Abstract
The genus Pneumocystis contains a family of fungal organisms that infect a wide variety of mammalian species. Although it is a cause of pneumonia in immunocompromised hosts, recent evidence suggests that these organisms colonize nonimmunosuppressed hosts. Detection of cryptic colonization with Pneumocystis becomes important in animal studies when infection-free animals are necessary. Provocation by chronic immunosuppression, histology, and serology has been widely used to detect the presence of Pneumocystis in rat colonies, requiring lengthy time periods and/or postmortem tissue. We conducted a study to evaluate the use of PCR amplification of oral swabs for the antemortem detection of Pneumocystis in 12 rat groups from three commercial vendors. Sera were collected upon arrival, and the oral cavity was swabbed for PCR analysis. Ten of these groups of rats were then housed in pairs under barrier and immunosuppressed to provoke Pneumocystis growth. Once moribund, the rats were sacrificed, and the lungs were collected to evaluate the presence of Pneumocystis by PCR and microscopic enumeration. DNA was extracted from oral swabs and lung homogenates, and PCR was performed using primers targeting a region within the mitochondrial large-subunit rRNA of Pneumocystis carinii f. sp. carinii. Upon receipt, 64% of rats were positive for P. carinii f. sp. carinii-specific antibodies, while P. carinii f. sp. carinii DNA was amplified from 98% of oral swabs. Postmortem PCR analysis of individual lungs revealed P. carinii f. sp. carinii DNA in all rat lungs, illustrating widespread occurrence of Pneumocystis in commercial rat colonies. Thus, oral swab/PCR is a rapid, nonlethal, and sensitive method for the assessment of Pneumocystis exposure.
Pneumocystis organisms are a group of fungi that infect the lung alveoli of mammals, including humans. In an immunosuppressed host, Pneumocystis spp. proliferate in the lung alveoli, causing a lethal pneumonia. The complete life cycle of these organisms has not been fully characterized, primarily due to a historical lack of a long-term culture system, although sexual and asexual stages have been described by light and electron microscopy studies (10, 18). Animal studies have shown that transmission occurs through an airborne route (9, 20), but the infectious form has not been identified. Recent studies suggest that nonimmunosuppressed hosts may play a more significant role in the Pneumocystis life cycle than previously believed (3, 16).
The immunosuppressed rat model of infection has been used extensively in Pneumocystis research and is the model evaluated in the present study. Many rat strains are known to harbor Pneumocystis spp. (4, 22), and these rats can develop fulminant Pneumocystis infection with chronic administration of corticosteroids. Genetic analyses have identified two Pneumocystis carinii populations that reside in rat lungs, P. carinii f. sp. carinii and P. carinii f. sp. ratti (2). The focus of the present study is the population most prevalent in commercial vendor colonies, P. carinii f. sp. carinii. Because Pneumocystis spp. have been found in a wide variety of commercial rat colonies (2, 22), it is necessary to be able to rapidly assess the presence of Pneumocystis in rats prior to their use in most studies. Studies that address immunological responses of primary Pneumocystis exposure, as well as studies in which defined Pneumocystis inocula are administered, require Pneumocystis-naive rats. Serology is one current antemortem method used to determine whether rats have been exposed to Pneumocystis (14, 21), but serology sensitivity is decreased by rat-to-rat variation in the time required for antibody production after an initial Pneumocystis exposure (21).
The purpose of the present study was to evaluate the use of oral swabs combined with PCR for the determination of Pneumocystis exposure in individual rat prior to immunosuppression. Previous studies showed that bronchoalveolar lavage fluid, oropharyngeal washes, or oral washes could be used to diagnose Pneumocystis infections in humans (5, 7, 12, 15), and in one study, nasopharyngeal aspirates were used for Pneumocystis detection in rats postmortem (11). These techniques have been adapted further for this investigation. In the present study, we asked if Pneumocystis DNA could be detected in the oral cavities of nonimmunosuppressed rats and if the presence of Pneumocystis-specific DNA produced by targeted PCR correlated with infection after chronic immunosuppression.
