Abstract
To determine the stability of pulsed-field gel electrophoresis (PFGE) patterns of methicillin-resistant Staphylococcus aureus in the nosocomial setting, we analyzed isolates from long-term carriers (>1 month) and from patients involved in well-defined nosocomial epidemics. The number of fragment differences between the first isolate and subsequent isolates in long-term carriers showed a bimodal distribution, with one group having 0 to 6 fragment differences and the other group having 14 to 24 fragment differences. The PFGE patterns of isolates involved in epidemics also presented a similar bimodal distribution of the number of fragment differences. Typing these isolates with another molecular method (inter-IS256 PCR) showed that isolates of the first group (i.e., with 1 to 6 fragment differences) were clonally related, whereas the second group (with 14 to 24 fragment differences) could be considered genetically different. Among long-term carriers with clonally related isolates, 74 of 84 (88%) of consecutive isolates showed indistinguishable patterns, whereas 10 of 84 (12%) showed related patterns differing by one to six fragments. Moreover, the frequency of apparition of related patterns is higher when the time between the first and the subsequent isolate is longer. During seven nosocomial epidemics lasting from 1 to 15 months, only 2 of 120 isolates (1.7%) showed a pattern which was different, although related, from the predominant one involved in each of these outbreaks.
Pulsed-field gel electrophoresis (PFGE) analysis of genomic macrorestriction DNA fragments is one of the most commonly used methods for the epidemiological typing of many bacteria. When the method is used for the investigation of an epidemic, it has been proposed that the so-called “related” isolates should be considered as probably (a 1- to 4-DNA-fragment difference) or possibly (a 5- to 7-DNA-fragment difference) part of the outbreak (6, 10, 12).
The mutation rate, including point mutations, genetic rearrangements, and horizontal transfers of mobile DNA elements such as phages and transposons, is different from one bacterial species to another. In a given typing system, the mutation rate will directly influence the stability of the typing patterns during the replication cycles of a given bacterial clone. Thus, for a given bacterium, the epidemiological interpretation of related PFGE patterns will depend on the time and space scales that one considers (2, 9, 11, 13). For a species which has a high mutation rate, isolates with related patterns observed over a short period of time should be considered as sharing recent epidemiological links. On the other hand, if the species has a low mutation rate, isolates with related patterns are likely to have more distant epidemiological links. To evaluate the evolution rate of typing patterns, long-term experiments using microbial in vitro cultures extended over several years have been proposed (11, 13). However, this may not properly reflect the in vivo conditions of microorganism multiplication and transmission, conditions that might influence the mutation rate of the organism, and investigations using natural conditions of growth of the microorganism should also be performed (9, 11, 13).
The purpose of the present study was to examine the stability of PFGE patterns of methicillin-resistant Staphylococcus aureus (MRSA) strains. This was investigated by the analysis of MRSA isolates of long-term carriers and of patients involved in well-defined epidemics observed in acute care hospitals with low (Switzerland) and high (Belgium) MRSA rates. The results allow criteria for the interpretation in epidemic and endemic nosocomial investigations of PFGE results to be set.
MATERIALS AND METHODS
Typing.
PFGE typing was carried out in each center with the same technique (3, 10). Plugs of genomic DNA were obtained by lysis with lysostaphin, followed by a treatment with proteinase K. The DNA was digested with the enzyme SmaI, according to the manufacturer's recommendations. Electrophoresis was performed by using a CHEF DR II or III system (Bio-Rad, Hercules, Calif.) in a TBE 1× buffer; the conditions were 6 V/cm at 12 to 14°C, with alternating pulses at a 120° angle in a 1- to 45-s pulse time gradient for 24 h (Lausanne) or a 5- to 15-s gradient for 10 h and then a 15- to 45-s gradient for 15 h (Brussels). The banding patterns of the different gels were analyzed with the GelCompar software (Applied Math, Kortrijk, Belgium). Inter-IS256 PCR typing was performed on bacterial lysate obtained in a three-step procedure with lysostaphin, proteinase K, and boiling, as already described (4). Amplification of the inter-IS256 region was performed using the Ready-To-Go RAPD beads (Amersham Pharmacia Biotech, Roosendaal, The Netherlands).
