. Appl Environ Microbiol. 1994;60:307–312. doi: 10.1128/aem.60.1.307-312.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowers J, Parke J. Epidemiology of Pythium damping-off and Aphanomyces root rot of peas after seed treatment with bacterial agents for biological control. Phytopathology. 1993;83:1466–1473. [Google Scholar]
- 10.Brett P J, DeShazer D, Woods D E. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 11.Brisse S, Verduin C M, Milatovic D, Fluit A, Verhoef J, Laevens S, Vandamme P, Tummler B, Verbrugh H A, van Belkum A. Distinguishing species of the Burkholderia cepacia complex and Burkholderia gladioli by automated ribotyping. J Clin Microbiol. 2000;38:1876–1884. doi: 10.1128/jcm.38.5.1876-1884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burbage D A, Sasser M. A medium selective for Pseudomonas cepacia. Phytopathology. 1982;72:706. [Google Scholar]
- 13.Burkholder W H. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;40:115–117. [Google Scholar]
- 14.Burns J L, Emerson J, Stapp J R, Yim D L, Krzewinski J, Louden L, Ramsey B W, Clausen C R. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- 15.Butler S L, Doherty C J, Hughes J E, Nelson J W, Govan J R W. Burkholderia cepacia and cystic fibrosis: do natural environments present a potential hazard? J Clin Microbiol. 1995;33:1001–1004. doi: 10.1128/jcm.33.4.1001-1004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell P W, III, Phillips J A, III, Heidecker G J, Krishnamani M R S, Zahorchak R, Stull T L. Detection of Pseudomonas (Burkholderia) cepacia using PCR. Pediatr Pulmonol. 1995;20:44–49. doi: 10.1002/ppul.1950200109. [DOI] [PubMed] [Google Scholar]
- 17.Carson L R, Tablan O C, Cusick L B, Jarvis W R, Favero M S, Bland L A. Comparative evaluation of selective media for isolation of Pseudomonas cepacia from cystic fibrosis patients and environmental sources. J Clin Microbiol. 1988;26:2096–2100. doi: 10.1128/jcm.26.10.2096-2100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaparro C, Maurer J, Gutierrez C, Krajden M, Chan C, Winston T, Keshavjee S, Scavuzzo M, Tullis E, Hutcheon M, Kesten S. Infection with Burkholderia cepacia in cystic fibrosis: outcome following lung transplantation. Am J Respir Crit Care Med. 2001;163:43–48. doi: 10.1164/ajrccm.163.1.9811076. [DOI] [PubMed] [Google Scholar]
- 19.Christenson J C, Welch D F, Mukwaya G, Muszynsky M J, Weaver R E, Brenner D J. Recovery of Pseudomonas gladioli from respiratory tract specimens of patients with cystic fibrosis. J Clin Microbiol. 1989;27:270–273. doi: 10.1128/jcm.27.2.270-273.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coenye T, Falsen E, Hoste B, Ohlén M, Goris J, Govan J R W, Gillis M, Vandamme P. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pmomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Microbiol. 2000;50:887–899. doi: 10.1099/00207713-50-2-887. [DOI] [PubMed] [Google Scholar]
- 21.Coenye T, Falsen E, Vancanneyt M, Hoste B, Govan J R W, Kersters K, Vandamme P. Classification of some Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int J Syst Bacteriol. 1999;49:405–413. doi: 10.1099/00207713-49-2-405. [DOI] [PubMed] [Google Scholar]
- 22.Coenye T, Holmes B, Kersters K, Govan J R W, Vandamme P. Burkholderia cocovenenans (van Damme et al. 1960) Gillis et al. 1995 and Burkholderia vandii Urakami et al. 1994 are junior subjective synonyms of Burkholderia gladioli (Severini 1913) Yabuuchi et al. 1993 and Burkholderia plantarii (Azegami et al. 1987) Urakami et al. 1994, respectively. Int J Syst Bacteriol. 1999;49:37–42. doi: 10.1099/00207713-49-1-37. [DOI] [PubMed] [Google Scholar]
- 23.Coenye T, Laevens S, Willems A, Ohlén M, Hannant W, Govan J R W, Gillis M, Falsen E, Vandamme P. Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int J Syst Evol Microbiol. 2001;51:1099–1107. doi: 10.1099/00207713-51-3-1099. [DOI] [PubMed] [Google Scholar]
- 24.Coenye T, LiPuma J J, Henry D, Hoste B, Vandemeulebroucke K, Gillis M, Speert D P, Vandamme P. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int J Syst Evol Microbiol. 2001;51:271–279. doi: 10.1099/00207713-51-2-271. [DOI] [PubMed] [Google Scholar]