We found that PCR analysis of oral swabs was a sensitive method for detection of Pneumocystis exposure and was correlated with the presence of organisms after chronic immunosuppression. Oral swabbing combined with PCR is a rapid, simple, and nonlethal method for determining Pneumocystis exposure in rats.
MATERIALS AND METHODS
Rat groups.
Twelve groups of 7 to 12 rats each were obtained from eight commercial rat colonies: Charles River (two groups from colony 064, Wilmington, Del.; colony areas 42 and 44, Hollister, Calif.; colony P03, Portage, Oreg.; and two groups from colony R09, Raleigh, N.C.), Taconic (two groups from colony MBU4 and two groups from IBU18, Germantown, N.Y.), and Harlan (Indianapolis, Ind.). All rats were maintained in pairs under barrier throughout their lives at the University of Cincinnati Department of Laboratory Animal Medicine, Cincinnati, Ohio. Barrier housing consisted of 3-μm exclusion microfilter-top cages supplied with HEPA-filtered air. All rats were fed sterile food and water and handled only under a sterile, horizontal laminar flow hood by personnel wearing sterile attire. Samples from the water, food, cage racks, and laminar flow hood were analyzed for the presence of P. carinii f. sp. carinii-specific DNA, as described below. Oral swabs and serum samples were collected from each rat under sterile conditions within 48 h after receipt into our facility.
One week after receipt, 10 of the 12 groups of rats received weekly, subcutaneous injections with 2 to 4 mg of methylprednisolone acetate (Upjohn, Kalamazoo, Mich.) to provoke the development of Pneumocystis infections if organisms were present. After 7 to 12 weeks of immune suppression, each moribund rat was sacrificed by administering an overdose of CO2. Morbidity was determined as a general decline in rat health, including significant weight loss (60 to 70%) and labored breathing. The remaining two groups of rats, which were not immunosuppressed, were sampled by oral swab, had sera collected, and were then sacrificed on the day of receipt into our facility. All handling and processing of these two groups were comparable to those for the other rat groups. Rats were handled according to Institutional Animal Care and Use Committee guidelines under University of Cincinnati protocol 90-05-15-01 (approval date, 12 June 2000).
Rat lung processing.
Rat lungs were removed from each rat using separate packages of sterile instruments, and the Pneumocystis organisms were extracted from the tissue by homogenization using a Stomacher Lab Blender 80 in 10 ml of sterile phosphate-buffered saline (Tekmar, Cincinnati, Ohio) (8). The homogenate was reduced in host cell contamination by treatment with aqueous 0.85% ammonium chloride at 37°C for 20 min, DNase I (Roche, Indianapolis, Ind.) treatment (0.2 mg/ml at 37°C for 30 min), and microfiltration with 10-μm White Mitex LCWP 25-mm filters (Millipore, Bedford, Mass.). The processed organisms were then suspended in 1 ml of DNA extraction buffer (100 mM Tris, 100 mM EDTA, 200 mM NaCl, 1% Sarkosyl) for organic-phase DNA extraction using Phase Lock Heavy Gel (Eppendorf, Westbury, N.Y.). DNA was stored in TE (1 mM EDTA, 10 mM Tris-Cl) at −20°C until analyzed by PCR. For most rats, three 10-μl drops of lung homogenate were heat fixed to glass microscope slides and then stained by cresyl echt violet (CEV) (1). Each slide was enumerated for Pneumocystis cysts, expressed as the concentration of cysts per lung. The limit of detection for microscopic enumeration of cysts was 1.8 × 104 cysts per lung.
Immunoblotting.
