Abstract
Rapidly growing yeasts with appropriate posttranslational modifications are favored hosts for protein production in the biopharmaceutical industry. However, limited production capacity and intricate transcription regulation restrict their application and adaptability. Here, we describe a programmable high-expression yeast platform, SynPic-X, which responds to defined signals and is broadly applicable. We demonstrated that a synthetic improved transcriptional signal amplification device (iTSAD) with a bacterial-yeast transactivator and bacterial-yeast promoter markedly increased expression capacity in Pichia pastoris. CRISPR activation and interference devices were designed to strictly regulate iTSAD in response to defined signals. Engineered switches were then constructed to exemplify the response of SynPic-X to exogenous signals. Expression of α-amylase by SynPic-R, a specific SynPic-X, in a bioreactor proved a methanol-free high-production process of recombinant protein. Our SynPic-X platform provides opportunities for protein production in customizable yeast hosts with high expression and regulatory flexibility.
This study developed a “plug and play” platform that can produce customized efficient yeast hosts responding to defined signals.
INTRODUCTION
High-level and controllable gene expression to produce proteins is required for the biopharmaceutical and biomanufacturing industries. Yeasts are the most favored eukaryotic host for recombinant protein production because of their rapid growth, ability to be genetically manipulated, and posttranslational modifications (1–3). However, their limited production capacity (compared to filamentous fungi) and intricate transcription regulation (compared to bacterial strains) restrict their range of applications (4–6). Scientists have continued to explore novel yeast hosts, yet a programmable gene expression system with high yields and regulatory flexibility remains undiscovered.
Baker’s yeast (Saccharomyces cerevisiae) was the earliest studied yeast host for recombinant protein production and industrial applications, but its use was limited because of hyperglycosylation of proteins, low protein yield, and plasmid instability (3). For nearly two decades, the methylotrophic and Crabtree-negative yeast Pichia pastoris (syn. Komagataella phaffii) has been a workhorse for protein production in both academic and industrial applications (7–13). High cell density, strong expression, genetic stability, and appropriate posttranslational modifications make it a popular system, and it has been used to express more than 5000 proteins (10), including the most recently approved drugs of monoclonal antibody eptinezumab Vyepti and nanobody caplacizumab Cablivi by the U.S. Food and Drug Administration.
Although P. pastoris has served as a standard yeast host for production of recombinant proteins (10, 14), it has limitations that prevent it from being a versatile “protein factory” with high productivity and adaptability. Few powerful transcription tools are currently available in P. pastoris (12), apart from promoters that mostly respond to methanol (toxic, flammable, and explosive) (15, 16). The narrow regulation mode of methanol-inducible promoters severely restricts the scope of application (10, 17). Efforts have been devoted to rewiring this expression host. For instance, promoter variants have been constructed to enable fine-tuning of expression (18–22). Transcription factor overexpression and deficiency have also been used to vary gene expression levels and regulation (23–25). These engineering strategies retained the cross-talk between the endogenous trans/cis-factors and the cell genetic background (18, 21). In recent years, the gene circuit design involving synthetic transcription factors with heterologous DNA binding and transactivation domains has allowed reconstruction of the transcriptional system with minimal interference from the cell genetic background (26, 27). However, none of these studies have generated a universal powerful yeast expression system that is responsive to extensive signals. This prevents yeast from becoming a pervasive protein production platform that accommodates a variety of production requirements.
Because of the fact that P. pastoris was used as an expression host but not a model organism, limited research was conducted to understand its fundamental functions. Recently, the progress made in transcriptional regulation mechanisms (28–31) and gene editing methods (32–37) by our and other groups has made it possible to construct a fully engineered expression platform for this host. Moreover, as a breakthrough in biotechnology, clustered regularly interspaced short palindromic repeats (CRISPR)–mediated gene transcriptional regulation has attracted much attention because of its flexibility, high efficiency, and programmability (38). CRISPR interference (CRISPRi) (38) and CRISPR activation (CRISPRa) (39, 40) systems used in different organisms provide flexible tools and ideas for the design of genetic circuits in P. pastoris.
Here, we describe a synthetic yeast platform based on P. pastoris (SynPic-X) that can precisely respond to user-defined signals. The expression system contains an improved transcriptional signal amplification device (iTSAD) composed of a fused bacterial-yeast transactivator and a hybrid bacterial-yeast promoter, which far exceeded the expression level of the most widely used methanol-inducible promoter PAOX1 (11, 12) in commercial yeast systems. To eliminate the high leaky expression induced by iTSAD, activation by a CRISPRa device (CRISPRaD) and iTSAD repression by a CRISPRi device (CRISPRiD), which are strictly responsive to defined signals, were integrated. SynPic-X represents a new breakthrough in the development of yeast expression hosts for both expression yield and regulatory flexibility. Theoretically, we provide a programmable high-expression yeast platform, which can respond to an abundance of defined factors and is broadly applicable simply by the input of user-defined promoters.
RESULTS
Engineered iTSAD for high-expression performance in P. pastoris
Previously, we identified a critical transcription activator, Mit1, which responds to methanol-inducible signaling and highly activates AOX1 transcription by binding to the PAOX1 promoter in P. pastoris (24, 30). A synthetic transcription device (scheme referred to Fig. 1A) was subsequently engineered, in which lacO (operator sequence from the Escherichia coli lac operon) and cPAOX1 (core promoter of PAOX1) were composed as a hybrid cis-promoter, and LacI (lacO binding protein) and Mit1AD [activation domain (AD) of Mit1] were fused as a transactivator (41). This newly constructed regulatory part was termed as TSAD, which amplified the transcription levels of inducible and constitutive promoters (41).
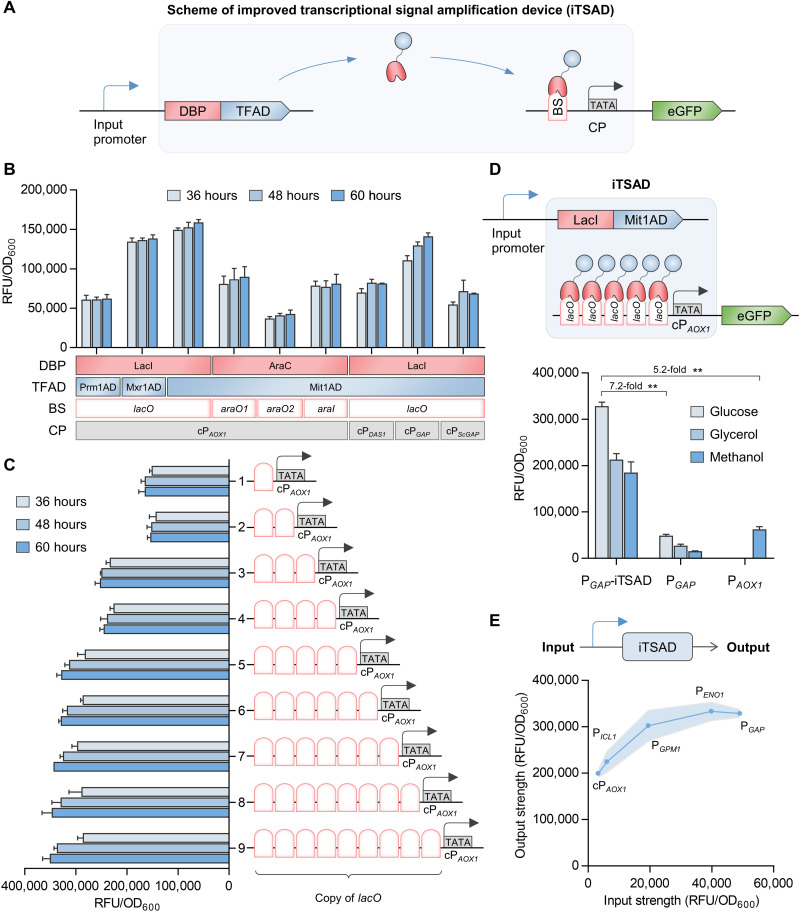
Fig. 1. Design and characterization of an iTSAD.
(A) Genetic circuit scheme of iTSAD. The chimeric transactivator composed of DNA binding proteins (DBPs) and transcription factor activation domains (TFADs) driven by input promoters targets the binding sequences (BS) upstream of the core promoter (CP) and recruits RNA polymerase to activate transcription of the target gene. (B) Function analysis of various biological elements used for the construction of iTSAD. Two DBPs, three TFADs, four BSs, and four CPs were tested in combinations. The combination of LacI-Mit1AD and lacO-cPAOX1 showed the greatest activation effect and was selected for subsequent construction. (C) Effects of lacO copy numbers on the output signal of iTSAD. One to nine copies of lacO were inserted upstream of cPAOX1 driving eGFP expression, and 5lacO-cPAOX1 was selected for subsequent experiments. (D) The eGFP fluorescence intensity of iTSAD with various carbon sources. The output signals of iTSAD driven by PGAP were measured. The most widely used constitutive promoter PGAP and methanol-inducible promoter PAOX1 in P. pastoris were compared. Cells were cultured and compared in glucose, glycerol, and methanol. Statistical significance of eGFP intensity of each strain in specific carbon sources is shown (**P < 0.01). (E) The relationship of input and output signals of iTSAD driven by selective input promoters with different strengths. Input strength, shown on the x axis, represents the evaluation of eGFP expression driven by specific input promoters. Output strength represents eGFP expression driven by iTSAD under specific input promoters. The regression model is shown in fig. S1. RFU, relative fluorescence unit.