- 25.Coenye T, Mahenthiralingam E, Henry D, LiPuma J J, Laevens S, Gillis M, Speert D P, Vandamme P. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int J Syst Evol Microbiol. 2001;51:1481–1490. doi: 10.1099/00207713-51-4-1481. [DOI] [PubMed] [Google Scholar]
- 26.Coenye T, Schouls L M, Govan J R W, Kersters K, Vandamme P. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int J Syst Bacteriol. 1999;49:1657–1666. doi: 10.1099/00207713-49-4-1657. [DOI] [PubMed] [Google Scholar]
- 27.Denton M, Kerr K G. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11:57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folsom B R, Chapman P J, Pritchard P H. Phenol and trichloroethylene degradation by Pseudomonas cepacia G4: kinetics and interactions between substrates. Appl Environ Microbiol. 1990;56:1279–1285. doi: 10.1128/aem.56.5.1279-1285.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gancheva A, Pot B, Vanhonacker K, Hoste B, Kersters K. A polyphasic approach towards the identification of strains belonging to Lactobacillus acidophilus and related species. Syst Appl Microbiol. 1999;22:573–585. doi: 10.1016/S0723-2020(99)80011-3. [DOI] [PubMed] [Google Scholar]
- 30.Gilligan P H. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilligan P H, Gage P A, Bradshaw L M, Schidlow D V, DeCicco B T. Isolation medium for the recovery of Pseudomonas cepacia from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 1985;22:5–8. doi: 10.1128/jcm.22.1.5-8.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillis M, Van Van T, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 33.Govan J R W, Balandreau J, Vandamme P. Burkholderia cepacia—friend and foe. ASM News. 2000;66:124–125. [Google Scholar]
- 34.Govan J R W, Brown P H, Maddison J, Doherty C, Nelson C J, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis patients. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 35.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 37.Govan J R W, Vandamme P. Agricultural and medical microbiology : a time for bridging gaps. Microbiology. 1998;144:2373–2375. doi: 10.1099/00221287-144-9-2373. [DOI] [PubMed] [Google Scholar]
- 38.Hagedorn C, Gould W D, Bardinelli T R, Gustavson D R. A selective medium for enumeration and recovery of Pseudomonas cepacia biotypes from soil. Appl Environ Microbiol. 1987;53:2265–2268. doi: 10.1128/aem.53.9.2265-2268.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hebbar P K, Martel M H, Heulin T. Suppression of pre- and postemergence damping-off in corn by Burkholderia cepacia. Eur J Plant Pathol. 1998;104:29–36. [Google Scholar]
- 40.Henry D A, Campbell M E, LiPuma J J, Speert D P. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry D, Campbell M, McGimpsey C, Clarke A, Louden L, Burns J L, Roe M H, Vandamme P, Speert D. Comparison of isolation media for recovery of Burkholderia cepacia complex from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 1999;37:1004–1007. doi: 10.1128/jcm.37.4.1004-1007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henry D A, Mahenthiralingam E, Vandamme P, Coenye T, Speert D P. Biochemical and molecular approaches for determining genomovar status of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:1073–1078. doi: 10.1128/JCM.39.3.1073-1078.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hobson R, Gould I, Govan J. Burkholderia (Pseudomonas) cepacia as a cause of brain abscesses secondary to chronic suppurative otitis media. Eur J Clin Microbiol Infect Dis. 1995;14:908–911. doi: 10.1007/BF01691499. [DOI] [PubMed] [Google Scholar]
- 44.Holmes A, Govan J, Goldstein R. Agricultural use of Burkholderia (Pseudomonas) cepacia: a threat to human health? Emerg Infect Dis. 1998;4:221–227. doi: 10.3201/eid0402.980209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huys G, Coopman R, Janssen P, Kersters K. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int J Syst Bacteriol. 1996;46:572–580. doi: 10.1099/00207713-46-2-572. [DOI] [PubMed] [Google Scholar]