P. carinii f. sp. carinii form 1 organisms were solubilized in lysis buffer (2% sodium dodecyl sulfate [SDS], 0.06 M Tris [pH 6.8], 1% glycerol, 5% 2-mercaptoethanol) at 100°C for 5 min in preparation for polyacrylamide gel electrophoresis (PAGE) (14, 21). This preparation was loaded on SDS–10% PAGE gels in equal concentrations and electrophoresed for 1 h at 200 V on an EI9001-Xcell II Minicell (Novel Experimental Techniques, San Diego, Calif.) (20). The separated proteins were transferred to nitrocellulose membranes for 1 h at 100 V using a Mini 2-D Trans Blot (Bio-Rad, Richmond, Calif.). Immunoblotting was performed as previously described (14, 21). Briefly, the transfer membrane strips were blocked in 1% nonfat milk at 4°C overnight and then incubated with individual rat sera at a dilution of 1:40 for 2 h at 4°C. Antibody presence was visualized using a horseradish peroxidase conjugate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
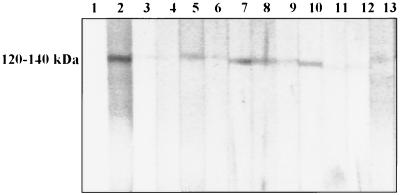
Each rat was assessed for the presence or absence of antibodies against the P. carinii f. sp. carinii form 1 major surface glycoprotein (MSG) group of antigens by the presence or absence of a band ranging from 120 to 140 kDa, as previously reported (6). Rats were also scored for the presence or absence of bands of 45 to 55 kDa (21), but these bands were more variable in size and intensity than the 120- to 140-kDa bands. The antigen group at 120 to 140 kDa produced unambiguous banding and was chosen as the hallmark for the presence of P. carinii f. sp. carinii-specific antibodies. Previous studies have shown no cross-reactivity between P. carinii f. sp. carinii and P. carinii f. sp. ratti in this region (17).
Oral swabs.
Oral swabs were collected from each rat before immunosuppression and, in some cases, at death using sterile cotton-tipped wooden applicators (Fisher, Pittsburgh, Pa.) moistened in DNA extraction buffer. Each swab sample was collected by rubbing the cotton swab over the hard palate, surface of the tongue, and buccal surface, and under the tongue of each rat. DNA was collected from the entire, intact cotton swab tip for each oral sample by organic-phase DNA extraction. Oral swab controls were extracted with known numbers of Pneumocystis organisms. No evidence for inhibitory factors was detected by these controls. DNA samples were stored in TE at −20°C until analyzed by PCR.
PCR analysis.
DNA samples from all oral swabs, lung homogenates, and controls were amplified with the Rcc primer set (13) using a GeneAmp PCR System 9700 thermocycler (PE Applied Biosystems, Norwalk, Conn.). These primers target a region of the mitochondrial large-subunit (mtLSU) rRNA specific for P. carinii f. sp. carinii. Each reaction used 1× Hot Start Taq Master Mix (Qiagen, Valencia, Calif.) (1.5 mM MgCl2, 50 mM KCl, 200 μM (each) deoxynucleoside triphosphate) to which 0.05 ng each of Rcc 1 and 2 primers, 1.0 μl of template DNA, and molecular-grade water were added. Each reaction was set up under a UV-treated laminar flow hood to prevent contamination of samples. Positive (P. carinii f. sp. carinii; DNA) and negative (water) controls were included in each experiment, and DNA extraction and PCR reagents were UV irradiated and tested for contamination. PCR conditions were 95°C hot start for 15 min, 94°C denaturing for 1 min, 54°C annealing for 30 s, 72°C extension for 30 s (up to 40 cycles total), and 72°C final extension for 10 min. Amplified DNA products were visualized by staining with ethidium bromide in 2% agarose gels run at 90 V for ∼1.5 h. Gel images were captured with NIH Image 1.6 software. Each rat was scored positive for P. carinii f. sp. carinii by the presence of a band at 137 bp.
Analytical PCR primer sensitivity.
The theoretical sensitivity of the primer set used in this study was determined. Templates for P. carinii f. sp. carinii were obtained by amplifying DNA extracted from organisms embedded in low-melt agarose. The organisms used to produce these templates were previously characterized by contour-clamped homogeneous electric field analysis as P. carinii f. sp. carinii form 1 (7). The DNA was extracted from the agarose plugs using Light Phase Lock gel DNA extraction (Eppendorf, Westbury, N.Y.) and then amplified using primers paz102H and paz102E (which target the mtLSU) (19) under the conditions described above. This amplification yielded 360-bp products that included the region to be amplified by the Rcc primers. These amplification products were cloned into the Topo 2 vector (Invitrogen, Carlsbad, Calif.), transformed into TOPO10 One Shot competent Escherichia coli cells (Invitrogen), and plated onto Luria-Bertani agar plates with kanamycin (50 μg/ml).