PFGE and inter-IS256 PCR patterns were each interpreted independently by two different persons. The number of fragment difference between two patterns was the sum of all fragments present in one pattern but absent in the other. For inter-IS256 PCR patterns, two isolates were considered identical when they differed by 0 or 1 fragment and different when they differed by more than 1 fragment (4).
Long-term MRSA carriers.
were defined as patients from whom two or more MRSA isolates were recovered during a period longer than 1 month, during either the same or different hospital stays. Only one isolate per quarter was analyzed (1 to 3, 3 to 6, 6 to 12, and >12 months [Table 1]). They were identified from the MRSA surveillance database of either the University Hospital of Lausanne (CHUV) or the Erasme University Hospital of Brussels (ULB).
TABLE 1.
Comparisons of PFGE patterns obtained in the first and subsequent isolates recovered from 71 long-term MRSA carriers (>1 month)a
| Period (mo.) | No. of pairs | No. indistinguishable | No. relatedb (%) |
|---|---|---|---|
| 1–3 | 40 | 39 | 1 (2.5) |
| 3–6 | 19 | 17 | 2 (10.5) |
| 6–12 | 15 | 11 | 4 (26.7) |
| >12 | 10 | 7 | 3 (30) |
| Total | 84 | 74 | 10 (11.9) |
In nine carriers, the first isolate was compared to more than one subsequent isolates (two for eight carriers and six for one carrier; the latter carrier had four identical isolates at days 70, 129, 286, and 450 and two different related isolates at days 472 and 517).
That is, a 1- to 6-fragment difference.
Epidemics.
Seven epidemics were investigated. An epidemic was defined as a significant increase in the number of patients harboring MRSA during a given period of time (maximum of 15 months) in the same institution. The duration and number of persons involved in each epidemic are indicated in Table 2. Epidemics were investigated by the infection control teams of either the CHUV or the ULB hospital.
TABLE 2.
Analysis of PFGE patterns of MRSA isolates recovered during seven hospital epidemics in Belgium and Switzerland
| Hospitala | Epidemic duration (mo) | No. of involved personsb | Classification of fragments
|
|
|---|---|---|---|---|
| No. indistinguishable | No. relatedc (%) | |||
| BE 3 | 1 | 6 | 6 | 0 (0) |
| CH 1 | 2 | 7 | 7 | 0 (0) |
| CH 1 | 2.5 | 19 | 19 | 0 (0) |
| BE 1 | 3 | 7 | 6 | 1 (14) |
| BE 2 | 4 | 20 | 20 | 0 (0) |
| CH 1 | 12 | 30 | 30 | 0 (0) |
| CH 2 | 15 | 31 | 30 | 1 (3) |
| All | 120 | 118 | 2 (1.5) | |
Hospitals were located in Belgium (BE) or Switzerland (CH).
One isolate per person.
That is, with a ≤6-fragment difference.
RESULTS
Analysis of PFGE patterns of MRSA isolates in long-term carriers and epidemics.