- 46.Ieven M, Goossens H. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin Microbiol Rev. 1997;10:242–256. doi: 10.1128/cmr.10.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 48.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 49.Jonsson V. Proposal of a new species Pseudomonas kingii. Int J Syst Bacteriol. 1970;20:255–257. [Google Scholar]
- 50.Kaitwatcharachai C, Silpapojakul K, Jitsurong S, Kalnauwakul S. An outbreak of Burkholderia cepacia bacteremia in hemodialysis patients: an epidemiologic and molecular study. Am J Kidney Dis. 2000;36:199–204. doi: 10.1053/ajkd.2000.8295. [DOI] [PubMed] [Google Scholar]
- 51.Karpati F, Jonasson J. Polymerase chain reaction for the detection of Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia in sputum of patients with cystic fibrosis. Mol Cell Probes. 1996;10:397–403. doi: 10.1006/mcpr.1996.0055. [DOI] [PubMed] [Google Scholar]
- 52.Kersters K, De Ley J. Genus Alcaligenes. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Wilkins; 1984. [Google Scholar]
- 53.Kersters K, Ludwig W, Vancanneyt M, De Vos P, Gillis M, Schleifer K H. Recent changes in the classification of the pseudomonads: an overview. Syst Appl Microbiol. 1996;19:465–477. [Google Scholar]
- 54.Kilbane J J, Chatterjee D K, Karns J S, Kellogg S T, Chakrabarty A M. Biodegradation of 2:4,5-trichlorophenoxyacetic acid by a pure culture of Pseudomonas cepacia. Appl Environ Microbiol. 1982;44:72–78. doi: 10.1128/aem.44.1.72-78.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiska D L, Kerr A, Jones M C, Caracciolo J A, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan P. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative, nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–891. doi: 10.1128/jcm.34.4.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kokotovic B, On S L W. High-resolution genomic fingerprinting of Campylobacter jejuni and Campylobacter coli by analysis of amplified fragment length polymorphisms. FEMS Microbiol Lett. 1999;173:77–84. doi: 10.1111/j.1574-6968.1999.tb13487.x. [DOI] [PubMed] [Google Scholar]
- 57.Krumme M L, Timmis K N, Dwyer D F. Degradation of trichloroethylene by Pseudomonas cepacia G4 and the constitutive mutant strain G4 5223 PR1 in aquifer microcosms. Appl Environ Microbiol. 1993;59:2746–2749. doi: 10.1128/aem.59.8.2746-2749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lacy D E, Spencer D A, Goldstein A, Weller P H, Darbyshire P. Chronic granulomatous disease presenting in childhood with Pseudomonas cepacia septicaemia. J Infect. 1993;27:301–304. doi: 10.1016/0163-4453(93)92271-w. [DOI] [PubMed] [Google Scholar]
- 59.Larsen G Y, Stull T L, Burns J L. Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. J Clin Microbiol. 1993;31:788–792. doi: 10.1128/jcm.31.4.788-792.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leff L G, Kernan R M, McArthur J V, Shimkets L J. Identification of aquatic Burkholderia (Pseudomonas) cepacia by hybridization with species-specific rRNA gene probes. Appl Environ Microbiol. 1995;61:1634–1636. doi: 10.1128/aem.61.4.1634-1636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liou T G, Adler F R, FitzSimmons S C, Cahill B C, Hibbs J R, Marshall B C. Predictive 5-year survivorship model of cystic fibrosis. Am J Epidemiol. 2001;153:345–352. doi: 10.1093/aje/153.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.LiPuma J J. Burkholderia cepacia: management issues and new insights. Clin Chest Med. 1998;19:473–486. doi: 10.1016/s0272-5231(05)70094-0. [DOI] [PubMed] [Google Scholar]
- 63.LiPuma J J. Burkholderia cepacia epidemiology and pathogenesis: implications for infection control. Curr Opin Pulm Med. 1998;4:337–441. doi: 10.1097/00063198-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 64.LiPuma J J, Dasen S E, Nielson D W, Stern R C, Stull T L. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet. 1990;336:1094–1096. doi: 10.1016/0140-6736(90)92571-x. [DOI] [PubMed] [Google Scholar]