Six clones were chosen for analysis and grown in selective broth overnight at 37°C. The plasmids were purified with a QIAspin miniprep kit (Qiagen), digested with EcoRI, and run on a 1% agarose gel to verify the proper insert size (360 bp). Two plasmids were chosen for sequencing to verify their identity (PE Applied Biosystems 373 long plate; University of Cincinnati DNA Core Facility). Plasmids with the expected sequences were amplified in PCRs using the Rcc primer set to determine the sensitivity and specificity of these primers. The weight for one plasmid was calculated to be 5.9 × 10−9 ng. Tenfold dilutions from 1.0 to 10−10/ng of the plasmids were analyzed under the PCR conditions described above (40 amplification cycles). Equal volumes of PCR products (5μl) were electrophoresed at 90 V for 1.5 h and visualized on 2% agarose gels plus ethidium bromide. Rcc primers were also used to amplify DNA template with an excess of P. carinii f. sp. ratti, resulting in the amplification of only P. carinii f. sp. carinii template, as determined by sequence analysis. NIH Image 1.6 was used to document each gel at 3×-frame integration. The sensitivity for these primers was determined by noting the lowest plasmid DNA concentration with a visible PCR product. The lowest plasmid DNA concentration was then divided by the weight of a single plasmid to determine the actual number of plasmids, and thus the actual number of templates, detected under these conditions.
RESULTS
Immunoblotting and oral swab results prior to immunosuppression.
Sera and oral swabs were collected from each rat upon receipt, prior to immunosuppression. Eighty-seven of 137 (64%) rats were positive for Pneumocystis-specific antibodies (Table 1 and Fig. 1). Oral swabs collected from each of the 137 rats showed that 134 of 137 (98%) were positive for P. carinii f. sp. carinii-specific DNA (Table 1). Figure 2A illustrates amplicons resulting from oral swabs of rats obtained from the Raleigh R09 colony prior to immunosuppression. Rats that were negative for P. carinii f. sp. carinii-specific antibodies and had no detectable P. carinii f. sp. carinii-specific DNA from oral swabs before immunosuppression represented 1% (2 of 137) of the total rat population. Rats that were negative for P. carinii f. sp. carinii-specific antibodies and positive for P. carinii f. sp. carinii-specific DNA from oral swabs before immunosuppression represented 35% (48 of 137) of the total rat population. One of 137 rats (<1%) was positive for P. carinii f. sp. carinii-specific antibodies and had no detectable P. carinii f. sp. carinii-specific DNA, whereas rats that were positive for both P. carinii f. sp. carinii-specific antibodies and DNA represented 63% (86 of 137) of the rat population. PCR of water, food, and laminar hood swabs was negative for P. carinii f. sp. carinii-specific DNA.
TABLE 1.
Summary of Pneumocystis-specific DNA, Pneumocystis-specific antibody presence, and Pneumocystis organism number in rat groups
| Rat colony | % Positive (no. positive/no. tested) for P. carinii f. sp. carinii-specific DNA
|
||||
|---|---|---|---|---|---|
| Oral swabs
|
Lung homogenates | Serology | Average no. of cysts lung ± SD | ||
| At arrival | At death | ||||
| Wilmington 064 | 100 (12/12) | 100 (10/10) | 100 (12/12) | 0 (0/12) | 1 × 106 ± 0.9 |
| Taconic MBU4 | 100 (5/5) | 100 (5/5) | 100 (5/5) | 80 (4/5) | 1 × 108 ± 1.0 |
| Hollister area 44 | 67 (6/9) | Not collected | 100 (9/9) | 22 (2/9) | 1 × 105 ± 0.4 |
| Hollister area 42 | 92 (11/12) | Not collected | 100 (12/12) | 100 (12/12) | 4 × 105 ± 0.2 |
| Portage P03 | 100 (12/12) | Not collected | 100 (12/12) | 100 (12/12) | 9 × 104 ± 0.3 |
| Raleigh R09 | 100 (24/24) | 82 (9/11) | 100 (24/24) | 46 (11/24) | 5 × 105 ± 0.6 |
| Harlan transfer | 100 (7/7) | 100 (5/5) | 100 (7/7) | 14 (1/7) | ≤2 × 104 ± 0.1 |
| Wilmington 064 | 100 (12/12) | Not collected | 100 (12/12) | 25 (3/12) | 3 × 104 ± 1.0 |
| Taconic MBU4 | 100 (24/24) | Not collected | 100 (24/24) | 100 (24/24) | 2 × 105 ± 0.9 |
| Taconic IBU18 | 100 (20/20) | Not collected | 100 (20/20) | 90 (18/20) | 1 × 106 ± 0.3 |
| All groups | 98 (134/137) | 94 (29/31) | 100 (137/137) | 64 (87/137) | 1 × 107 ± 0.4 |
FIG. 1.