To further improve TSAD, we elaborately evaluated the combined effects of exogenous DNA binding proteins (DBPs)/binding sequences (BSs) and ADs of endogenous transcription factors (TFADs) (Fig. 1, A and B). Three chimeric activators were engineered by fusion of the AD of P. pastoris transcription activators Mit1, Mxr1, and Prm1 (30) to the C terminus of the E. coli LacI protein by a GGGGS linker. Exogenous general activators, such as yeast Gal4AD or viral VP16, were not involved because our previous study proved that they had a weaker activation effect compared with endogenous activators in P. pastoris (41). LacI-Mit1AD presented the strongest activation effect with lacO-cPAOX1 (Fig. 1B), consistent with the high activation capacity of native Mit1 on PAOX1 (30). The results from various groups of exogenous DBP/BS indicated that LacI-lacO had the best activation function (Fig. 1B). Four core regions of strong yeast promoters were tested, and cPAOX1 worked better than cPDAS1, cPGAP, and cPScGAP (Fig. 1B). Moreover, a synthetic upstream activating sequence composed of five to nine copies of lacO achieved high expression levels of enhanced green fluorescent protein (eGFP) (Fig. 1C). On the basis of our results, we constructed and defined the device equipped with LacI-Mit1AD/5lacO-cPAOX1, which had five copies of lacO, as the iTSAD (Fig. 1D). The eGFP output was amplified tremendously (7.2-fold) by iTSAD from the strong constitutive PGAP (PGAP versus PGAP-iTSAD) with glucose, and it even amplified output 5.2-fold from the strong methanol-inducible PAOX1 (PAOX1 versus PGAP_iTSAD) in P. pastoris.
We further investigated the relationship between input strength and iTSAD output strength. Endogenous promoters of P. pastoris—that is, cPAOX1, PICL1, PGPM1, PENO1, and PGAP, categorized from weak to strong with glucose—were selected as input promoters of iTSAD to build the response model (fig. S1A). The highest levels of input and output signals of each promoter were used to plot the correlation curves (Fig. 1E). The regression curve fit well with the Michaelis-Menten equation (fig. S1B). The results revealed that iTSAD can give a high output when driven by a very weak input promoter, which allows for versatile high-expression performance that is compatible with a broad spectrum of input promoters.
CRISPRi-based repressive control of iTSAD
For practical use, inducible gene expression systems that can separate growth and production are favored to relieve the metabolic burden from recombinant proteins (or their catalyzing products) (42–45). The fact that iTSAD can be triggered by a low input signal (Fig. 1E) poses the problem that excessive expression leakage cannot be eluded when adapted with an inducible input promoter. Therefore, additional regulatory strategies for the “ON/OFF” control of iTSAD were needed.
We first integrated CRISPRi-mediated transcriptional repression of iTSAD (Fig. 2A). Since the initial report in 2013 (38, 39), CRISPRi-mediated gene regulation has been applied successfully in various species, including P. pastoris (36, 37). We designed a CRISPRiD with expression of a human codon-optimized dCas9 protein (Streptococcus pyogenes Cas9 nuclease deficient of D10A and H840A) (38) from a strong constitutive promoter PGAP and giRNA (single-guide RNA for interference) from defined input promoters (PR) in P. pastoris. The SV40 nuclear localization sequence (NLS) was fused to the N terminus of dCas9 for nuclear targeting, and two self-cleaving ribozymes, HH and HDV, were added to both ends of the giRNA to remove redundant sequences and prevent nuclear export of giRNA (34, 35). The dCas9/giRNA complex was directed to the cPAOX1, which was used to not only drive the iTSAD but also control the output signal by iTSAD (Fig. 2A). Therefrom, a dual suppression of iTSAD is expected from the steric effect of dCas9 against transcriptional preinitiation complex. We designed six giRNAs that guide dCas9 to various sites with a 5′-NGG-3′ protospacer-adjacent motif (PAM) across cPAOX1 (Fig. 2B, fig. S2A, and table S1). The repressive effects of six giRNAs transcribed by PGAP on intact PAOX1 were first tested, and four of six giRNAs targeting cPAOX1 knocked down eGFP expression by 30 to 60% (fig. S2B). Following this, combinations of three giRNAs (giRNA_F1 and giRNA_R1 targeting the TATA box and giRNA_R2 targeting the transcriptional starting site) were tested for dual suppression of iTSAD. We observed that all six combinations tested knocked down eGFP expression by at least 90% (Fig. 2B). Last, giRNA_F1 was selected for the construction of CRISPRiD considering its high repression efficiency (~96%) and operational convenience as a single giRNA.
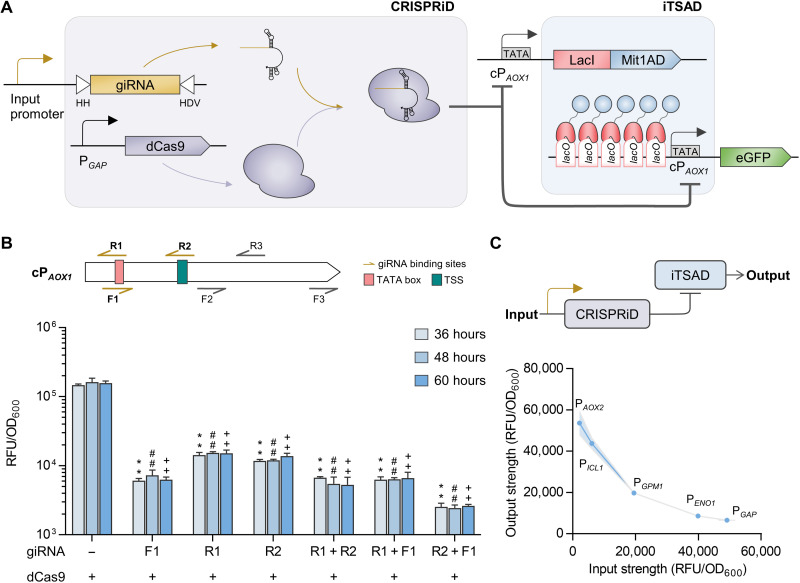
Fig. 2. Development of CRISPRiD to suppress the leaky expression of iTSAD.
(A) Schematic diagram illustrating repression of CRISPRiD on iTSAD. The giRNA was designed to target cPAOX1, which drives both iTSAD and eGFP, thereby recruiting dCas9 to achieve dual suppression of iTSAD by CRISPRiD. (B) Repressive effect of CRISPRiD with different giRNAs on iTSAD. Six giRNAs targeting various sites with a 5′-NGG-3′ PAM sequence across the cPAOX1 were designed to suppress cPAOX1 activity. Detailed information of the binding sites and repressive effects of the giRNAs is elaborated in fig. S2. The three best giRNAs (giRNA_F1, giRNA_R1, and giRNA_R2) and their pairwise combinations were selected to suppress the leaky expression of iTSAD. Statistical significance of eGFP expression in strains with various giRNAs relative to the parent strain without a giRNA at each time point is shown (**P < 0.01 at 36 hours; ##P < 0.01 at 48 hours; ++P < 0.01 at 60 hours). (C) The relationship of input strength and output strength of the CRISPRiD-iTSAD tandem system. A simplified diagram of CRISPRiD-iTSAD system is shown. Five promoters with various strengths were set as input promoters to test the repressive function of CRISPRiD on iTSAD. The regression model is shown in fig. S2. RFU, relative fluorescence unit.
We then evaluated the relationship between the input strength of giRNA_F1 and the amplified output strength using iTSAD. Input promoters of giRNA_F1—that is, PAOX2, PICL1, PGPM1, PENO1, and PGAP, categorized from weak to strong with glucose (fig. S2C)—were used to build the response model. Correlation curves of the input and output signal strengths of each promoter were plotted. The repression intensity continued to increase with the increase in input strength (Fig. 2C). Regression analysis suggested that it fit well with the competitive enzyme inhibition model (fig. S2D). From this model, a weak expression of giRNA_F1 (PAOX2 as input promoter) can notably suppress the output of iTSAD (6-fold decrease compared with the control without giRNA), and a strong input promoter, PGAP, markedly repressed the output by 50-fold. In general, the CRISPRiD-iTSAD tandem system can easily avoid leaky expression by iTSAD; however, it can decrease the upper limit of output.
RNA interaction–mediated derepression regulation against CRISPRiD
As mentioned above, the control of the leaky expression of iTSAD was successfully achieved by CRISPRiD. Therefore, we needed to implement controllable derepression against CRISPRiD to enhance the upper limit of output intensity. RNA-based genetic controllers have been widely recognized owing to their small size (<200 nucleotides), specific binding to ligands, ease of design, and flexible programmability (46). For CRISPRiD, giRNA_F1 is essential for guiding the dCas9 protein for targeted gene regulation. Thus, we attempted to inactivate giRNA_F1 (or its mutant) by RNA interaction to reduce its repressive effect (fig. S3A).
First, two trigger RNAs were designed to interact with the giRNA (fig. S3B and table S1). A short antisense RNA complementary to the DNA-targeting region of giRNA_F1 decreased repression by ~15% (fig. S3C). A cis-hairpin was added to the 5′ end of giRNA_F1 to generate giRNA_F1c, and a riboswitch-like RNA was designed to dimerize with giRNA_F1c (fig. S3B). The newly formed stem-loop obstructed the DNA-targeting region, thereby achieving ~5% derepression of the output signal (fig. S3C). Thus, these trigger RNAs only slightly counteract the repressive effect of CRISPRiD.