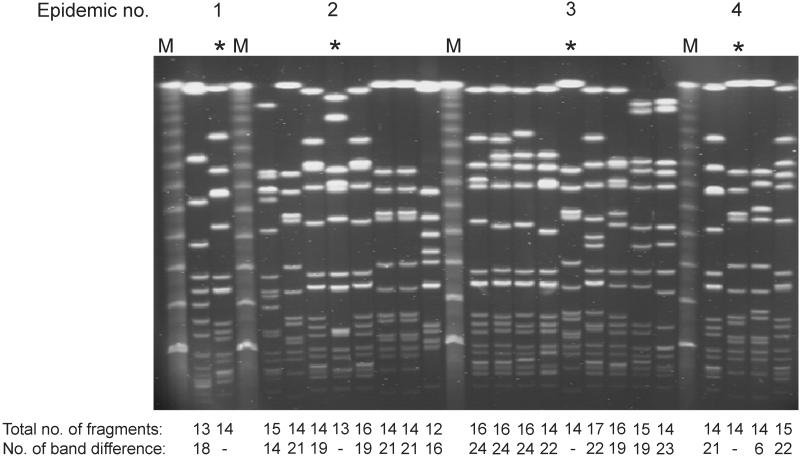
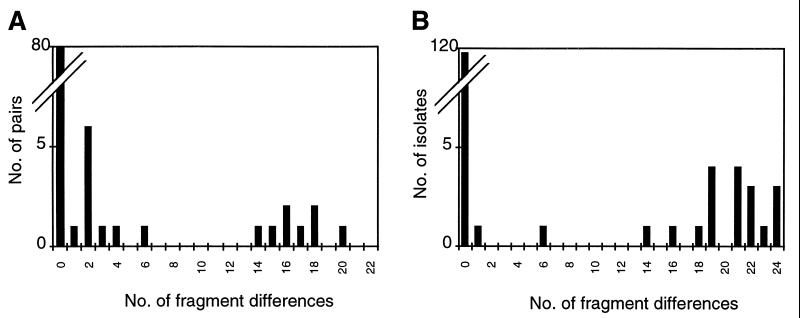
A total of 165 MRSA isolates from 77 long-term carriers and 138 isolates from seven epidemics were analyzed by PFGE. The fragment differences were counted between patterns of the first and subsequent isolate(s) of long-term carriers and between the predominant epidemic pattern and other patterns observed during each epidemic (see Fig. 1 for examples). The distributions of the number of fragment differences are shown in Fig. 2. Both figures show a bimodal distribution: a first group with 0 to 6 fragment differences and another with 14 to 24 fragment differences. A perfect concordance between the results of comparison of PFGE patterns by the two persons were obtained when the two patterns differed by six fragments or less. Variations of only 1 to 4 fragments in the interpretation of fragment difference were observed when patterns were compared that differed by more than 14 fragments. These variations did not affect the results.
FIG. 1.
Examples of PFGE patterns observed in four epidemics. The asterisk denotes the most predominant patterns of the epidemic. The total number of fragments per pattern and the number of fragment differences compared to the epidemic pattern are indicated.
FIG. 2.
Distribution of the number of fragment differences observed between PFGE patterns. A comparison of the first and subsequent MRSA isolates from long-term MRSA carriers (A) and a comparison between the epidemic pattern (most predominant) and other patterns recovered during epidemics (B) is shown.
Analysis of clonality of MRSA isolates showing different PFGE patterns.
To assess whether two isolates of the first group were clonally related and whether two isolates of the second group should not be considered clonally related, a selection of 97 isolates was typed by another molecular method: inter-IS256 PCR. This sample included isolates presenting PFGE patterns showing a ≥1-fragment difference collected from 16 long-term carriers (n = 31) and from four epidemics (n = 20). In addition, we selected isolates showing indistinguishable PFGE patterns: 10 pairs of isolates in long-term carriers (n = 20) and five to seven isolates per epidemic (n = 26). In long-term carriers, the first and last available identical isolates were selected. In epidemics, identical isolates were randomly selected over the whole period of the epidemic. Comparison between PFGE and inter-IS256 PCR showed that all 45 isolates with a 0- to 6-fragment difference in PFGE were identical to their reference as determined by the inter-IS256 PCR typing method, whereas the 27 isolates with 14- to 24-fragment differences by PFGE analysis were different. Thus, isolates producing “related” PFGE patterns (differing by 1 to 6 fragments) were clonally related, whereas those with 14- to 24-fragment differences were genetically different. Consequently, isolates of this latter group should not be considered as part of the epidemic or, in long-term carriers, as a colonization with a different strain.
Analysis of the stability of PFGE patterns of MRSA based on the analysis of clonal isolates.