Immunoblotting results for Hollister (area 42) serum samples. Lane 1 is the negative control (Tween-Tris-buffered saline [0.02 M Tris, 0.5 M NaCl, 0.05% Tween 20]), lane 2 is the positive control (Pneumocystis antibody-positive rat serum), and lanes 3 to 13 are Hollister area 42 rat sera. All rats have an antibody reaction with the 120- to 140-kDa MSG of P. carinii f. sp. carinii form 1 surface antigen preparation.
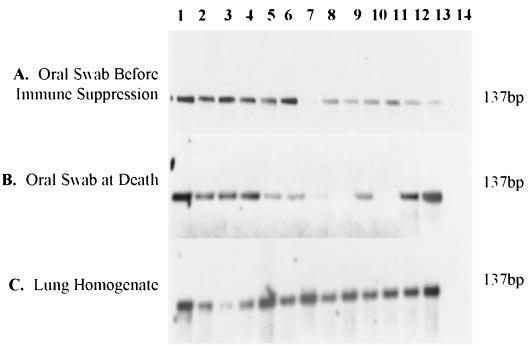
FIG. 2.
PCR products showing the presence or absence of P. carinii f. sp. carinii DNA in nonimmunosuppressed Raleigh R09 rats. Lanes 1 to 12, individual rat samples; lane 13, a positive control (Pneumocystis DNA); lane 14, a negative control (water). (A) Oral swabs collected before rats were immunosuppressed. (B) Oral swabs collected at rat death. (C) Lung homogenate samples for each rat.
Lung homogenate analysis prior to immunosuppression.
Two groups of 12 rats (Raleigh R09 and Taconic MBU4) were sacrificed upon receipt to determine the presence or absence of P. carinii f. sp. carinii-specific DNA in lung tissue before immunosuppression. All 24 rats were positive for P. carinii f. sp. carinii-specific DNA in oral swabs and lung homogenates upon receipt into our facility (data not shown). Serology showed that 50% (12 of 24) were positive for P. carinii f. sp. carinii-specific antibodies (data not shown).
Oral swab and lung homogenate analysis at rat death.
At the time of sacrifice, each rat lung was removed and analyzed by PCR to assess the presence of Pneumocystis spp. A limited number of rats were orally swabbed to correlate the presence of Pneumocystis in the oral cavity with that in the lung at the time of death. For the total lung homogenates examined at death, all 137 (100%) rats were positive for P. carinii f. sp. carinii DNA in lung tissue homogenates (Table 1). Shown in Fig. 2C are amplicons resulting from lung homogenates of rats from the Hollister area 42 colony. Of the 31 swabs collected at the time of death, 94% (29 of 31) were positive for P. carinii f. sp. carinii DNA, with 6% (2 of 31) of these rats having no detectable Pneumocystis DNA (Table 1 and Fig. 2B).
Pneumocystis cyst enumeration.
For most rats, Pneumocystis cysts were enumerated by microscopic analysis of CEV-stained samples. The data obtained are shown in Table 1. All groups of rats had detectable cysts in their lung homogenates.
Summary of oral swab findings.