We further designed six activator RNAs to disable CRISPRiD by blocking the DNA-targeting region or Cas handle region of the giRNA (Fig. 3A and table S1). The activator RNAs also had Cas protein–recruiting regions that could be used for the design of CRISPRa later. To this end, two CRISPR regulatory proteins orthogonal to dCas9 were selected to avoid cross-talk between CRISPRa and CRISPRi, namely VRER (47) and dCpf1 (48). The VRER protein (mutant of dCas9 with D1135A, G1218R, R1335E, and T1337R) recognizes the PAM sequences 5′-NGCG-3′, which distinguishes it from 5′-NGG-3′ recognized by dCas9. Cpf1, as a class II CRISPR-associated protein, binds to target DNA in the direction of CRISPR RNA (crRNA) without requiring an additional transactivating crRNA, which cleaves target DNA preceded by a short T-rich PAM compared with the G-rich PAM for Cas9 (49). Therefore, dCpf1 (Lachnospiraceae bacterium Cpf1 nuclease deficient with E832A), which recognizes a PAM of 5′-TTTV-3′, was used (Fig. 3B).
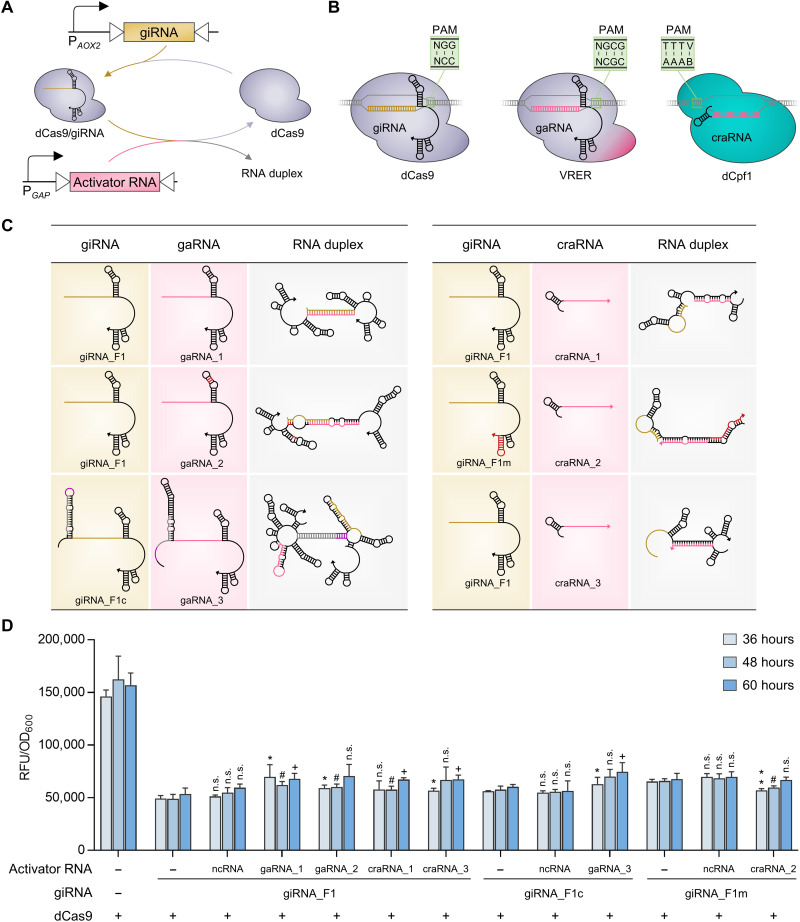
Fig. 3. Design of activator RNAs and their derepression effects on CRISPRiD.
(A) Schematic diagram of activator RNA–mediated interference of CRISPRiD repression. The activator RNA and giRNA were driven by PGAP and PAOX2, respectively. PGAP is much stronger than the PAOX2. (B) dCas regulators orthogonal to dCas9, which recognizes 5′-NGG-3′ PAM sequences, were selected, which were adapted to activator RNAs. These dCas regulators were selected keeping the design of CRISPRa, which is depicted in Fig. 4, in mind. Two dCas regulators were selected to avoid cross-talk between CRISPRa and CRISPRi. The S. pyogenes dCas9 mutant VRER (D1135V/G1218R/R1335E/T1337R), which recognizes 5′-NGCG-3′ PAM sequences, and L. bacterium–derived dCpf1 (E832A), which recognizes 5′-TTTV-3′ PAM sequences, were used. (C) Design of activator RNAs and secondary structure predictions for the duplex giRNAs and activator RNAs (left, gaRNA; right, craRNA). Three gaRNAs and three craRNAs were designed to dimerize with the corresponding giRNA, thereby interfering with the repression of CRISPRiD. The arrow points to the 3′ end of the RNA. Details of RNA design and interactions are described in the Supplementary Materials. (D) Derepression effects on CRISPRiD by the interaction of activator RNA with giRNA. The ncRNA refers to a short RNA without an interaction region with giRNA_F1, giRNA_F1c, or giRNA_F1m. Statistical significance of eGFP expression of each strain with activator RNA relative to the parent strain without activator RNA is shown for each time point (**P < 0.01 and *P < 0.05 at 36 hours; #P < 0.05 at 48 hours; +P < 0.05 at 60 hours; n.s., not significant). RFU, relative fluorescence unit.
Three gaRNAs (guide RNAs for activation) were designed to bind VRER (Fig. 3C). gaRNA_1 and gaRNA_2 interacted with giRNA_F1, and gaRNA_3 interacted with giRNA_F1c. In addition, three craRNAs (crRNAs for activation) were designed to bind dCpf1 (Fig. 3C). craRNA_1 and craRNA_3 interacted with giRNA_F1, and craRNA_2 interacted with giRNA_F1m (a 3′-end stem-loop–mutated giRNA_F1 complementary to the stem-loop of craRNA_2). The activator RNAs driven by PGAP were used for repression interference of the giRNAs driven by PAOX2. The repressive effect of CRISPRiD was slightly relieved by gaRNA_1 (~14%), gaRNA_2 (~16%), gaRNA 3 (~15%), craRNA_1 (~13%), and craRNA_3 (~12%), but not craRNA_2 (failed to function) (Fig. 3D).
For all three giRNAs—that is, giRNA_F1, giRNA_F1c, and giRNA_F1m—knock-in of negative control RNA (ncRNA) that did not interact with the giRNAs showed little effect on the output signal, validating the functions of the regulatory RNAs that were designed to target the giRNAs (Fig. 3D and fig. S3C). Nevertheless, the derepressive effect was quite limited even with high expression levels of activator RNA (by PGAP) but low expression levels of giRNA (by PAOX2) (Fig. 3D). Therefore, high-expression performance of iTSAD by RNA interaction–based derepression of CRISPRiD in this yeast host was not achieved.
gaRNA/craRNA-based CRISPRaD for activation of iTSAD
We further attempted to antagonize CRISPRiD using CRISPRaD. The feasibility of CRISPRa on cPAOX1 was first tested. Chimeric activators were first constructed by fusion of dCas9 with Mit1AD, Mxr1AD, and a codon-optimized viral activator VP16. The SV40 NLS was fused to the N terminus of the synthetic transactivators for nuclear localization. We then targeted these activators to various binding sites with PAM (5′-NGG-3′) sequences that were 0 to 972 base pairs (bp) upstream of the TATA box of cPAOX1 by designing various guide RNAs (gRNA_A1 to gRNA_A8). Bacterial fapO motifs were inserted upstream of cPAOX1 to adjust the distance between the TATA box and gRNA binding sites (fig. S4A). For dCas9-Mit1AD and dCas9-Mxr1AD, we observed no activation effect when directed to any site by gRNA. For dCas9-VP16, only 2 of 14 binding sites functioned, and the site that was 26 bp upstream of the TATA box worked best with this chimeric activator (fig. S4B). This location was then selected as the reference binding region for CRISPRaD with VRER-VP16 or dCpf1-VP16 (Fig. 4A). A preliminary test verified the activity of dCpf1 in P. pastoris, and dCpf1 directed by a crRNA upstream of the TATA box strongly repressed iTSAD (fig. S5).
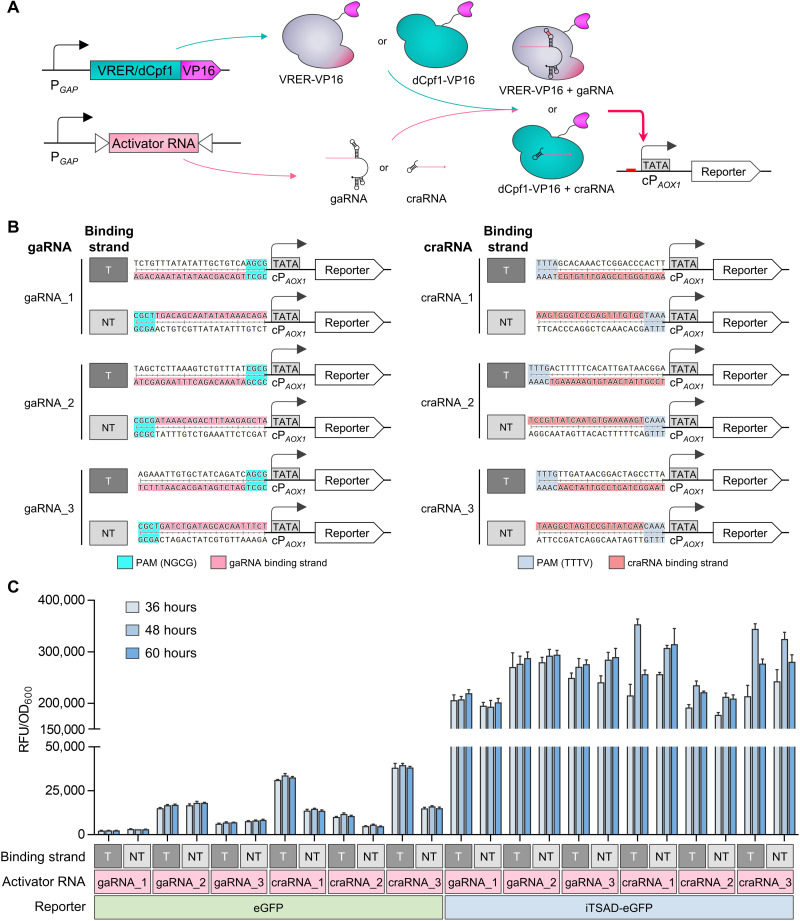
Fig. 4. Design and characterization of CRISPRaD based on VRER or dCpf1.