Pairs of isolates from long-term MRSA carriers colonized with clonally related strains were analyzed. The majority of subsequent isolates (74 of 84 [88.1%]) had a pattern indistinguishable from that of the first isolate (Table 1). The 10 (11.9%) remaining clonal isolates showed variant patterns differing by one to six DNA fragments. These 10 variants were isolated from nine persons, with one person exhibiting two different variants. Moreover, the frequency of apparition of related patterns was higher when the time between the first and the subsequent isolates was longer (2.5% for 1 to 3 months to >26% for >6 months).
Seven nosocomial epidemics with clonally related isolates were analyzed. They involved 6 to 31 persons over periods varying between 1 and 15 months (Table 2). During these outbreaks, only 2 of 120 (1.7%) persons harbored a MRSA strain with a PFGE pattern distinct but related to the predominant epidemic pattern. The first isolate had two-fragment difference compared to the predominant pattern (epidemic of 3 months' duration), whereas the second isolate had a six-fragment difference and was isolated 15 months after the beginning of the epidemic.
DISCUSSION
Analysis of MRSA isolates with two different typing methods (PFGE and inter-IS256 PCR) strongly suggests that S. aureus isolates producing PFGE patterns differing by 6 fragments or fewer should be considered clonally related, whereas isolates with patterns differing by 14 or more fragments should be considered genetically different. This is in agreement with the theoretical presumptions (6, 12). These findings allow us (i) to differentiate persistent colonization from colonization with a new strain in long-term carriers and (ii) to ascertain clonally related isolates in epidemics. Moreover, in long-term MRSA carriers, 9 of 10 related patterns had a ≤4-fragment difference and 1 of 10 had a 6-fragment difference. This is consistent with the theoretical consideration that one mutation (suggested by a difference of ≤4 fragments) should be more frequently observed than two mutations (as suggested by a difference of 4 to 7 fragments) (6).
Only 11.9% of the subsequent isolates of long-term MRSA carriers showed a pattern different but related to the first one. Our results are in agreement with those of another study, which investigated sequential MRSA isolates (30 to 228 days) from colonized patients and showed that 80% of the subsequent isolates had a PFGE pattern indistinguishable from that of the initial one and only 20% showed a related PFGE pattern (1- or 2-fragment difference) (7). Furthermore, our results show that the frequency of apparition of related patterns is higher when the time between the first isolate and the subsequent isolate is longer (Table 1). However, patterns were relatively stable in pairs of isolates recovered within <3 months, with only 2.5% of the isolates showing pattern variation. In the isolates of well-defined hospital epidemics examined in the present study, the stability was even greater because only 1.5% of the involved patients harbored a variant pattern. This is in agreement with a recent study showing a low rate of mutation in MRSA strains recovered during a hospital outbreak (14).
Indistinguishable patterns observed in isolates of two patients do not demonstrate a close epidemiological link. MRSA strains with an indistinguishable pattern were often isolated at intervals of several years from different countries where these strains are endemic (1, 2, 5, 8, 15). Thus, in a setting where an MRSA strain is highly endemic it is possible that encounters with other patients or healthcare personnel harboring this strain during previous hospitalizations were responsible for the transmission.
Our results present the intriguing feature that the rate of related patterns observed over time was different in long-term carriers compared to patients involved in epidemics. Indeed, a third of the long-term carriers already showed MRSA isolates with subtype pattern variation after 6 months, whereas only 1.5% of persons harbored isolates with variant patterns in the two epidemics lasting more than 1 year. Further work is needed to understand this difference.
In conclusion, since the epidemiological use of a typing method requires interpretation criteria to differentiate clonally related isolates from unrelated isolates, the rate of evolution of the patterns obtained by this particular typing method should be investigated according to the species of bacteria, as well as with reference to the time and space scale under study. The present investigation showed that MRSA strains produce PFGE patterns that are relatively stable over periods of weeks to months.
ACKNOWLEDGMENTS
We are grateful to Michel Bernard and Arlette Cruchon for technical assistance.