Every rat from all colonies surveyed was found to be positive for P. carinii-specific DNA upon conclusion of this investigation. There was wide geographic distribution among the 12 groups of rats tested: two groups from Delaware, two groups from California, four groups from New York, one group from Oregon, one group from Indiana, and two groups from North Carolina. The strain of rat was also variable, with seven groups of Sprague Dawley rats, two groups of Wistar rats, two groups of Long Evans, and one group of Brown Norway rats. These findings support previous karyotypic studies that revealed the wide prevalence of P. carinii f. sp. carinii in commercial rat vendors and across rat strains (2).
Analytical PCR primer sensitivity.
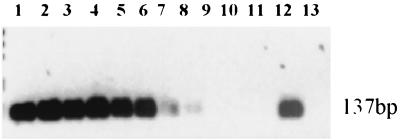
Plasmid templates were used to ascertain the sensitivity for the Rcc primers used in this study. The sensitivity for Rcc primers was calculated to be 17 plasmid templates by identifying the lowest concentration of plasmid template able to yield a visible PCR product under specific conditions. Template number was calculated by dividing the lowest plasmid concentration with a visible PCR product (10−7 ng) by the weight of an individual plasmid (5.9 × 10−9 ng), yielding a result of 17 plasmid templates/Pneumocystis-specific insert (Fig. 3). The specificity of the Rcc primers was also demonstrated by the amplification of P. carinii f. sp. carinii template from a sample containing an excess of P. carinii f. sp. ratti template.
FIG. 3.
PCR analysis showing Rcc primer sensitivity for P. carinii f. sp. carinii plasmid template. Lanes 1 to 11, P. carinii f. sp. carinii plasmid concentrations ranging from 1 to 10−10 ng (theoretical one plasmid). Lane 12 is the positive control (Pneumocystis DNA). Lane 13 is the negative control (water).
DISCUSSION
We asked two questions in this study: could Pneumocystis DNA be detected in the oral cavity of rats by oral swab/targeted PCR, and could the presence or absence of Pneumocystis-specific DNA predict infection after chronic immunosuppression. The results of this study showed that 98% of the rats were positive for P. carinii f. sp. carinii-specific DNA upon receipt into our facility, and thus organism DNA was detectable within the oral cavity of the nonimmunosuppressed rat. All of the rats eventually developed P. carinii f. sp. carinii infection with chronic immunosuppression, demonstrating that this technique can be used to predict the exposure of rats to P. carinii f. sp. carinii. The presence of anti-MSG antibodies, determined by the immunoblotting technique using the 120- to 140-kDa band representing MSG antigens, was determined to be a less reliable predictor of Pneumocystis outcome than the results of oral swab-PCR analysis (64 versus 98%).
Many researchers rely on the detection of Pneumocystis-specific antibodies in the sera of their test animals to determine a prior exposure to Pneumocystis (14, 21). Because the time required for Pneumocystis-specific antibody production is not known, serology is not a reliable method for screening individual animals. If serology were used, a series of samplings over time would likely provide a more accurate assessment of Pneumocystis exposure. Based on the data presented here, serological methods do not identify all rats that have been exposed to Pneumocystis within a single sampling. Of those rats that were positive for P. carinii f. sp. carinii-specific antibodies, 99% (86 of 87) were also positive for P. carinii f. sp. carinii-specific DNA. Thus, if the antibody response is positive, it is likely that there will be detectable Pneumocystis DNA in the oral cavity, indicating a previous exposure. However, rats negative for P. carinii f. sp. carinii-specific antibodies and positive for P. carinii f. sp. carinii-specific DNA represented 96% (48 of 50) of the rat oral cavities evaluated. This observation suggests that the oral swab-PCR method is more sensitive than immunoblotting for detection of P. carinii f. sp. carinii. Swabbing of the oral cavity is less stressful to rats than serum collection and can be predictive of Pneumocystis exposure with the collection of only one sample. Serological methods are useful to assess the presence of Pneumocystis in a given colony, but would likely produce false-negatives due to a lag in antibody production. Also, the presence of antibody does not necessarily predict that a fulminant infection will develop, only that there has been an exposure.