(A) Schematic diagram illustrating the activation effect of CRISPRaD on cPAOX1-driven reporters. The binding sequences of gaRNAs or craRNAs were inserted upstream of cPAOX1. The viral activator VP16 was fused to the C terminus of VRER or dCpf1, and it recruited RNA polymerase. The reporters are eGFP or iTSAD-driven eGFP. (B) Design of various sequences inserted upstream of the cPAOX1 for the recognition and binding of activator RNA (gaRNA and craRNA). T represents activator RNA binding to the transcribed strand, and NT represents activator RNA binding to the nontranscribed strand. The sequences of gaRNA or craRNA binding sites are highlighted in pink and red, respectively. The PAM sequences of gaRNA and craRNA are highlighted in cyan and blue, respectively. (C) Activation effects of CRISPRaDs on the cPAOX1-driven reporters with different activator RNAs. Each activator RNA was designed to bind with the T-strand and NT-strand by targeting the same base-pairing region. The fluorescence intensities of eGFP and iTSAD-driven eGFP as reporters are shown. RFU, relative fluorescence unit.
With our designed gaRNAs and craRNAs, VRER-VP16 and dCpf1-VP16, respectively, will bind to the upstream target of the TATA box in cPAOX1. Then, the synthetic transactivator will recruit RNA polymerase to cPAOX1, which drives iTSAD. The three craRNAs and three gaRNAs designed for RNA interaction (Fig. 3C) were further tested for their guidance of dCpf1-VP16 and VRER-VP16, respectively. For each craRNA or gaRNA, we designed both a forward (T-strand) and a reverse (NT-strand) sequence of the DNA-targeting region linked with cPAOX1 (Fig. 4B). We first tested the activation function of CRISPRaD on cPAOX1 expressing eGFP. VRER-VP16, targeted by gaRNA_2, functioned well and promoted the expression of eGFP. dCpf1-VP16 worked best when targeted by craRNA_3. Notably, the dCpf1-VP16 targeted by craRNAs at the NT-strand activated cPAOX1 much more than their reverse complements (Fig. 4C), showing binding direction–dependent activation at specific sites. iTSAD was then equipped with a CRISPRaD control. The eGFP output signal was amplified by CRISPRaD with each of the activator RNAs, especially gaRNA_2, craRNA_1, and craRNA_3 (Fig. 4C). In addition, for each craRNA, the cPAOX1-eGFP output with dCpf1-VP16 targeted at the T-strand was stronger than that at the NT-strand, whereas iTSAD-eGFP output with dCpf1-VP16 targeted at the NT-strand was similar to that at the T-strand (Fig. 4C). This illustrated that LacI-Mit1AD expressed with dCpf1-VP16 targeted at the NT-strand was sufficient to support high-level output of eGFP. We lastly selected gaRNA_2 at the NT-strand and craRNA_3 at the T-strand for CRISPRaD in the following constructions.
Reactivation of iTSAD by CRISPRaD from CRISPRiD repression
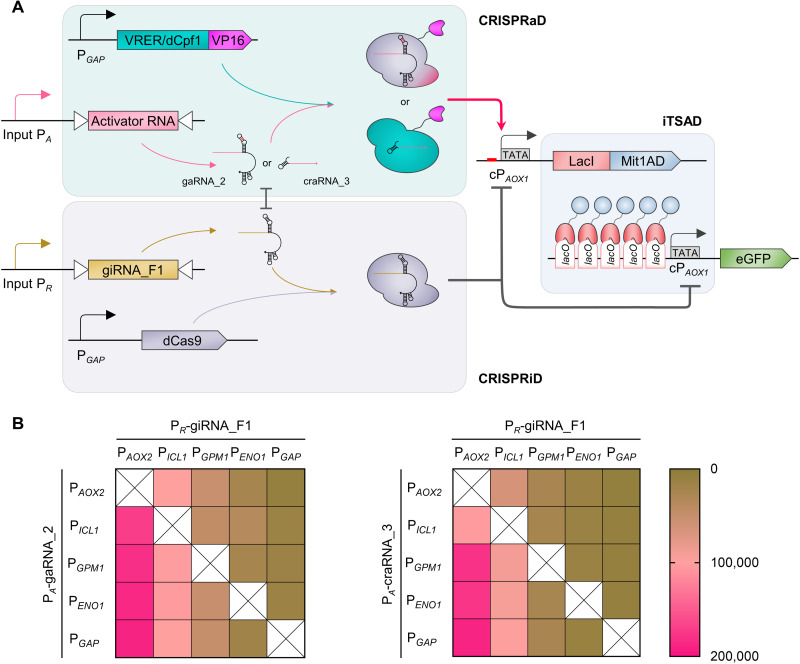
We further aimed to use CRISPRaD to reactivate iTSAD from the repression status caused by CRISPRiD. As mentioned above, both gaRNA_2 and craRNA_3 carried specific sequences complementary to giRNA_F1 (Fig. 3C), which attenuated repression mediated by CRISPRiD (Fig. 3D). Meanwhile, CRISPRaD equipped with gaRNA_2 or craRNA_3 effectively recovered the output intensity of iTSAD (Fig. 4C). Therefore, craRNA_3 and gaRNA_2 serve a dual function of alleviating repression and activation expression. Thus, CRISPRaD mediated by gaRNA_2 or craRNA_3 was assembled with CRISPRiD and iTSAD to construct two sets of synthetic transcription regulatory systems (Fig. 5A).
Fig. 5. Characterization of a synthetic transcription regulatory system integrating iTSAD, CRISPRiD, and CRISPRaD.
(A) Schematic diagram illustrating the assembly of iTSAD, CRISPRiD, and CRISPRaD. Two kinds of CRISPRaD, mediated by VRER-VP16/gaRNA_2 and dCpf1-VP16/craRNA_3, were tested. For this integrated system, the cPAOX1-driven iTSAD was activated by PA-driven CRISPRaD and repressed by PR-driven CRISPRiD. The PA and PR can be selected to respond to various signals, so they can be user-defined. As shown in Fig. 4B, gaRNA_2 was designed to bind the NT-strand upstream of cPAOX1, and craRNA_3 was designed to bind the T-strand upstream of cPAOX1. Activator RNA (gaRNA_2 or craRNA_3) of CRISPRaD mildly interfered with giRNA_F1 of CRISPRiD as shown in Fig. 3D. (B) Heatmaps showing the output strength of the synthetic transcription regulatory system with various input strengths of PA and PR. Left, system with CRISPRaD based on VRER-VP16/gaRNA_2; right, system with CRISPRaD based on dCpf1-VP16/craRNA_3. Five promoters with increasing native strength—that is, PAOX2, PICL1, PGPM1, PENO1, and PGAP—were used as PA and PR. A total of 20 pairwise combinations of these promoters for PA and PR were designed, and 40 strains with different synthetic transcription regulatory systems were constructed. The eGFP expression of the different strains was measured after being cultured for 48 hours in YPD medium.
For the whole construct, we used a relatively strong promoter, PGAP, to constitutively express the chimeric activators and the repressor (dCas9). In contrast, we retained the flexible choice for input promoters that drive the expression of gaRNA_2 or craRNA_3 (defined as PA) and giRNA_F1 (defined as PR). Promoters with increasing strength with glucose—that is, PAOX2, PICL1, PGPM1, PENO1, and PGAP—were adapted for PA and PR. Therefore, a total of 20 constructs with various combinations of PA and PR were obtained and tested for each system (Fig. 5B). The change in PR showed a more obvious influence on the output signal than that of PA. When the PR used was weaker than PICL1, the signal output turned “ON,” indicating that CRISPRiD was not effectively repressing expression. In contrast, when PR was stronger than PGPM1, the signal output changed to “OFF,” indicating that repression by CRISPRiD was sufficient. This was true for both dCpf1-VP16/craRNA_3 and VRER-VP16/gaRNA_2 (Fig. 5B). In general, dCpf1-VP16 worked orthogonal to dCas9 for both repression and activation processes (fig. S6), so it was selected preferentially for the following synthetic expression platform.