- 65.LiPuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LiPuma J J, Mahenthiralingam E. Commercial use of Burkholderia cepacia. Emerg Infect Dis. 1999;5:305–306. doi: 10.3201/eid0502.990226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.LiPuma J J, Marks-Austin K A, Holsclaw D S, Winnie G B, Gilligan P H, Stull T S. Inapparent transmission of Pseudomonas (Burkholderia) cepacia among patients with cystic fibrosis. Pediatr Infect Dis J. 1994;13:716–719. doi: 10.1097/00006454-199408000-00007. [DOI] [PubMed] [Google Scholar]
- 68.LiPuma J J, Spilker T, Gill L H, Campbell III P W, Liu L, Mahenthiralingam E. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am J Resp Crit Care Med. 2001;164:92–96. doi: 10.1164/ajrccm.164.1.2011153. [DOI] [PubMed] [Google Scholar]
- 69.Liu P-Y F, Dhi Z-Y, Lau Y-J, Hu B-S, Shyr J-M, Tsai W-S, Lin Y-H, Tseng C-Y. Comparison of different PCR approaches for characterization of Burkholderia (Pseudomonas) cepacia isolates. J Clin Microbiol. 1995;33:3304–3307. doi: 10.1128/jcm.33.12.3304-3307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mahenthiralingam E, Bischof J, Byrne S K, Radomski C, Davies J E, Av-Gay Y, Vandamme P. DNA-based diagnostic approaches for the identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38:3165–3173. doi: 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahenthiralingam E, Coenye T, Chung J W, Speert D P, Govan J R W, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mardiney M, Jackson S H, Spratt S K, Li F, Holland S M, Malech H L. Enhanced host defense after gene transfer in the murine p47phox-deficient model of chronic granulomatous disease. Blood. 1997;89:2268–2275. [PubMed] [Google Scholar]
- 73.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology : application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 74.McLoughlin T J, Quinn J P, Bettermann A, Bookland R. Pseudomonas cepacia suppression of sunflower wilt fungus and role of antifungal compounds in controlling the disease. Appl Environ Microbiol. 1992;58:1760–1763. doi: 10.1128/aem.58.5.1760-1763.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McMenamin J D, Zaccone T M, Coenye T, Vandamme P, LiPuma J J. Misidentification of Burkholderia cepacia in U.S. cystic fibrosis treatment centers: an analysis of 1051 recent sputum isolates. Chest. 2000;117:1661–1665. doi: 10.1378/chest.117.6.1661. [DOI] [PubMed] [Google Scholar]
- 76.Millar-Jones L, Paull A, Saunders Z, Goodchild M C. Transmission of Pseudomonas cepacia among cystic fibrosis patients. Lancet. 1992;340:491. doi: 10.1016/0140-6736(92)91817-r. [DOI] [PubMed] [Google Scholar]
- 77.Morris M B, Roberts J B. A group of pseudomonads able to synthesize poly-β-hydroxybutyric acid. Nature. 1959;183:1538. doi: 10.1038/1831538a0. [DOI] [PubMed] [Google Scholar]
- 78.O'Callaghan E M, Tanner M S, Boulnois G J. Development of a PCR probe test for identifying Pseudomonas aeruginosa and Pseudomonas (Burkholderia) cepacia. J Clin Pathol. 1994;47:222–226. doi: 10.1136/jcp.47.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palleroni N J. Genus I. Pseudomonas Migula 1894, 237AL (nom. cons. opin. 5, Jud. Comm. 1952, 237) In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Wilkins; 1984. p. 141. [Google Scholar]
- 81.Palleroni N J, Holmes B. Pseudomonas cepacia, sp. nov., nom. rev. Int J Syst Bacteriol. 1981;31:479–481. [Google Scholar]
- 82.Palleroni N J, Kunisawa R, Contopoulo R, Doudoroff M. Nucleic acid homologies in the genus Pseudomonas. Int J Syst Bacteriol. 1973;23:333–339. [Google Scholar]
- 83.Parke J L, Rand R E, Joy A E, King E B. Biological control of Pythium damping-off and Aphanomyces root rot of peas by application of Pseudomonas cepacia or P. fluorescens to seed. Plant Dis. 1991;75:987–992. [Google Scholar]
- 84.Pegues D A, Carson L A, Tablan O C, FitzSimmons S C, Roman S B, Miller J M, Jarvis W R the Summer Camp Study Group. Acquisition of Pseudomonas cepacia at summer camps for patients with cystic fibrosis. J Pediatr. 1994;124:694–702. doi: 10.1016/s0022-3476(05)81357-5. [DOI] [PubMed] [Google Scholar]
- 85.Rademaker J L W, Hoste B, Louws F J, Kersters K, Swings J, Vauterin L, Vauterin P, de Bruijn F J. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol. 2000;50:665–677. doi: 10.1099/00207713-50-2-665. [DOI] [PubMed] [Google Scholar]