Our findings show that Pneumocystis is widespread in commercial rat colonies, supporting previous reports of the presence widespread of Pneumocystis in commercial rat colonies (2, 22). It is necessary for researchers who require Pneumocystis-naive rats to assess the presence or absence of Pneumocystis in commercial rat colonies prior to their use in projects that are sensitive to an initial Pneumocystis exposure. The technique presented here is an ideal method to determine whether a particular animal harbors Pneumocystis organisms. The sensitivity of this technique permits relatively low numbers of sentinel animals to be sampled to determine the presence of P. carinii f. sp. carinii.
The findings of this study also have implications in understanding the life cycle of Pneumocystis. Recent studies suggest that nonimmunosuppressed hosts may be reservoirs of Pneumocystis infection (3, 16). This observation is supported by the present study through the detection of Pneumocystis-specific DNA in the oral cavities of nonimmunosuppressed rats. Although it is not known whether the DNA present represents intact or viable organisms at this time, these findings suggest that organisms may traffic to the oral cavity. It is known that Pneumocystis is transmitted via airborne routes, and thus it is possible that the P. carinii f. sp. carinii-specific DNA isolated from the oral cavities of these rats is associated with viable organisms. If viable Pneumocystis organisms do travel to the oral cavity, this could provide a means of transmission via coughing, contact with the oral mucosa, and/or grooming. Further investigations using the techniques presented here should provide additional insights into the Pneumocystis life cycle.
The application of oral swabs combined with PCR was able to detect P. carinii f. sp. carinii-specific DNA in the rat oral cavity, predict Pneumocystis exposure, and confirm the widespread prevalence of Pneumocystis in commercial rat colonies. Oral swab-PCR detection of Pneumocystis is a valuable tool for investigators involved in the study of rat Pneumocystis, providing a rapid, reliable, noninvasive, and nonlethal method for its detection. The proposed oral swab technique may also be applied to the detection of other respiratory pathogen models, and may provide a useful method for similar studies in humans.
ACKNOWLEDGMENT
This research was supported by NIH grant RO1 AI32436.
REFERENCES
- 1.Bowling M, Smith I, Wescott S. A rapid staining procedure for Pneumocystis carinii. Am J Med Tech. 1973;39:267–268. [PubMed] [Google Scholar]
- 2.Cushion M T, Kaselis M, Stringer S L, Stringer J R. Genetic stability and diversity of Pneumocystis carinii infecting rat colonies. Infect Immun. 1993;61:4801–4813. doi: 10.1128/iai.61.11.4801-4813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumoulin A, Mazars E, Seguy N, Gargallo-Viola D, Vargas S, Cailliez J, Aliouat E, Wakefield A E, Dei-Cas E. Transmission of Pneumocystis carinii disease from immunocompetent contacts of infected hosts to susceptible hosts. Eur J Clin Microbiol Infect Dis. 2000;19:671–678. doi: 10.1007/s100960000354. [DOI] [PubMed] [Google Scholar]
- 4.Frenkel J, Good J, Shultz J. Latent Pneumocystis infection of rats, relapse, and chemotherapy. Lab Investig. 1966;15:1559–1577. [PubMed] [Google Scholar]
- 5.Graves D, Chary-Reddy S, Becker-Hapak M. Detection of Pneumocystis carinii in induced sputa from immunocompromised patients using a repetitive DNA probe. Mol Cell Probes. 1997;11:1–9. doi: 10.1006/mcpr.1996.0070. [DOI] [PubMed] [Google Scholar]
- 6.Graves D, McNabb S, Worley M, Downs T, Ivey M. Analyses of rat Pneumocystis carinii antigens recognized by human and rat antibodies by using Western immunoblotting. Infect Immun. 1986;54:96–103. doi: 10.1128/iai.54.1.96-103.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helweg-Larsen J, Tsolaki A, Miller R, Lundgren B, Wakefield A E. Clusters of Pneumocystis carinii pneumonia: analysis of person-to-person transmission by genotyping. Q J Med. 1998;91:813–820. doi: 10.1093/qjmed/91.12.813. [DOI] [PubMed] [Google Scholar]