Synthetic expression platform intensely and precisely responsive to defined signals
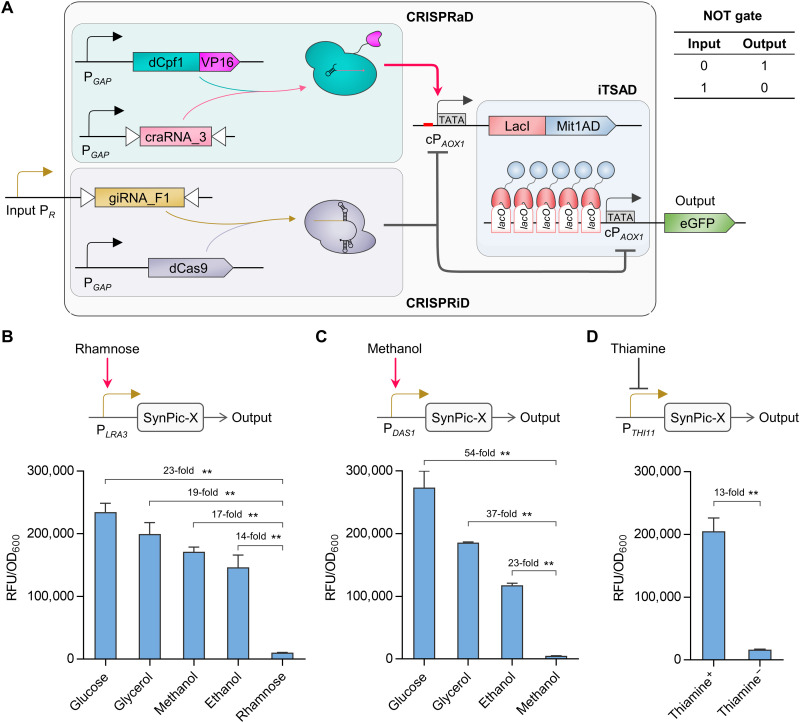
The behavior of the entire system enabled us to design a universal expression platform, SynPic-X, which is responsive to defined signals. The construct with dCpf1-VP16/craRNA_3 was used to create this expression platform. We set PA as PGAP to have a high expression capacity. Then, PR was set as a programmable input promoter in response to a specific factor. Accordingly, a logic “NOT” gate will be implemented between the input of PR and the output of the SynPic-X (Fig. 6A). Theoretically, with the amplification effect of iTSAD, SynPic-X can easily achieve high-level expression that far exceeds the natural P. pastoris expression system. In addition, the combined action of CRISPRiD and CRISPRaD can ensure the precise regulation of SynPic-X (50).
Fig. 6. Development of the synthetic P. pastoris system (SynPic) responsive to defined signals.
(A) Overview of the SynPic-X platform. CRISPRaD mediated by dCpf1-VP16/craRNA_3, CRISPRiD mediated by dCas9/giRNA_F1, and iTSAD were used to set up the SynPic-X platform. A relatively strong promoter PGAP was used to constitutively drive dCpf1-VP16/craRNA_3 in CRISPRaD and dCas9 in CRISPRiD. The input promoter PR was used to drive giRNA_F1 in CRISPRiD. The whole genetic circuit logic of the SynPic-X platform conforms to a logic NOT gate. (B) SynPic-R system derived from SynPic-X with input promoter PLRA3. P. pastoris endogenous rhamnose-inducible promoter, PLRA3, was selected as PR to construct a rhamnose-responsive switch. The output of SynPic-R was repressed with rhamnose but activated with glucose (23-fold), glycerol (19-fold), methanol (17-fold), and ethanol (14-fold). (C) SynPic-M system derived from SynPic-X with input promoter PDAS1. P. pastoris endogenous methanol-inducible promoter, PDAS1, was selected as PR to produce a methanol-responsive switch, which was repressed with methanol but activated with glucose (54-fold), glycerol (37-fold), and ethanol (23-fold). (D) SynPic-T system derived from SynPic-X with input promoter PTHI11. P. pastoris endogenous promoter, PTHI11, which is repressed by thiamine, was used to construct a thiamine-responsive switch. The SynPic-T remained OFF in the absence of thiamine but turned ON after thiamine was added (13-fold). Statistical significance of eGFP expression of each strain in specific carbon sources is shown (**P < 0.01). RFU, relative fluorescence unit.
To further explore the programmability of SynPic-X, we selected three independent endogenous promoters as input promoters. A rhamnose-responsive promoter, PLRA3 (50), was first assembled into SynPic-X as the PR, thereby obtaining a rhamnose-responsive switch designated as SynPic-R. SynPic-R was OFF with rhamnose but ON with other substrates tested, including glucose (23-fold activation), glycerol (19-fold activation), methanol (17-fold activation), and ethanol (14-fold activation) (Fig. 6B). SynPic-R also showed dose-dependent behavior with a 24-fold expression difference in response to rhamnose concentrations ranging from 10−5 to 20 g/liter (fig. S7A). Similarly, PDAS1, which was activated by methanol but repressed by other carbon sources (15), was assembled to produce SynPic-M. It was OFF with methanol but ON with glucose (54-fold activation), glycerol (37-fold activation), and ethanol (23-fold activation) (Fig. 6C). As cells grew slowly (<0.2 g/liter) or were impaired with methanol (>1.0 g/liter), dose-dependent expression was not tested, but it presented a carbon source dose-independent expression mode. This indicated that SynPic-M was rigorously regulated by methanol repression and nonmethanol derepression (fig. S7B). In addition, a thiamine (vitamin B1)–repressible promoter, PTHI11 (51), was used to construct SynPic-T. As expected, SynPic-T showed thiamine-inducible performance and remained OFF in the absence of thiamine and turned ON upon the addition of thiamine (Fig. 6D). SynPic-T showed a good “S” curve induction mode with 13-fold activation in response to thiamine concentrations ranging from 4 × 10−8 to 4 mM. Notably, the eGFP expression levels of SynPic-R, SynPic-M, and SynPic-T reached up to 3.8-fold, 4.4-fold, and 3.3-fold of PAOX1, respectively, which is the most widely used methanol-inducible promoter in commercial P. pastoris systems (Figs. 6, B to D, and 1D). In summary, we built a synthetic P. pastoris expression platform SynPic-X, which can achieve an intense and precise response to various defined signals by adjusting PR.
Protein production by SynPic-R in bioreactor fermentation
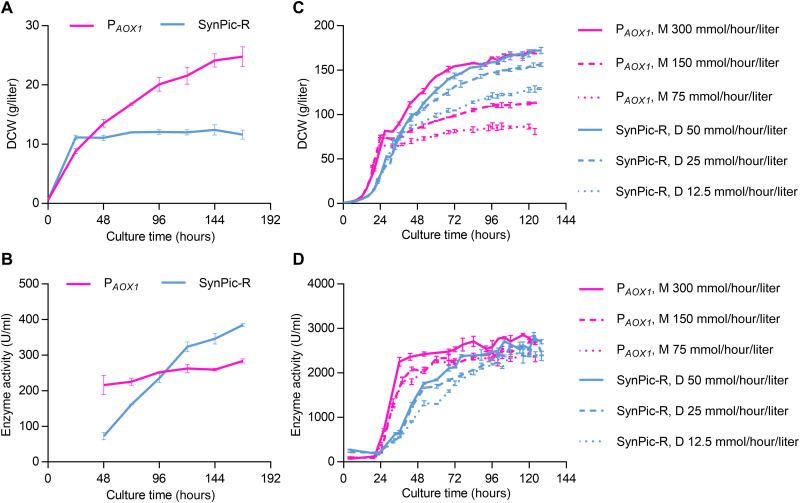
We lastly selected SynPic-R to express industrial α-amylase (52) to evaluate the practical performance of SynPic-X in protein synthesis applications. One copy of the α-amylase–coding gene was expressed with SynPic-R in comparison to the commercial system (GS115 host/methanol-inducible PAOX1). In shake flasks, SynPic-R cells grew weakly with glucose compared to GS115 growth with methanol (Fig. 7A), whereas α-amylase production by SynPic-R was 1.4-fold higher than that of the GS115/PAOX1 system (Fig. 7B). In the bioreactor, however, SynPic-R cells grew better with glucose than GS115 with methanol with an identical feeding rate of carbon (methanol of 75 mmol/hour per liter versus glucose of 12.5 mmol/hour per liter; methanol of 150 mmol/hour per liter versus glucose of 25 mmol/hour per liter), and cell growth reached similar levels with sufficient supply of carbon sources (methanol of 300 mmol/hour per liter versus glucose of 50 mmol/hour per liter) (Fig. 7C). Therefore, the involvement of multiple biological components in SynPic-R did not affect cell growth at the bioreactor scale, indicating its potential for industrial application. The production of α-amylase by SynPic-R with glucose reached a level comparable to that of GS115/PAOX1 with methanol (Fig. 7D). As a tightly regulated promoter, PAOX1 was strongly repressed by fermentable carbon sources but was highly induced by methanol (10–12). This was also true for α-amylase expression under the control of this promoter. Comparatively, SynPic-R also showed strong rhamnose repression and glucose induction in the bioreactor (Fig. 7D). After glucose feeding, the production of α-amylase increased quickly until it reached a saturation level, which was similar to that of GS115/PAOX1 under methanol feeding.
Fig. 7. Comparison of α-amylase production driven by PAOX1 and SynPic-R.
(A) Growth and (B) enzyme activity of yeast strains expressing α-amylase by GS115/PAOX1 or SynPic-R in shake flasks. Methanol and glucose were used as the sole carbon source to induce GS115/PAOX1 and SynPic-R, respectively. (C) Growth and (D) enzyme activity of yeast strains expressing α-amylase by GS115/PAOX1 or SynPic-R in 3-liter bioreactors fermented under different feeding rates. For α-amylase expression by PAOX1, the fermentation strategy included a glycerol-fed batch phase and a methanol (M) induction phase. For α-amylase expression by SynPic-R, the fermentation strategy included a rhamnose-fed batch phase and a glucose (D) induction phase. The yeast strains were grown in the basal salt medium with glycerol or rhamnose feeding to a dry cell weight of about 67 g/liter and then were switched to the induction phase. Three different feeding rates of methanol and glucose were used with consistent carbon content in the induction phase. Broth samples were collected every 24 hours in a shake flask and every 4 to 8 hours in a bioreactor. Error bars at some time points are small and covered by the time curves. Detailed medium and culture methods are described in Materials and Methods. DCW, dry cell weight.