- 86.Samuels S B, Moss C W, Weaver R E. The fatty acids of Pseudomonas multivorans (Pseudomonas cepacia) and Pseudomonas kingii. J Gen Microbiol. 1973;74:275–279. doi: 10.1099/00221287-74-2-275. [DOI] [PubMed] [Google Scholar]
- 87.Savelkoul P H M, Aarts J J M, De Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker J L W, Schouls L M, Lenstra J A. Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol. 1999;37:3083–3091. doi: 10.1128/jcm.37.10.3083-3091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shelly D B, Spilker T, Gracely E J, Coenye T, Vandamme P, LiPuma J J. Utility of commercial systems for identification of Burkholderia cepacia complex from cystic fibrosis sputum culture. J Clin Microbiol. 2000;38:3112–3115. doi: 10.1128/jcm.38.8.3112-3115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shreve M R, Butler S, Kaplowitz H J, Rabin H R, Stokes D, Light M, Regelmann W E for North American Scientific Advisory Group and Investigators for the Epidemiologic Study of Cystic Fibrosis. Impact of microbiology practice on cumulative prevalence of respiratory tract bacteria in patients with cystic fibrosis. J Clin Microbiol. 1999;37:753–757. doi: 10.1128/jcm.37.3.753-757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simpson I N, Finlay J, Winstanley D J, Dehwurst N, Nelson J W, Butler S L, Govan J R W. Multi-resistance isolates possessing characteristics of both Burkholderia (Pseudomonas) cepacia and Burkholderia gladioli from patients with cystic fibrosis. J Antimicrob Chemother. 1994;34:353–361. doi: 10.1093/jac/34.3.353. [DOI] [PubMed] [Google Scholar]
- 92.Sinsabaugh H A, Howard G W. Emendation of the description of Pseudomonas cepacia Burkholder (synonyms: Pseudomonas multivorans Stanier et al., Pseudomonas kingae Jonsson; EO-1 Group) Int J Syst Bacteriol. 1975;25:187–201. [Google Scholar]
- 93.Skerman V B D, McGowan V, Sneath P A H. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. [Google Scholar]
- 94.Smith D L, Smith E G, Gumery L B, Stableforth D E. Pseudomonas cepacia infection in cystic fibrosis. Lancet. 1992;339:252. doi: 10.1016/0140-6736(92)90063-9. [DOI] [PubMed] [Google Scholar]
- 95.Snell J J S, Hill L R, Lapage S P, Curtis M A. Identification of Pseudomonas cepacia Burkholder and its synonymy with Pseudomonas kingii Jonsson. Int J Syst Bacteriol. 1972;22:127–138. [Google Scholar]
- 96.Speert D P, Bond M, Woodmann R C, Curnutte J T. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J Infect Dis. 1994;170:1524–1531. doi: 10.1093/infdis/170.6.1524. [DOI] [PubMed] [Google Scholar]
- 97.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 98.Stead D E. Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int J Syst Bacteriol. 1992;42:281–295. [Google Scholar]
- 99.Sun L, Jiang R Z, Steinbach S, Holmes A, Campanelli C, Forstner J, Tan Y, Riley M, Goldstein R. The emergence of a highly transmissible lineage of Cbl+Pseudomonas (Burkholderia) cepacia causing CF center epidemics in North America and Britain. Nat Med. 1995;1:661–666. doi: 10.1038/nm0795-661. [DOI] [PubMed] [Google Scholar]
- 100.Tabacchioni S, Visca P, Chiarini L, Bevivino A, Di Serio C, Fancelli S, Fani R R. Molecular characterisation of rhizosphere and clinical isolates of Burkholderia cepacia. Res Microbiol. 1995;146:531–542. doi: 10.1016/0923-2508(96)80559-6. [DOI] [PubMed] [Google Scholar]
- 101.Tyler S D, Strathdee C A, Rozee K R, Johnson W M. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequence of genes coding for 16S–23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Urakami T, Ito-Yoshida C, Araki H, Kijima T, Suzuki K, Komagata K. Transfer of Pseudomonas plantarii and Pseudomonas glumae to Burkholderia as Burkholderia spp. and description of Burkholderia vandii sp. nov. Int J Syst Bacteriol. 1994;44:235–245. [Google Scholar]
- 103.Ursing J B, Rossello-Mora R A, Garcia-Valdes E, Lalucat J. Taxonomic note: a pragmatic approach to the nomenclature of phenotypically similar genomic groups. Int J Syst Bacteriol. 1995;45:604. [Google Scholar]