This study was supported by grant 32-45820.95 from the Swiss National Research Foundation.
REFERENCES
- 1.Aires de Sousa M, Santos Sanches I, Ferro M L, Vaz M J, Saraiva Z, Tendeiro T, Serra J, de Lencastre H. International spread of multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc D S, Hauser P M, Francioli P, Bille J. Molecular typing methods and their discriminatory power. Clin Microbiol Infect. 1998;4:61–63. doi: 10.1111/j.1469-0691.1998.tb00356.x. [DOI] [PubMed] [Google Scholar]
- 3.Blanc D S, Petignat C, Moreillon P, Entenza J, Eisenring M C, Kleiber H, Wenger A, Troillet N, Blanc C H, Francioli P. Unusual spread of a penicillin-susceptible methicillin-resistant Staphylococcus aureus clone in a geographic area of low incidence. Clin Infect Dis. 1999;29:1512–1518. doi: 10.1086/313522. [DOI] [PubMed] [Google Scholar]
- 4.Deplano A, Vaneechoutte M, Verschraegen G, Struelens M J. Typing of Staphylococcus aureus and Staphylococcus epidermidis strains by PCR analysis of inter-IS256 spacer length polymorphisms. J Clin Microbiol. 1997;35:2580–2587. doi: 10.1128/jcm.35.10.2580-2587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deplano A, Witte W, van Leeuwen W J, Brun Y, Struelens M J. Clonal dissemination of epidemic methicillin-resistant Staphylococcus areus in Belgium and neighbouring countries. Clin Microbiol Infect. 2000;6:239–245. doi: 10.1046/j.1469-0691.2000.00064.x. [DOI] [PubMed] [Google Scholar]
- 6.Goering R V. The molecular epidemiology of nosocomial infection: an overview of principles, application, and interpretation. In: Specter S, et al., editors. Rapid detection of infectious agents. New York, N.Y: Plenum Press; 1998. pp. 131–157. [Google Scholar]
- 7.Hartstein A I, Phelps C L, Kwok R Y, Mulligan M E. In vivo stability and discriminatory power of methicillin-resistant Staphylococcus aureus typing by restriction endonuclease analysis of plasmid DNA compared with those of other molecular methods. J Clin Microbiol. 1995;33:2022–2026. doi: 10.1128/jcm.33.8.2022-2026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mato R, Sanches S, Venditti M, Platt D J, Brown A, Chung M, de Lencastre H. Spread of the multiresistant Iberian clone of methicillin-resistant Staphylococcus aureus (MRSA) to Italy and Scotland. Microb Drug Resist. 1998;4:107–112. doi: 10.1089/mdr.1998.4.107. [DOI] [PubMed] [Google Scholar]
- 9.Struelens M J, De Gheldre Y, Deplano A. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect Control Hosp Epidemiol. 1998;19:565–569. doi: 10.1086/647874. [DOI] [PubMed] [Google Scholar]
- 10.Struelens M J, Deplano A, Godard C, Maes N, Serruys E. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J Clin Microbiol. 1992;30:2599–2605. doi: 10.1128/jcm.30.10.2599-2605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Struelens M J The European Study Group on Epidemiological Markers. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2:2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 12.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995;36:48–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen W, van Belkum A, Kreiswirth B, Verbrugh H. Genetic diversification of methicillin-resistant Staphylococcus aureus as a function of prolonged geographic dissemination and as measured by binary typing and other genotyping methods. Res Microbiol. 1998;149:497–507. doi: 10.1016/s0923-2508(98)80004-1. [DOI] [PubMed] [Google Scholar]
- 15.Witte W, Kresken M, Braulke C, Cuny C. Increasing incidence and widespread dissemination of methicillin-resistant Staphylococcus aureus in hospitals in central Europe, with special reference to German hospitals. Clin Microbiol Infect. 1997;3:414–422. doi: 10.1111/j.1469-0691.1997.tb00277.x. [DOI] [PubMed] [Google Scholar]