DISCUSSION
Yeast expression hosts are well known for their versatility in biopharmaceutical and biomanufacturing applications. Therefore, the methylotrophic yeast P. pastoris has become a preferred host for protein production in academia and industry. For a long time, scientists have been pursuing a universal P. pastoris chassis with a high gene expression capacity and flexible regulation mode; however, ideal results have not been achieved. Promoter engineering of the strong PAOX1 has produced a series of promoter libraries that achieve 6 to 160% intensity of the wild type, but it is generally difficult to override the regulation mode present in P. pastoris (18, 20, 21, 53, 54). Modifications of specific transcription factors altered the regulation mode, but it was still difficult to markedly increase gene expression in wild-type P. pastoris (23, 24, 55). The rapid development of synthetic biology technology has led to new ideas and powerful tools to fully rebuild this workhorse. Accordingly, a synthetic expression system, SynPic-X, was constructed, which can achieve impressive levels of transcription in response to various defined signals (Fig. 5). In particular, a series of derived regulatory switches—that is, SynPic-R, SynPic-M, and SynPic-T (Fig. 6)—exemplified the programmable regulation mode of SynPic-X.
The function of iTSAD is critical for the extraordinary transcriptional activity of SynPic-X, which can be attributed to the performance of the chimeric activator LacI-Mit1AD. As we previously reported, Mit1 is an essential transcription activator that acts downstream of the methanol regulatory network and is crucial for PAOX1 activity (30). Mit1AD has the outstanding ability to recruit RNA polymerase, thereby allowing high output levels by iTSAD. This represents a successful case of transcriptional clarification–based synthetic biological systems. The compatibility of iTSAD with other yeasts and eukaryotic cells is worth testing in the future. Our previous study showed that LacI fused with the versatile viral transactivator VP16 led to very weak transcriptional activity in comparison to LacI-Mit1AD (41). However, in this case, the activation effect of CRISPRa on cPAOX1 was achieved only when VP16 was used. Neither Mit1AD nor Mxr1AD fused with dCas9 showed an activation effect (fig. S4). Therefore, the compatibility between DBP and TFAD is crucial for the efficacy of synthetic biological components. Updated versions of DBP and TFAD may bring further breakthroughs in transcriptional intensity, which is worthy of in-depth exploration.
In addition to iTSAD, our constructed CRISPRiD and CRISPRaD worked together efficiently to suppress the leaky expression of iTSAD and have a high level of output simultaneously (Figs. 5 and 6). The combined use of the three devices ensured precise and efficient expression control in SynPic-X. For engineered biological systems, it is usually an inevitable contradiction to improve the expression ability and reduce the leakage level simultaneously. This study demonstrates an efficient de novo assembly strategy to solve this problem and provides valuable reference and ideas for the design and construction of genetic circuits in P. pastoris and other chassis hosts. In addition, RNA-mediated interactions between CRISPRiD and CRISPRaD were used to improve the fineness and strictness of signal regulation. Although similar strategies have been reported to notably interfere with the function of gRNA in E. coli (56–58), RNA interaction had a weak effect in this study. This could be ascribed to inefficient pairing or an insufficient difference in quantity of the two types of RNAs. As RNA regulation provides a quick and flexible response, the RNA regulatory strategy can be further investigated to explore sister platforms of SynPic-X in the future.
The behavior of SynPic-X is consistent with the logic NOT gate, in which input signal is inverted and amplified for output. The regulation mode of SynPic-X can be determined by screening natural promoters that are responsive to specific factors as input signals. In the present study, three different endogenous promoters were used for proof of concept, and the derived SynPic-R, SynPic-M, and SynPic-T all showed high expression and desired performance. The expression level of SynPic-R can be fine-tuned by changing rhamnose input, and the expression level of SynPic-T can be sharply increased by feeding a small amount of thiamine. Heterologous expression of industrial α-amylase by SynPic-R at the bioreactor scale allowed tight induction regulation, better cell growth, and comparable production levels to the commercial system of P. pastoris GS115 with methanol-inducible PAOX1. It represents a green and safe process with glucose instead of methanol. In addition, multiple-copy gene expression has been proven as an effective strategy to increase production capacity of recombinant proteins in P. pastoris. This strategy is theoretically compatible with our SynPic-X platform, which can be achieved via integration of multiple expression cassettes of gene of interest (5lacO-cPAOX1-GOI) into the genome. Protein folding and modification in endoplasmic reticulum and Golgi apparatus could also be limiting factors for the production of functional proteins. To extend the industrial use of SynPic-X, it is necessary to test more complicated proteins with multiple disulfide bonds or posttranslational modifications. The SynPic-X may be further updated by improving protein folding and processing in the future.
P. pastoris has been used over the years because of its potency as a synthetic biological chassis for small-molecule production and one-carbon assimilation (9, 42). The reported genome of P. pastoris has allowed for the identification of open reading frames and promoters (8, 59, 60). Synthetic promoters also provide more choices for broader applications (15, 53, 61). It is expected that an increasing number of promoters responding to various signals will be identified and characterized using bioinformatics and biochemical technologies. These promoters may be assembled into SynPic-X to generate updating expression systems for various application scenarios. In conclusion, a powerful and flexible yeast gene expression system was proposed and constructed in this study, which represents a previously unidentified “plug and play” platform to produce customized high-level expression hosts that respond to defined signals.
MATERIALS AND METHODS
Strains and growth conditions
The strains and plasmids used in this study are listed in table S2. Yeast recombinant strains were constructed from P. pastoris GS115 (Invitrogen) and Δku70 (KU70 deficient in GS115 for the nonhomologous end joining defective purpose) (32). P. pastoris GS_PAOX1-G (21), GS_PAOX1-Amy (52), and S. cerevisiae BY4741 (41) were stored in our laboratory. All strains were stored at −80°C in medium with 20% (v/v) glycerol. E. coli Top10 was used for the construction and propagation of plasmids. Yeast expression vectors pPIC3.5 K and pGAPZ B were purchased from Invitrogen. The plasmid BB3eN_14 was purchased from Addgene (#98549). The pAG32 (30), pP-PAOX1G (pP-GFP) (21), pGZB_cPAOX1-GFP (62), pP-PGAPG (pPGG), pZ_PICL1-LacI-VP16 (41), 3.5k-TEF1-gRNA1, pDTg1-npgA (32), and pPIC9k-Amy (52) were constructed in our laboratory. The plasmids p414-TEF1p-Cas9-CYC1t and pET28TEV-LbCpf1 were provided by F. Wang and G. Tan in our university, respectively.
MGY (0.67% YNB and 1% glycerol) or YPD (2% tryptone, 1% yeast extract, and 2% glucose) medium was used for screening P. pastoris transformants, and the appropriate antibiotics [zeocin (100 μg/ml), hygromycin B (750 μg/ml), and nourseothricin (100 μg/ml)] were added when required. P. pastoris transformants were cultured and incubated at 30°C (200 rpm). E. coli cells were cultured at 37°C (200 rpm) in low-salt LB (LLB) medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl) supplemented with ampicillin (100 μg/ml) or zeocin (50 μg/ml). Cells were cultivated in an incubator shaker (type of MQD B2R, Shanghai Minquan Instrument Co. Ltd., China). In addition, YPG (2% tryptone, 1% yeast extract, and 2% glycerol), YPR (2% tryptone, 1% yeast extract, and 2% rhamnose), YNE (0.67% YNB and 1% ethanol), and YNM (0.67% YNB and 1% methanol) were used to verify the function of SynPic-R and SynPic-M. For SynPic-T, the recombinant strain was cultured in a synthetic medium [2% glycerol, 2% (NH4)2SO4, 1.2% KH2PO4, 0.47% MgSO4·7H2O, and 0.036% CaCl2, plus trace elements: 0.2 μM CaSO4·5H2O, 1.25 μM NaI, 4.5 μM MnSO4·4H2O, 2 μM Na2MoO4·2H2O, 0.75 μM H3BO3, 17.5 μM ZnSO4·7H2O, and 44.5 μM FeCl3·6H2O (pH 5.5)] and different concentrations of thiamine in the form of thiaminechloride-hydrochloride. BMMY and BMDY media (pH 6.0) contained 2% tryptone, 1% yeast extract, 1.34% YNB, biotin (0.4 μg/ml), and 100 mM potassium phosphate buffer, plus either 1% methanol (BMMY) or 2% glucose (BMDY), which were used for α-amylase expression in shake flasks.
Plasmid construction
Oligonucleotides used for construction of plasmids were synthesized by Suzhou Genewiz Biotech Co. Ltd., China and are listed in table S3. Plasmids used in this study were constructed using conventional restriction enzyme cloning and/or Gibson Assembly (ClonExpress II one-step cloning kit, VazymeBiotech Co. Ltd., China). The sequences of all plasmids were verified using Sanger sequencing. Other molecular biology operations of E. coli and P. pastoris were performed as described previously (32, 41, 42).