- 104.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 105.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Laer F, Raes D, Vandamme P, Lammens C, Sion J P, Vrints C, Snoeck J, Goossens H. An outbreak of Burkholderia cepacia with septicemia on a cardiology ward. Infect Control Hosp Epidemiol. 1998;19:112–113. [PubMed] [Google Scholar]
- 108.van Pelt C, Verduin C M, Goessens W H F, Vos M C, Tümmler B, Segonds C, Reubsaet F, Verbrugh H, van Belkum A. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J Clin Microbiol. 1999;37:2158–2164. doi: 10.1128/jcm.37.7.2158-2164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Viallard V, Poirier I, Cournoyer B, Haurat J, Wiebkin S, Ophel-Keller K, Balandreau J. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassesment of [Pseudomonas] phenazinium, [Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int J Syst Bacteriol. 1998;48:549–563. doi: 10.1099/00207713-48-2-549. [DOI] [PubMed] [Google Scholar]
- 110.Vidaver A K, Doyle M P, Gerone P J, Gonzalez C F, Hall P, Hunter-Cereva J C, Loria R, Newsome R L, Shore S H, Wilkins T. Burkholderia cepacia—friend or foe? ASM News. 1999;65:587. [Google Scholar]
- 111.von Graevenitz A. Acinetobacter, Alcaligenes, Moraxella, and other nonfermentative gram-negative bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 112.Welch D F. Application of cellular fatty acid analysis. Clin Microbiol Rev. 1991;4:422–438. doi: 10.1128/cmr.4.4.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Welch D F, Muszynski M J, Pai C H, Marcon M J, Hribar M M, Gilligan P H, Matsen J M, Ahlin P G, Hilman B C, Chartrand S A. Selective and differential medium for the recovery of Pseudomonas cepacia from the respiratory tract of patients with cystic fibrosis. J Clin Microbiol. 1987;25:1730–1734. doi: 10.1128/jcm.25.9.1730-1734.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Whitby P W, Carter K B, Hatter K L, LiPuma J J, Stull T L. Identification of members of the Burkholderia cepacia complex by species-specific PCR. J Clin Microbiol. 2000;38:2962–2965. doi: 10.1128/jcm.38.8.2962-2965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Whitby P W, Pope L C, Carter K B, LiPuma J J, Stull T L. Species-specific PCR as a tool for the identification of Burkholderia gladioli. J Clin Microbiol. 2000;38:282–285. doi: 10.1128/jcm.38.1.282-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wilsher M, Kolbe J, Morris A J, Welch D F. Nosocomial acquisition of Burkholderia gladioli in patients with cystic fibrosis. Am J Resp Crit Care Med. 1997;155:1136–1140. doi: 10.1164/ajrccm.155.4.9105090. [DOI] [PubMed] [Google Scholar]
- 117.Wittke R, Ludwig W, Peiffer S, Kleiner D. Isolation and characterisation of Burkholderia norimbergensis sp. nov., a mildly alkaliphilic sulfur oxidizer. Syst Appl Microbiol. 1997;20:549–553. [Google Scholar]
- 118.Yabuuchi E, Kawamura Y, Ezaki T, Ikedo M, Dejsirilert S, Fujiwara N, Naka T, Kobayashi K. Burkholderia uboniae sp. nov., L-arabinose asimilating but different from Burkholderia thailandensis and Burkholderia vietnamiensis. Microbiol Immunol. 2000;44:307–317. doi: 10.1111/j.1348-0421.2000.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 119.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov; and transfer of seven species of the Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 120.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 121.Yohalem D S, Lorbeer J W. Multilocus isoenzyme diversity among strains of Pseudomonas cepacia isolated from decayed onions, soils, and clinical sources. Syst Appl Microbiol. 1994;17:116–124. doi: 10.1007/BF00871753. [DOI] [PubMed] [Google Scholar]
- 122.Zhang H, Hanada S, Shigematsu T, Shibuya K, Kamagata Y, Kanagawa T, Kurane R. Burkholderia kururiensis sp. nov., a trichloroethylene (TCE)-degrading bacterium isolated from an aquifer polluted with TCE. Int J Syst Evol Microbiol. 2000;50:743–749. doi: 10.1099/00207713-50-2-743. [DOI] [PubMed] [Google Scholar]
- 123.Zhao N, Qu C, Wang E, Chen W. Phylogenetic evidence for the transfer of Pseudomonas cocovenenans (van Damme et al. 1960) to the genus Burkholderia as Burkholderia cocovenenans (van Damme et al. 1960) comb. nov. Int J Syst Bacteriol. 1995;45:600–603. doi: 10.1099/00207713-45-3-600. [DOI] [PubMed] [Google Scholar]