- 8.Hong S, Steel P, Cushion M T, Walzer P D, Stringer S L, Stringer J E. Pneumocystis carinii karyotypes. J Clin Microbiol. 1990;28:1785–1795. doi: 10.1128/jcm.28.8.1785-1795.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes W. Natural mode of acquisition for de novo infection with Pneumocystis carinii. J Infect Dis. 1982;145:842–848. doi: 10.1093/infdis/145.6.842. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto Y, Yoshida Y. Sporogony in Pneumocystis: synaptonemal complexes and meiotic nuclear division observed in precysts. J Protozool. 1984;31:420–428. doi: 10.1111/j.1550-7408.1984.tb02989.x. [DOI] [PubMed] [Google Scholar]
- 11.Oz H, Hughes W. DNA Amplification of nasopharyngeal aspirates in rats: a procedure to detect Pneumocystis carinii. Microb Pathog. 1999;27:119–121. doi: 10.1006/mpat.1999.0292. [DOI] [PubMed] [Google Scholar]
- 12.Oz H, Hughes W. Search for Pneumocystis carinii DNA in upper and lower respiratory tract of humans. Diagn Microbiol Infect Dis. 2000;37:161–164. doi: 10.1016/s0732-8893(00)00146-2. [DOI] [PubMed] [Google Scholar]
- 13.Palmer R J, Cushion M T, Wakefield A E. Discrimination of rat-derived Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. ratti using the polymerase chain reaction. Mol Cell Probes. 1999;13:147–155. doi: 10.1006/mcpr.1999.0229. [DOI] [PubMed] [Google Scholar]
- 14.Peglow S L, Smulian A G, Linke M J, Pogue C L N, Crisler S J, Phair J, Gold J W, Armstrong D, Walzer P D. Serologic responses to Pneumocystis carinii antigens in health and disease. J Infect Dis. 1990;161:296. doi: 10.1093/infdis/161.2.296. [DOI] [PubMed] [Google Scholar]
- 15.Smith J, Wiggins C. Identification of Pneumocystis carinii in sputum. N Engl J Med. 1973;289:1254–1255. doi: 10.1056/NEJM197312062892320. [DOI] [PubMed] [Google Scholar]
- 16.Vargas S, Ponce C, Gigliotti F, Ulloa A, Prieto S, Munoz M, Hughes W. Transmission of Pneumocystis carinii DNA from a patient with P. carinii pneumonia to immunocompetent contact health care workers. J Clin Microbiol. 2000;38:1536–1538. doi: 10.1128/jcm.38.4.1536-1538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasquez J, Smulian A G, Linke M J, Cushion M T. Antigenic differences associated with genetically distinct Pneumocystis carinii from rats. Infect Immun. 1996;64:290–297. doi: 10.1128/iai.64.1.290-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vossen M, Beckers P, Meuwissen J, Stadhouders A. Developmental biology of Pneumocystis carinii, an alternative view on the life cycle of the parasite. Z Parasitenkd. 1978;55:101–118. doi: 10.1007/BF00384826. [DOI] [PubMed] [Google Scholar]
- 19.Wakefield A E, Pixley F, Sinclair K, Miller R, Moxon E, Hopkin J. Detection of Pneumocystis carinii with DNA amplification. Lancet. 1990;336:451–453. doi: 10.1016/0140-6736(90)92008-6. [DOI] [PubMed] [Google Scholar]
- 20.Walzer P D, Schnelle V, Armstrong D, Rosen P. Nude mouse: a new experimental model for Pneumocystis carinii infection. Science. 1977;197:177–179. doi: 10.1126/science.301657. [DOI] [PubMed] [Google Scholar]
- 21.Walzer P D, Stanforth D, Linke M J, Cushion M T. Pneumocystis carinii: immunoblotting and immunofluorescent analyses of serum antibodies during experimental rat infections and recovery. Exp Parasitol. 1987;63:319–328. doi: 10.1016/0014-4894(87)90179-2. [DOI] [PubMed] [Google Scholar]
- 22.Weisbroth S, Geistfeld J, Weisbroth S, Williams B, Feldman S, Linke M J, Orr S, Cushion M T. Latent Pneumocystis carinii infection in commercial rat colonies: comparison of inductive immunosuppressants plus histopathology, PCR, and serology as detection methods. J Clin Microbiol. 1999;37:1441–1446. doi: 10.1128/jcm.37.5.1441-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]