Genetic device assembly
As shown in Fig. 1, the plasmids harboring the egfp gene were constructed from pP-PAOX1G and integrated into the P. pastoris genome at the HIS4 locus. The plasmids expressing DBP-TFAD fusion protein were constructed using pGAPZ B and integrated into the P. pastoris genome at the PGAP locus. In Fig. 1E and fig. S1, the plasmids harboring different input promoters were constructed using pGZB_cPAOX1-GFP and integrated into the P. pastoris genome at the PGAP locus for the detection of input and output strength. As shown inFigs. 2 to 4, the LacI-Mit1AD cassette driven by cPAOX1 was inserted into the plasmid pPlacO5cAG, and the resulting plasmid pPcALMO5 was integrated into the P. pastoris genome at the HIS4 locus. The cassettes of CRISPR regulators (dCas9, VRER-VP16, and Cpf1-VP16) were inserted into pGAPZ B, and the resulting plasmids were integrated into the P. pastoris genome at the PGAP locus. Plasmids harboring various giRNAs and/or activator RNA cassettes were constructed using the pAG32 plasmid and integrated into the P. pastoris genome at the PAOX1 locus. InFigs. 1 to 4, all strains were constructed on the basis of the wild-type P. pastoris GS115, and inFigs. 5 to 7, the Δku70 strain (knockout of KU70 in P. pastoris GS115) was used to construct the yeast strains. Plasmids containing the α-amylase cassette were integrated into the P. pastoris genome by the PENO1 locus. The α-factor signal peptide was fused to α-amylase for secretion expression. Construct details for plasmids and strains are described in the Supplementary Materials.
Microplate reader experiments
Yeast strains were transferred from −80°C stock cultures and incubated in 2 ml of YPD medium for 2 days. The cells were inoculated into fresh YPD medium for preculture to an optical density at 600 nm (OD600) of 6.0. The cells were harvested by centrifugation (5000g, 5 min), washed twice with sterile water, and then transferred to the required medium at an OD600 of 1.0 in a 24-well plate. Culture samples were collected at the desired time points for fluorescence detection. The samples were washed twice with sterile water and stored at −80°C. Reporter eGFP (intracellular expression) fluorescence (normalized with OD600) with various samples was analyzed using a multimode microplate reader (Synergy 2, BioTek Instruments, USA) at an excitation wavelength of 485 nm and an emission wavelength of 525 nm (gain, 60).
Production and activity assays of α-amylase
In shake flask cultures, the strain producing α-amylase (52) driven by PAOX1 was precultured in YPD medium and induced in BMMY medium, and methanol was added every 24 hours to a final concentration of 1% (v/v). The strain producing α-amylase driven by SynPic-R was precultured in YPR medium and induced in BMDY medium, and glucose was added every 24 hours to a final concentration of 2% (w/v). Fermentation in 3-liter bioreactors was adapted from the previous study (24, 41). The strain producing α-amylase driven by PAOX1 grew in basal salt medium (24) with glycerol (40 g/liter). After the initial glycerol was exhausted, it continued to feed at a rate of 10 g/hour per liter until the dry cell weight reached about 67 g/liter and then stopped until glycerol was used up (~0.5 hours). Afterward, it was switched to the methanol-feeding phase. Methanol was initially fed at a low level for cellular adaptation and then increased to the required feeding rates gradually (table S4). The strain producing α-amylase driven by SynPic-R grew in the basal salt medium with rhamnose (40 g/liter). After the initial rhamnose was exhausted, it continued to feed at a rate of 10 g/hour per liter until the dry cell weight reached about 67 g/liter and then was switched to the glucose-feeding phase with the required feeding rates. The pH was kept at 5.0, using ammonia, and the dissolved oxygen level was kept above 30% saturation. For both experiments, in shake flasks and 3-liter bioreactors, the culture supernatants were collected at specific time points, and the enzyme activity of α-amylase was determined by the dinitrosalicylic acid (DNS) method as previously reported (63).
Prediction of RNA secondary structure
The secondary structures of various RNAs used in this study were predicted using the RNAstructure software version 6.0.1 (http://rna.urmc.rochester.edu/RNAstructureWeb/) (64). The secondary structure of RNA duplex was predicted using the biofold tool. The main parameters were set as follows: nucleic acid type = RNA and temperature = 30°C. The full sequences of various RNAs are given in table S1.
Statistical analyses
Data were obtained from three biological replicates from at least three experimental batches and presented as means ± SD. Data were analyzed and fitted using GraphPad Prism (version 7.04). The unpaired, two-tailed Student’s t test was used to assess the differences among the grouped data. Statistical significance was set at P < 0.05 and P < 0.01.
Acknowledgments
We thank G. Tan and F. Wang in our university for providing the plasmids of pET28TEV-LbCpf1 and p414-TEF1p-Cas9-CYC1t, respectively.
Funding: This work was funded by the National Natural Science Foundation of China (31870073), National Key R&D Program of China (2018YFC1706200, 2020YFA0907800), Shanghai Rising-Star Program, China (19QA1402700), and the Fundamental Research Funds for Central Universities (2222012016019).
Author contributions: Conceptualization: M.C. and Q.L. Funding acquisition: M.C. Experiments: Q.L., L.S., Q.P., Q.Z., X.S., and M.X. Project administration: M.C. Supervision: M.C. Writing—original draft: M.C. and Q.L. Writing—review and editing: M.C., Q.W., and Y.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S7
Tables S1 to S4
References
REFERENCES AND NOTES
- 1.Çelik E., Çalık P., Production of recombinant proteins by yeast cells. Biotechnol. Adv. 30, 1108–1118 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Ergün B. G., Hüccetoğulları D., Öztürk S., Çelik E., Çalık P., Established and upcoming yeast expression systems. Methods Mol. Biol. 1923, 1–79 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Baghban R., Farajnia S., Rajabibazl M., Ghasemi Y., Mafi A. A., Hoseinpoor R. H. R., Rahbarnia L., Aria M., Yeast expression systems: Overview and recent advances. Mol. Biotechnol. 61, 365–384 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Ward O. P., Production of recombinant proteins by filamentous fungi. Biotechnol. Adv. 30, 1119–1139 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Wang Q., Zhong C., Xiao H., Genetic engineering of filamentous fungi for efficient protein expression and secretion. Front. Bioeng. Biotechnol. 8, 293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davy A. M., H. F. K., Andersen M. R., Cell factory engineering. Cell Syst. 4, 262 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Cregg J. M., Madden K. R., Development of the methylotrophic yeast, Pichia pastoris, as a host system for the production of foreign proteins. Dev. Ind. Microbiol. 29, 33–41 (1988). [Google Scholar]
- 8.de Schutter K., Lin Y. C., Tiels P., Hecke A. V., Glinka S., Weber-Lehmann J., Rouzé P., van de Peer Y., Callewaert N., Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 27, 561–566 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Gassler T., Sauer M., Gasser B., Egermeier M., Steiger M. G., The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 38, 210–216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarzhans J. P., Luttermann T., Geier M., Kalinowski J., Friehs K., Towards systems metabolic engineering in Pichia pastoris. Biotechnol. Adv. 35, 681–710 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Peña D. A., Gasser B., Zanghellini J., Steiger M. G., Mattanovich D., Metabolic engineering of Pichia pastoris. Metab. Eng. 50, 2–15 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Yang Z., Zhang Z., Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: A review. Biotechnol. Adv. 36, 182–195 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Perez-Pinera P., Han N., Cleto S., Cao J., Purcell O., Shah K. A., Lee K., Ram R., Lu T. K., Synthetic biology and microbioreactor platforms for programmable production of biologics at the point-of-care. Nat. Commun. 7, 12211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bill R. M., Playing catch-up with Escherichia coli: Using yeast to increase success rates in recombinant protein production experiments. Front. Microbiol. 5, 85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogl T., Sturmberger L., Kickenweiz T., Wasmayer R., Schmid C., Hatzl A. M., Gerstmann M. A., Pitzer J., Wagner M., Thallinger G. G., Geier M., Glieder A., A toolbox of diverse promoters related to methanol utilization: Functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth. Biol. 5, 172–186 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Gasser B., Steiger M. G., Mattanovich D., Methanol regulated yeast promoters: Production vehicles and toolbox for synthetic biology. Microb. Cell Factories 14, 196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasser B., Mattanovich D., A yeast for all seasons—Is Pichia pastoris a suitable chassis organism for future bioproduction? FEMS Microbiol. Lett. 365, 365 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Hartner F. S., Ruth C., Langenegger D., Johnson S. N., Glieder A., Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 36, e76 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redden H., Alper H. S., The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 6, 7810 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogl T., Glieder A., Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnol. 30, 385–404 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Xuan Y., Zhou X., Zhang W., Xiao Z., Song Z., Zhang Y., An upstream activation sequence controls the expression of AOX1 gene in Pichia pastoris. FEMS Yeast Res. 9, 1271–1282 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Yuan T., Guo Y., Dong J., Li T., Zhou T., Sun K., Zhang M., Wu Q., Xie Z., Cai Y., Cao L., Dai J., Construction, characterization and application of a genome-wide promoter library in Saccharomyces cerevisiae. Front. Chem. Sci. Eng. 11, 107–116 (2017). [Google Scholar]
- 23.Vogl T., Sturmberger L., Fauland P. C., Hyden P., Fischer J. E., Schmid C., Thallinger G. G., Geier M., Glieder A., Methanol independent induction in Pichia pastoris by simple derepressed overexpression of single transcription factors. Biotechnol. Bioeng. 115, 1037–1050 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Wang X., Shi L., Qi F., Zhang P., Zhang Y., Zhou X., Song Z., Cai M., Methanol-independent protein expression by AOX1 promoter with trans-acting elements engineering and glucose-glycerol-shift induction in Pichia pastoris. Sci. Rep. 7, 41850 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cámara E., Monforte S., Albiol J., Ferrer P., Deregulation of methanol metabolism reverts transcriptional limitations of recombinant Pichia pastoris (Komagataella spp) with multiple expression cassettes under control of the AOX1 promoter. Biotechnol. Bioeng. 116, 1710–1720 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Zhao E. M., Zhang Y., Mehl J., Park H., Lalwani M. A., Toettcher J. E., Avalos J. L., Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 555, 683–687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rantasalo A., Landowski C. P., Kuivanen J., Korppoo A., Reuter L., Koivistoinen O., Valkonen M., Penttilä M., Jäntti J., Mojzita D., A universal gene expression system for fungi. Nucleic Acids Res. 46, e111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin-Cereghino G. P., Godfrey L., de la Cruz B. J., Johnson S., Khuongsathiene S., Tolstorukov I., Yan M., Lin-Cereghino J., Veenhuis M., Subramani S., Cregg J. M., Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol. Cell. Biol. 26, 883–897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahu U., Rao K. K., Rangarajan P. N., Trm1p, a Zn(II)2Cys6-type transcription factor, is essential for the transcriptional activation of genes of methanol utilization pathway, in Pichia pastoris. Biochem. Biophys. Res. Commun. 451, 158–164 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Wang X., Wang Q., Wang J., Bai P., Shi L., Shen W., Zhou M., Zhou X., Zhang Y., Cai M., Mit1 transcription factor mediates methanol signaling and regulates the Alcohol Oxidase 1 (AOX1) promoter in Pichia pastoris. J. Biol. Chem. 291, 6245–6261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., Cai M., Shi L., Wang Q., Zhu J., Wang J., Zhou M., Zhou X., Zhang Y., PpNrg1 is a transcriptional repressor for glucose and glycerol repression of AOX1 promoter in methylotrophic yeast Pichia pastoris. Biotechnol. Lett. 38, 291–298 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Liu Q., Shi X., Song L., Liu H., Zhou X., Wang Q., Zhang Y., Cai M., CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Factories 18, 144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prielhofer R., Barrero J. J., Steuer S., Gassler T., Zahrl R., Baumann K., Sauer M., Mattanovich D., Gasser B., Marx H., GoldenPiCS: A Golden Gate-derived modular cloning system for applied synthetic biology in the yeast Pichia pastoris. BMC Syst. Biol. 11, 123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weninger A., Hatzl A. M., Schmid C., Vogl T., Glieder A., Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 235, 139–149 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Weninger A., Fischer J. E., Raschmanová H., Kniely C., Vogl T., Glieder A., Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J. Cell. Biochem. 119, 3183–3198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y., Liu G., Chen X., Liu M., Zhan C., Liu X., Bai Z., High efficiency CRISPR/Cas9 genome editing system with an eliminable episomal sgRNA plasmid in Pichia pastoris. Enzym. Microb. Technol. 138, 109556 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Baumschabl M., Prielhofer R., Mattanovich D., Steiger M. G., Fine-tuning of transcription in Pichia pastoris using dCas9 and RNA scaffolds. ACS Synth. Biol. 9, 3202–3209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., Lim W. A., Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., Stern-Ginossar N., Brandman O., Whitehead E. H., Doudna J. A., Lim W. A., Weissman J. S., Qi L. S., CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng A. W., Wang H., Yang H., Shi L., Katz Y., Theunissen T. W., Rangarajan S., Shivalila C. S., Dadon D. B., Jaenisch R., Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Bai C., Liu Q., Xu Q., Qian Z., Peng Q., Yu J., Xu M., Zhou X., Zhang Y., Cai M., Engineered ethanol-driven biosynthetic system for improving production of acetyl-CoA derived drugs in Crabtree-negative yeast. Metab. Eng. 54, 275–284 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Liu Y., Tu X., Xu Q., Bai C., Kong C., Liu Q., Yu J., Peng Q., Zhou X., Zhang Y., Cai M., Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab. Eng. 45, 189–199 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Blazeck J., Alper H. S., Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 8, 46–58 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Heyland J., Fu J., Blank L. M., Schmid A., Carbon metabolism limits recombinant protein production in Pichia pastoris. Biotechnol. Bioeng. 108, 1942–1953 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Cámara E., Landes N., Albiol J., Gasser B., Mattanovich D., Ferrer P., Increased dosage of AOX1 promoter-regulated expression cassettes leads to transcription attenuation of the methanol metabolism in Pichia pastoris. Sci. Rep. 7, 44302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jang S., Jang S., Yang J., Seo S. W., Jung G. Y., RNA-based dynamic genetic controllers: Development strategies and applications. Curr. Opin. Biotechnol. 53, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleinstiver B. P., Prew M. S., Tsai S. Q., Topkar V. V., Nguyen N. T., Zheng Z., Gonzales A. P., Li Z., Peterson R. T., Yeh J. R., Aryee M. J., Joung J. K., Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lian J., HamediRad M., Hu S., Zhao H., Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat. Commun. 8, 1688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zetsche B., Gootenberg J. S., Abudayyeh O. O., Slaymaker I. M., Makarova K. S., Essletzbichler P., Volz S. E., Joung J., van der Oost J., Regev A., Koonin E. V., Zhang F., Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu B., Zhang Y., Zhang X., Yan C., Zhang Y., Xu X., Zhang W., Discovery of a rhamnose utilization pathway and rhamnose-inducible promoters in Pichia pastoris. Sci. Rep. 6, 27352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landes N., Gasser B., Vorauer-Uhl K., Lhota G., Mattanovich D., Maurer M., The vitamin-sensitive promoter PTHI11 enables pre-defined autonomous induction of recombinant protein production in Pichia pastoris. Biotechnol. Bioeng. 113, 2633–2643 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Huang M., Gao Y., Zhou X., Zhang Y., Cai M., Regulating unfolded protein response activator HAC1p for production of thermostable raw-starch hydrolyzing α-amylase in Pichia pastoris. Bioprocess Biosyst. Eng. 40, 341–350 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Portela R. M., Vogl T., Kniely C., Fischer J. E., Oliveira R., Glieder A., Synthetic core promoters as universal parts for fine-tuning expression in different yeast species. ACS Synth. Biol. 6, 471–484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Portela R. M. C., Vogl T., Ebner K., Oliveira R., Glieder A., Pichia pastoris alcohol oxidase 1 (AOX1) core promoter engineering by high resolution systematic mutagenesis. Biotechnol. J. 13, e1700340 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Chang C.-H., Hsiung H.-A., Hong K.-L., Huang C.-T., Enhancing the efficiency of the Pichia pastoris AOX1 promoter via the synthetic positive feedback circuit of transcription factor Mxr1. BMC Biotechnol. 18, 81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins S. P., Rostain W., Liao C., Beisel C. L., Sequence-independent RNA sensing and DNA targeting by a split domain CRISPR-Cas12a gRNA switch. Nucleic Acids Res. 49, 2985–2999 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siu K. H., Chen W., Riboregulated toehold-gated gRNA for programmable CRISPR-Cas9 function. Nat. Chem. Biol. 15, 217–220 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Lee Y. J., Hoynes-O’Connor A., Leong M. C., Moon T. S., Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system. Nucleic Acids Res. 44, 2462–2473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Küberl A., Schneider J., Thallinger G. G., Anderl I., Wibberg D., Hajek T., Jaenicke S., Brinkrolf K., Goesmann A., Szczepanowski R., Pühler A., Schwab H., Glieder A., Pichler H., High-quality genome sequence of Pichia pastoris CBS7435. J. Biotechnol. 154, 312–320 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Sturmberger L., Chappell T., Geier M., Krainer F., Day K. J., Vide U., Trstenjak S., Schiefer A., Richardson T., Soriaga L., Darnhofer B., Birner-Gruenberger R., Glick B. S., Tolstorukov I., Cregg J., Madden K., Glieder A., Refined Pichia pastoris reference genome sequence. J. Biotechnol. 235, 121–131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogl T., Kickenweiz T., Pitzer J., Sturmberger L., Weninger A., Biggs B. W., Köhler E. M., Baumschlager A., Fischer J. E., Hyden P., Wagner M., Baumann M., Borth N., Geier M., Ajikumar P. K., Glieder A., Engineered bidirectional promoters enable rapid multi-gene co-expression optimization. Nat. Commun. 9, 3589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wen J., Tian L., Xu M., Zhou X., Zhang Y., Cai M., A synthetic malonyl-CoA metabolic oscillator in Komagataella phaffii. ACS Synth. Biol. 9, 1059–1068 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Ghose T. K., Measurement of cellulase activities. Pure Appl. Chem. 59, 257–268 (1987). [Google Scholar]
- 64.Mathews D. H., Using the RNAstructure software package to predict conserved RNA structures. Curr. Protoc. Bioinformatics 46, 12.4.1–12.4.22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isaacs F., Dwyer D., Ding C., Pervouchine D. D., Cantor C. R., Collins J. J., Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22, 841–847 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Goldstein A. L., McCusker J. H., Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 (1999). [DOI] [PubMed] [Google Scholar]
- 67.DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., Church G. M., Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang M., Li Z., Wang W., Liu J., Liu L., Zhu G., Karthik L., Wang M., Wang K., Wang Z., Yu J., Shui Y. T., Yu J., Zhang L., Yang Z., Li C., Zhang Q., Shi T., Zhou L., Xie F., Dai H., Liu X., Zhang J., Liu G., Zhuo Y., Zhang B., Liu C., Li S., Xia X., Tong Y., Liu Y., Alterovitz G., Tan G., Zhang L., Zhang L.-X., A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 10, 3672 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen W., Xue Y., Liu Y., Kong C., Wang X., Huang M., Cai M., Zhou X., Zhang Y., Zhou M., A novel methanol-free Pichia pastoris system for recombinant protein expression. Microb. Cell Factories 15, 178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S7
Tables S1 to S4
References